Abstract
Efficient methods to immobilize small molecules under continuous-flow microfluidic conditions would greatly improve label-free molecular interaction studies using biosensor technology. At present, small-molecule immobilization chemistries require special conditions and in many cases must be performed outside the detector and microfluidic system where real-time monitoring is not possible. Here, we have developed and optimized a method for on-chip bioorthogonal chemistry that enables rapid, reversible immobilization of small molecules with control over orientation and immobilization density, and apply this technique to surface plasmon resonance (SPR) studies. Immobilized small molecules reverse the orientation of canonical SPR interaction studies, and also enable a variety of new SPR applications including on-chip assembly and interaction studies of multicomponent structures such as functionalized nanoparticles, and measurement of bioorthogonal reaction rates. We use this approach to demonstrate that on-chip assembled functionalized nanoparticles show a preserved ability to interact with their target protein, and to measure rapid bioorthogonal reaction rates with k2 > 103 M−1 s−1. This method offers multiple benefits for microfluidic biological applications, including rapid screening of targeted nanoparticles with vastly decreased nanoparticle synthetic requirements, robust immobilization chemistry in the presence of serum, and a continuous flow technique that mimics biologic contexts better than current methods used to measure bioorthogonal reaction kinetics such as NMR or UV-vis spectroscopy (e.g., stopped flow kinetics). Taken together, this approach constitutes a flexible and powerful technique for evaluating a wide variety of reactions and intermolecular interactions for in vitro or in vivo applications.
Introduction
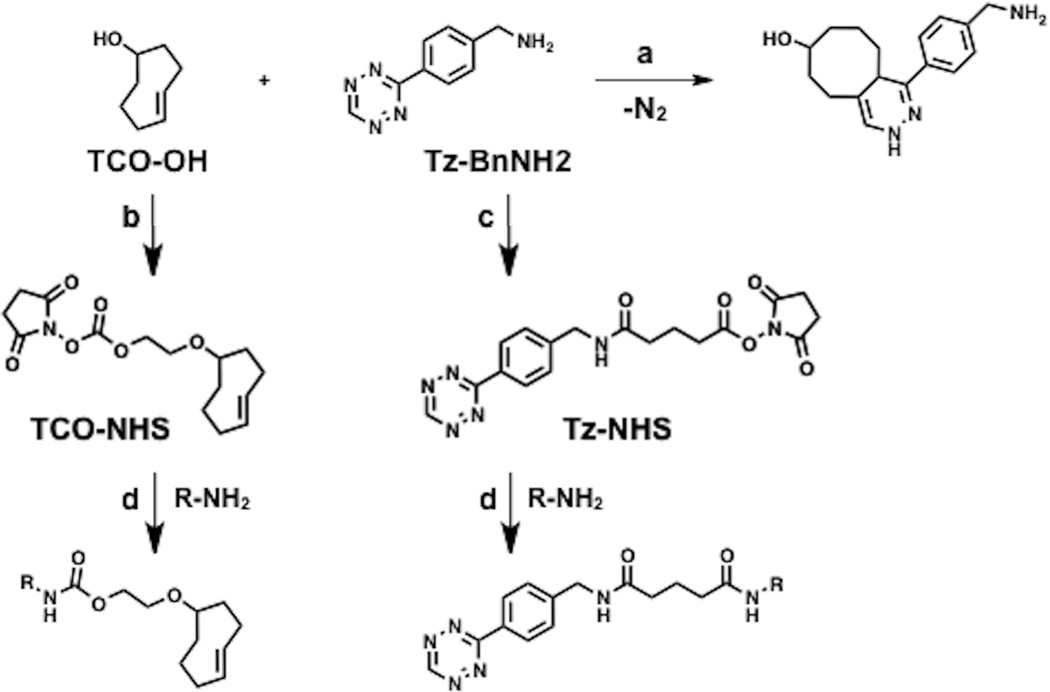
Bioorthogonal reactions (such as the Huisgen 1,3-cycloaddition reaction, [4+2] Diels-Alder cycloaddition, and variations thereof)1–5 are increasingly used to monitor and interrogate biological systems.6–9 For instance, cycloaddition reactions have been used to label cell surfaces,10,11 subcellular structures,12,13 antibodies and nanoparticles;14,15 the resulting conjugates have been used as cellular imaging probes in vitro and in vivo.16,17 Here we describe a rapid, reversible method to immobilize small molecules and nanoparticles onto Surface Plasmon Resonance (SPR) sensor chips by using the [4+2] cycloaddition reaction between trans-cyclooctene (TCO) and 1,2,4,5-tetrazine (Tz) coupled with antibody-antigen capture (Fig. 1).4,18,19
Fig. 1.
Bioorthogonal conjugation chemistry and derivatization of reagents for labeling molecules. (a) [4+2] cycloaddition reaction between trans-cyclooctene (TCO) and 1,2,4,5-tetrazine (Tz) moieties to give the 1,4-dihydropyridazine adduct. (b) Derivatization of TCO-OH to the corresponding TCO-NHS carbonate. (c) Derivatization of Tz-BnNH2 to the corresponding Tz-NHS ester. (d) Labeling of amine-containing molecules with Tz or TCO (R = MNP, peptide, small-molecule, etc.).
Using this chemistry, we demonstrate the on-chip immobilization and small-molecule derivatization of magnetic nanoparticles (MNP). This enables on-chip synthesis and label-free interaction analysis of modified nanoparticles, which promises to be more rapid and efficient than conventional nanoparticle synthesis, purification and screening.20–22 Much smaller nanoparticle quantities are required, with resulting benefits for experimental throughput, cost, synthesis requirements, and potential environmental concerns about nanoparticle use and disposal.23
More broadly, the straightforward immobilization of small molecules (with the capability for surface regeneration) is now possible. Current immobilization chemistries require high concentrations of small molecules to achieve electrostatic pre-concentration, which may be difficult to achieve in water without the use of substantial organic co-solvent; in most cases, organic solvents are not compatible with the flow system and immobilizations must be performed outside the instrument where real-time monitoring of the success or failure of the coupling process is not possible.24
Immobilizing small molecules to the sensor surface enables interaction studies that reverse the orientation of canonical SPR studies, providing facile access to study direct binding of soluble macromolecular targets and clarify phase-specific (immobilized vs. soluble) artefacts.25,26 The ability to analyze binding interactions by immobilizing either reacting partner (with the capability for surface regeneration) enables detailed kinetic analyses of binding interactions under carefully controlled conditions (e.g., surface density, buffer composition, flow rate, and temperature).27
Finally, we use this approach to monitor and quantitate the kinetics of bioorthogonal cycloaddition reactions in real-time under continuous flow. As the application of bioorthogonal reactions grows, and as even more suitable reaction partners are discovered, the need for a flexible, rapid and general method to quantitatively evaluate fast reaction rates of potential bioorthogonal conjugation reactions has also increased.28–31 The immobilization chemistry is robust in the presence of serum, rendering this approach suitable for evaluating a wide range of reactions intended for in vitro and in vivo cellular applications.
Results and discussion
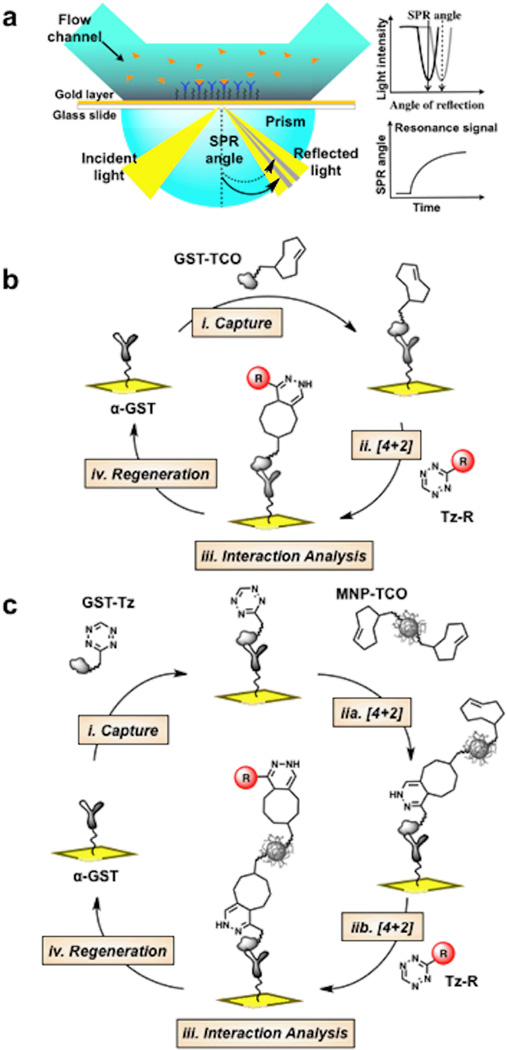
We first conjugated bioorthogonally reactive groups such as TCO directly to free amines on the glutathione-S-transferase (GST) protein surface using the corresponding TCO-NHS ester (Fig. 1). The GST protein amino acid sequence contains 20 lysine residues, which based on the crystal structure are located in both the interior and exterior surfaces (see Electronic Supporting Information (ESI) Fig. S1).32,33 Experimentally, we verified by mass spectrometry that ~10 TCO or Tz groups readily attached to the GST surface (ESI Fig. S2). Then, antibody mediated capture of GST functionalized with TCO groups (GST-TCO; Fig. 2a and b) provides a reactive surface for subsequent cycloaddition reactions with Tz derivatives (Fig. 2b, step ii). An advantage of this strategy is the ability to conjugate more than 1 TCO per GST, which increases the number of reactive sites ten-fold (maximum cycloaddition reaction capacity, Rmax in Resonance units) per captured GST. Furthermore, by modulating the concentration and contact time of injected GST-TCO, control over small-molecule immobilization density can be achieved. Regeneration of the surface is easily accomplished by disrupting the GST/anti-GST interaction, making many cycles of immobilization and interaction studies possible (Fig. 2b).
Fig. 2.
(a) Schematic of the SPR sensor surface and detection system. (b) Experimental scheme for reversible immobilization of tetrazine (Tz)/trans-cyclooctene (TCO) tagged molecules (c) Experimental scheme for reversible nanoparticle immobilization and functionalization.
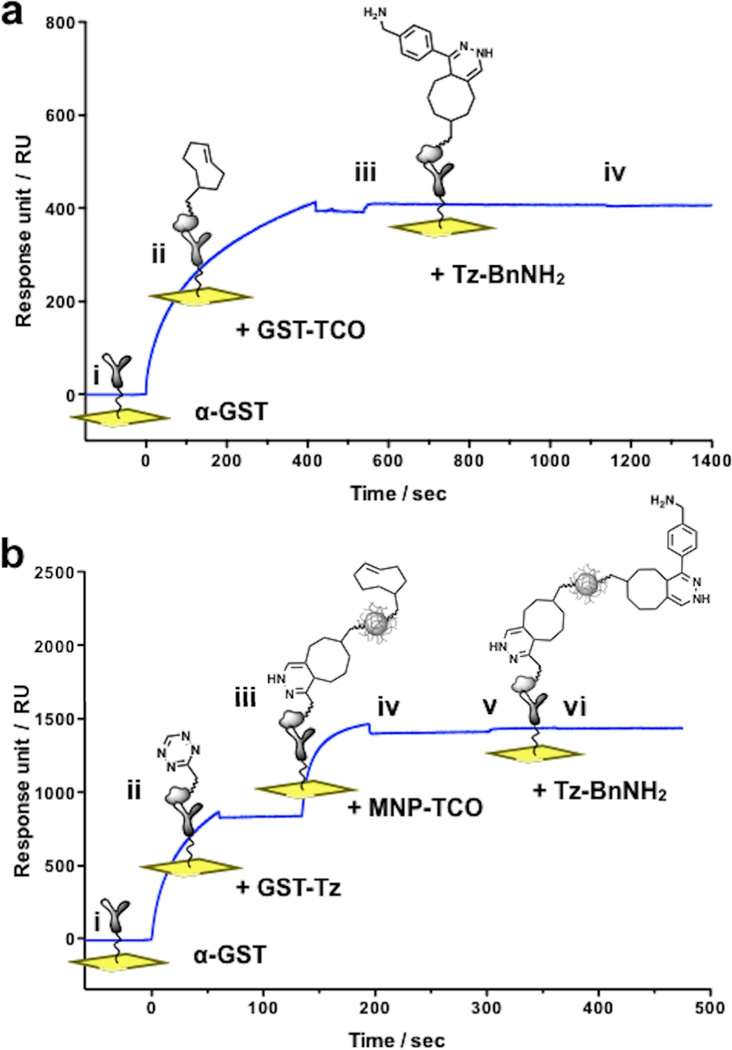
The stepwise assembly of a GST-TCO surface and subsequent cycloaddition with Tz was monitored in real-time by SPR as shown in Fig. 3a. We found that TCO derivatization of GST (~10 TCO/GST) had no discernible effect on antibody recognition, binding kinetics or stability when compared to the non-derivatized GST protein. Upon injection of a 10 µM solution of benzylamino-1,2,4,5-tetrazine (Tz-BnNH2) in PBS we observed a fast rate of cycloaddition (covalent association, ka) reaching completion (saturation) within seconds (Fig. 3a, step iii). As expected, the Rmax correlated with a higher reaction capacity created through multivalent TCO modification of GST. No dissociation was observed after switching from analyte solution to running buffer (kd = 0 for covalent capture) (Fig. 3a, step iv).
Fig. 3.
(a) Real-time monitoring of on-chip small molecule immobilization. i. Baseline: pre-immobilized anti-GST. ii. Capture of injected GST-TCO by anti-GST antibody (t = 0 – 420 sec), followed by injection of running buffer (t = 420 – 540 sec) showing persistence of interaction. iii. Cycloaddition between GST-TCO and benzylamino-tetrazine (Tz-BnNH2; ~15 RU rise in response; t = 540 – 1140 sec). iv. No signal decay occurs despite switching the mobile phase to running buffer (t = 1140 – 1820 sec). (b) Nanoparticle immobilization and functionalization. i. Baseline. ii. Capture of injected solution of GST-Tz (t = 0 – 60 sec). iii. Cycloaddition between immobilized GST-Tz and injected solution of MNP-TCO (t = 135 – 195 sec). iv. Injection of running buffer with no decay in signal (t = 195 – 300 sec). v. Cycloaddition between immobilized MNP-TCO and an injected solution of Tz-BnNH2 (~22 RU rise in response, t = 300 – 360 sec). vi. No signal decay despite switching the mobile phase to running buffer (t = 360 – 480 sec).
We have used the bioorthogonal cycloaddition reaction to demonstrate the on-chip capture and functionalization of nanoparticles used for molecular imaging (Fig. 2c).34 In this case, we prepared a surface composed of benzylamino-tetrazine linked to GST (GST-Tz), immobilized via anti-GST antibodies. Cycloaddition with TCO-modified magnetic nanoparticles (MNP-TCO, Fig. 2c step iia) immobilizes nanoparticles to the surface (while leaving the majority of TCO moieties on the nanoparticle unreacted). Importantly, a second round of cycloadditions using substituted benzylamino-tetrazine (Tz-R, Fig. 2c step iib) can now conjugate small molecules to the immobilized nanoparticles, which can be used in situ for interaction studies, vide infra (Fig. 2c, steps i–iv). The step-by-step on-chip capture of MNP-TCO and derivatization with Tz-BnNH2 was monitored in real-time as shown in Fig. 3b.
To demonstrate that both the small-molecule capture and on-chip nanoparticle conjugation techniques preserve the ability of the small molecules to interact with their target, we evaluated the interaction between FKBP12 (FK506 binding protein 12) and two different small molecules that bind FKBP12 (Fig. 4a)35,36 (sensorgrams are displayed in the ESI). For both small molecules, the equilibrium binding constant KD for FKBP12 binding was comparable whether the small molecule (solid blue symbol) or FKBP12 protein (open orange symbol) was immobilized on the sensor surface (Fig. 4b). However, reversing the orientation of the SPR experiment did affect the measured ka and kd, though by an order of magnitude or less. These effects are summarized on a rate map in Fig. 4c.
Fig. 4.
SPR studies of small molecule/protein interactions in varying configurations. (a) Tetrazine tagged synthetic derivatives of FK506. (b) Kinetic and equilibrium rate constants derived from binding data. (c) Rate map summarizing binding affinity and kinetics. Dashed lines indicate different combinations of ka and kd that result in the same KD.
Of note, the interaction between the immobilized small-molecule-conjugated nanoparticle and its target protein (FKBP12, as the analyte) results in KD’s that are comparable to the small-molecule protein interaction, without an increase in avidity (solid green symbols, Figures 4b and 4c). This is because the interaction between analyte FKBP12 and immobilized nanoparticles is monovalent (1:1), in contrast to the reverse experiment, where simultaneous interaction of a multivalent nanoparticle in solution with an immobilized protein surface results in a marked avidity effect.37 Monitoring the nanoparticle-protein interaction in both orientations allows experimental deconvolution of the intrinsic affinity of the small molecule for its protein target (including how affinity is altered by conjugating the small molecule to the nanoparticle) from the avidity effects from a multivalent nanoparticle. This underscores how different insights into binding interactions can be obtained from the ability to rapidly perform SPR experiments using both orientations.
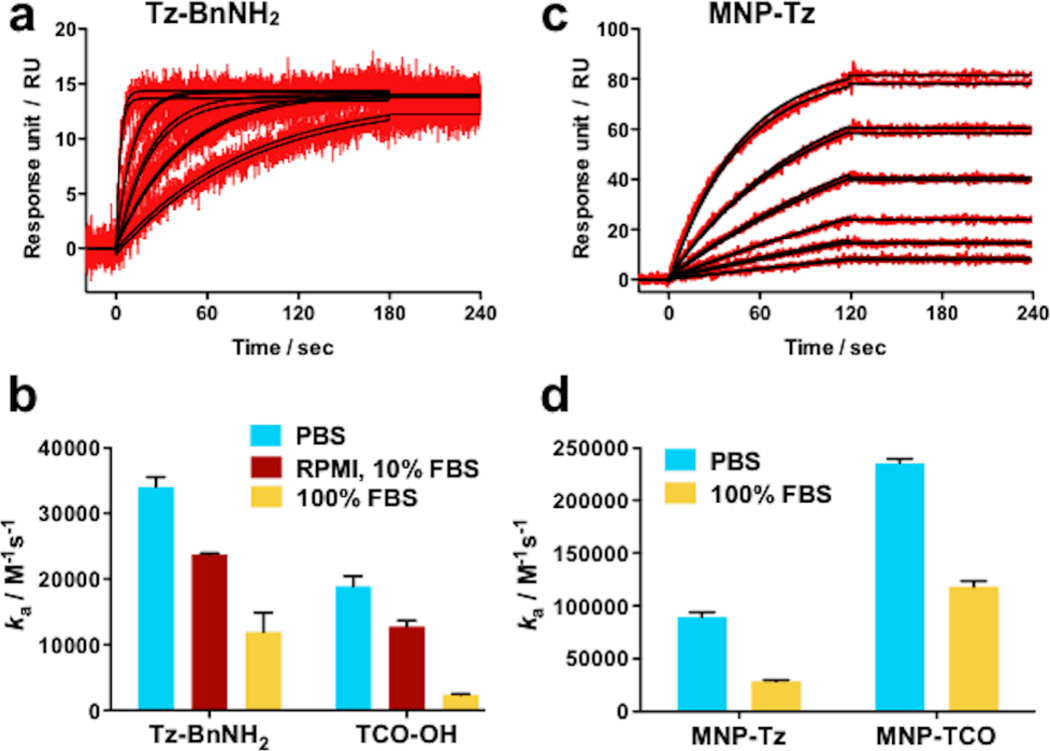
To further explore the kinetics of the on-chip cycloaddition reaction, covalent association data for different concentrations of Tz-BnNH2 in PBS at 25 °C were recorded (Fig. 5a, red lines). Fitting of the experimental data to a 1:1 Langmuir binding model (black lines)38 revealed an exceptionally fast rate constant ka = 34,062 M−1s−1. Reflecting the modular and flexible nature of this strategy, the same anti-GST surface can also be generated to display Tz instead of TCO (see Fig. 2c, step i). Following a similar set of cycloaddition experiments using TCO-OH as the analyte we calculated a rate constant ka = 18,880 M−1s−1 (see ESI). We were also interested in measuring the cycloaddition rates in cell culture media (containing 10% fetal bovine serum (FBS)) and even 100% FBS to assess whether these reactions could be applied for labeling and imaging studies in complex biological fluids. The results when either TCO or Tz are immobilized on the sensor surface are displayed in Figure 5b. In both formats the rate of cycloaddition, ka decreases as the running buffer complexity increases from PBS to cell culture media to 100% FBS.30 Notably however, even at extreme serum concentrations, Rmax saturation is achieved within minutes using analyte concentrations of 10 µM or less (SPR sensorgrams and association rates are displayed in ESI).
Fig. 5.
Representative binding data and kinetic analyses for the cycloaddition reaction between Tz and TCO tagged molecules. (a) Binding of Tz-BnNH2 to GST-TCO surfaces (b) Bar graph summarizing covalent association rate constants for free small molecule analytes in a variety of solvent conditions (c) Binding of MNP-Tz to GST-TCO surfaces (d) Bar graph summarizing rate constants for MNPs derivatized with Tz or TCO in a variety of solvent conditions. Error bars = standard error of the mean.
On closer analysis of the covalent nanoparticle immobilization (step iia) in Fig. 2c, we find a marked increase in ka compared to the analogous on-chip cycloaddition reactions (not in the presence of nanoparticles): MNP-Tz showed a ka of 89,573 M−1s−1 (Fig. 5c vs. 5a; a 2.6-fold increase in ka), and MNP-TCO showed a dramatic 12.5-fold increase in ka (235,440 M−1s−1) when compared to free TCO-OH. As expected, these reaction rates were lower in 100% FBS, but still the fastest reported to date (Fig. 5d). These results show that the fast rate of Tz/TCO cycloaddition and enhanced substrate availability on the nanoparticles allow the on-chip derivatization of MNP’s in very short periods of time and in complex biological media, an essential criteria for screening assays.
Conclusions
In summary, we have developed an approach that utilizes fast, highly specific cycloaddition reactions between 1,2,4,5-tetrazine (Tz) and trans-cyclooctene (TCO) to enable rapid, reversible immobilization of modified nanoparticles and small molecules for label-free chip-based interaction and kinetic studies. The capture-cycloaddition method requires short contact times with < 10 µM concentrations of small-molecule ligands while providing controlled display of ligands for improved experimental design. Our data show that on-chip bioorthogonal reactions preserve the ability of in situ functionalized nanoparticles or immobilized small molecules to interact with their targets, and illustrate how this approach can extend the range of applications for SPR studies. Regeneration of the surface extends the experimental lifetime of sensor surfaces making screening of different combinations of small molecules and nanoparticles fast, convenient and cost effective.
Screening of nanoparticle libraries is a potentially powerful approach to identify nanoparticles specifically targeted to specific proteins or cell types.20,39,40 However, library synthesis typically requires relatively large quantities of both the core nanoparticle (~1 mg for each library member) as well as individual targeting small molecules (~1 mg). Purification of individual library members is often a rate-limiting step of library synthesis. In contrast, the on-chip, microfluidic method described here vastly decreases the amounts of input materials required (with additional benefits related to potential environmental concerns of nanomaterials), eliminates the need for an explicit purification step, and allows combined synthesis and screening in one experiment. Furthermore, the method allows screening based on direct monitoring of the interaction with the desired target, rather than indirect measures such as accumulation of fluorescently-labeled nanoparticles. Importantly, nanoparticle functionalization reactions are robust to the presence of serum concentrations typically used for cell culture experiments, and even to extremely high serum concentrations. This feature can facilitate structure-activity relationships for nanoparticle design in the presence of serum (which can affect the protein corona and surface properties of the nanoparticle).41,42
We have also utilized this approach to characterize bimolecular cycloaddition rates between two small molecules, with molecular weights < 200 Da. This high sensitivity is achieved through multivalent functionalization of the GST capture protein resulting in enhanced signal through increased Rmax. To our knowledge this is the first time covalent cycloaddition reaction rates have been characterized using SPR technology. We also anticipate that this method may be applicable for parallel comparison of fast bioorthogonal rates (ka > 102 M−1s−1) and other studies (e.g., hybridization,43 supramolecular assemblies,44 covalent inhibitors,45 and immunosensing)46 under biologically relevant conditions in a straightforward manner. This technique will be especially useful when no suitable or chromophoric band is available to monitor the reaction using UV-Vis spectroscopy (stopped flow absorption or fluorescence)30 or the rates are too fast to be conveniently measured by NMR.29 As new reactions are discovered, the need for a common method to measure and compare rates within the timescale and environment of biological interactions is desirable. SPR instruments are ideal for this purpose as their performance currently spans a ka range of 103–107 M−1s−1 and kd of 10−6–100 s−1.
Our ability to reversibly immobilize the small molecule on the sensor changes the orientation of the canonical SPR experiment (in which the protein is typically immobilized). When the protein is immobilized, this introduces a series of subtle constraints to the protein’s rotational and diffusional freedom, influencing the thermodynamics of the binding interaction.47–49 While this may be appropriate for some cellular contexts (e.g., a cell surface receptor), it may less faithfully recapitulate the in vivo situation for other contexts (e.g., cytosolic proteins or enzymes). Additionally, when small molecules are in solution (i.e., analytes) and the protein is immobilized, analyte binding can be difficult to assess due to several factors including size (SPR signal is dependent on mass change at the sensor surface), promiscuous binding (nonspecific aggregation)50 and covalent modification (covalent inhibitors).45,51
We envision the modular and flexible nature of this approach will broadly facilitate construction of stable sensor surfaces for a wide variety of high-throughput and multiplex microfluidic screening applications:52,53 on-chip nanoparticle derivatization; an expanded repertoire of label-free interaction studies between proteins and either nanoparticles or small molecules; and analyses of intermolecular reaction rates.
Experimental section
Materials and methods
Commercial reagents were used without further purification and solvents were dispensed from a solvent purification system. Reactions were monitored by thin layer chromatography using silica gel 60 F-254 plates from Silicycle and were visualized by UV light or anisaldehyde stain. Compound purification was carried out by flash chromatography using silica gel 60 (40–63 µm) or preparatory HPLC. Analytical and preparatory LC-MS chromatography: LC-ESI-MS-ELSD analysis and HPLC-purifications were performed on a Waters (Milford, MA) LC-MS system operated by Fractionlynx 4.0 or Masslynx software. HPLC solvent A is H2O and solvent B is CH3CN, both with 0.1% formic acid. Analytical analyses were run at a flow rate of 5 mL/min using a Waters XTerra® MS C18 5 µm column. Solvent gradient: 0–1.5 min, 5–100%B; 1.5–2.0 min, 100%B. Preparative HPLC were run at a flow rate of 15 mL/min using an XTerra® Prep MS C18 OBD 5 µM column. Solvent gradient: 0–9.5 min, 5–100%B; 9.5–10.0 min, 100%B. MALDI-TOF: Measurements were performed on a Voyager-DE BioSpectrometry Workstation (Applied Biosystems) MALDI-TOF mass spectrometer. Single use microtube of sinapinic acid (Thermo Fisher) was used as the MALDI matrix and diluted with 70/30 acetonitrile/water with 0.1% TFA according to the manufacturers protocol. Surface Plasmon resonance (SPR): Measurements were performed on a Biacore T100 (GE Healthcare) instrument using a Sensor Chip CM5 (carboxymethylated dextran matrix immobilized on a gold surface). Surfactant P-20, CM5 sensor chips and immobilization reagents (amine coupling kit: ethanolamine, N-ethyl-N’-(3-dimethylamino propyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS)), regeneration solution (10 mM glycine-HCl pH 2.0) were purchased from GE Healthcare. Cycloaddition rates (ka) and stability were measured at 25°C using three different aqueous solutions, phosphate buffered saline (Gibco) pH 7.4 containing 0.005% P-20 surfactant (PBS-P), RPMI media 1640 (Gibco) supplemented with 10% FBS and 1% pen-strep and 100 % fetal bovine serum (Cellgro). Soluble FKBP12 binding experiments were measured at 25°C using PBS-P. Reversed orientation binding experiments (immobilized FKBP12) were measured at 25°C using PBS-P with 2% DMSO. Glutathione S-Transferase (GST) Schistosoma japonicum was purchased from Genscript. Recombinant human FKBP12 was purchased from R&D Systems. Immobilization of anti-GST and BSA: In a typical assay, carboxyl groups on the surface dextran matrix of CM5 sensor chips were activated with a 1:1 solution of 0.4 M EDC/0.1M NHS (480 second injection at 10 µL/min) followed by a solution of anti-GST (18 µg/mL in acetate buffer, pH 5.0, 420 second injection at 10 µL/min) or BSA (20 µg/mL in acetate buffer, pH 5.0). The remaining NHS-ester groups on the sensor surface were quenched with ethanolamine (420 second injection at 10 µL/min). Using this general method anti-GST was immobilization on flow cells (Fc), Fc1 = 14,527 RU, Fc2 = 17,372 RU, Fc4 = 17,463 RU and BSA on Fc3 = 11,057 RU. Data analysis: All data was analyzed using the T100 evaluation software. Data were double reference subtracted and kinetic analyses were performed using the 1:1 Langmuir binding models provided with the T100 evaluation software. Results of fitting the models to the experimental data were assessed using chi2 (χ2) values and residual plots (difference between experimental and fitted curves for each data point). Scatter in residual plots is a visual measure of how closely the results fit the experimental data. Representative sensorgrams and fitted curve data were exported to Prism to generate higher quality graphs for clarity in the main text. Original sensorgrams, fitted curves generated by the T100 evaluation software and residual scatter plots are presented in the Electronic Supplementary Information (ESI).
Preparation of GST conjugates
(E)-2-(cyclooct-4-en-1-yloxy)ethyl (2,5-dioxopyrrolidin-1-yl) carbonate (TCO-NHS) was prepared following a published procedure.19 (E)-2-(cyclooct-4-en-1-yloxy)ethanol,54 (29 mg, 0.17 mmol) was dissolved in 1.0 mL of CH3CN and 72 µL of Et3N (0.52 mmol) was added followed by 66 mg of N,N’-Disuccinimidyl carbonate (0.26 mmol). After stirring a total of 48 h the reaction was concentrated under reduced pressure. The residue was purified by flash chromatography (silica gel, 1:1 EtOAc/hexanes) to afford 42 mg (79%) of TCO-NHS as colorless oil. A solution of TCO-NHS (8 µL, 50mM) in DMSO was added to 100 µL of GST (1 mg/mL in PBS) and shaken at RT for 1 hr. Excess reagent was removed by spin desalting column (7k MWCO) equilibrated with PBS. The recovered filtrate containing the GST-TCO conjugate is stored at 4°C and stable for >3 months. Similarly, 2,5-dioxopyrrolidin-1-yl 5-((4-(1,2,4,5-tetrazin-3-yl)benzyl)amino)-5-oxopentanoate (Tz-NHS) was prepared according to literature procedure.14 A solution of Tz-NHS (6 µL, 25 mM) in DMF was added to 75 µL of GST (1 mg/mL in PBS) and shaken at RT for 1 hr. The mixture was diluted with 25 µL of PBS and purified by spin desalting column (7k MWCO) equilibrated with PBS. The recovered filtrate containing the GST-Tz conjugate is stored at 4°C and stable for >3 months. To verify GST-conjugate capture and stability, a dilution series of each conjugate (1.25 to 20 µg/mL in 1:2 dilution) was injected over anti-GST and reference flow cells (FC) in increasing concentration (FC2 = anti-GST, FC1 = BSA, 180 second contact, 5 µL/min, 180 second dissociation). Overlay of the reference subtracted sensorgrams show increasing levels of capture with increasing concentrations of conjugate injected (Figure S1, ESI). At 180 seconds the analyte flow was switched to running buffer. Negligible dissociation after 180 seconds of buffer flow indicated antibody/antigen complex stability. Surface regeneration between binding cycles was accomplished with two short pulses (30 second at 30 µL/min) of regeneration solution (10 mM glycine, pH = 2.0). The number of small molecules attached to GST protein was calculated as before by measuring the mass of the protein before and after small molecule modification. The difference in mass is then used to calculate the number of Tz or TCO molecules attached to each GST assuming 284 and 197 Da net masses added, respectively. MALDI-TOF mass spectra for GST shows the singly ionized peak (z=1) has an average molecular weight of ~27,886 Da with a doubly ionized peak (z = 2) at ~13,950 Da (Fig. S2, ESI). The modified protein, GST-Tz shows the singly ionized peak (z=1) has an average molecular weight of ~30,618 Da with a doubly ionized peak (z = 2) at ~15,400 Da. A change in mass of ~2,732 Da corresponds to ~9.6 Tz molecules per GST. Similarly, The modified protein, GST-TCO shows an average molecular weight of ~29,797 Da which corresponds to ~9.7 TCO molecules per GST.
Preparation of MNP conjugates
Magnetic nanoparticles (MNP) used in this study were prepared according to literature procedure.55 and consist of a cross-linked dextran-coated iron oxide core (CLIO, 3 nm diameter, 8000 Fe per nanoparticle) that was aminated by addition of epichlorohydrin and ammonia to provide an average of 90 primary amine groups (CLIO-NH2) for surface modification.56 A solution of TCO-NHS (100 µL. 50 mM) was added to 150 µL of CLIO-NH2 (8.7 mg Fe/mL, PBS, pH 7.4) and shaken at RT for 1h. At the end of this period, excess reagent was removed by gel filtration using a sephadex G-25 column eluting with PBS and the filtrate containing MNP-TCO concentrated to 150 µL using an Amicon Ultra 100K centrifugal filter device. Then, succinic anhydride (50 µL, 0.1 M DMSO) was added to the solution of MNP-TCO to acylate any remaining free amines. After 1h of shaking at RT, the desired product was purified by gel filtration using a sephadex G-25 column eluting with PBS. MNP-Tz was prepared using the same procedure as for MNP-TCO using 75 µL of CLIO-NH2 (8.7 mg Fe/mL, PBS) and 25 µL of Tz-NHS (35 mM) in DMF. Both conjugates are stable at 4°C.
Real-time monitoring of on-chip immobilization of nanoparticles and small molecules
Complex stability of immobilized molecules: Using the instrument manual run method, flow rate was set to 5 µL/min (PBS-P running buffer) and flow path to Fc2. GST-TCO (20 µg/mL) was then captured on the Fc2. Flow path was changed to Fc2, Fc1 and Tz-BnNH2 (10 µM) was injected as described in the main text, Fig. 3. Complex stability was verified by monitoring signal decay after the cycloaddition reaction. Signal response remained flat during the time allotted. Similarly, GST-Tz (20 µg/mL) was captured on the Fc2, followed by MNP-TCO (100 µg Fe/mL), flow path was changed to Fc2, Fc1 and Tz-BnNH2 (10 µM) was injected as described in the main text Fig 4. By monitoring signal decay after the second cycloaddition reaction we assessed complex stability. The signal response remained flat during the time allotted.
Measurements of small-molecule and MNP-conjugate cycloaddition rates
The reference flow cell (Fc1) was prepared by either capturing GST (20 µg/mL in PBS-P injected for 420 second at 5 µL/min) over immobilized anti-GST, left unmodified (anti-GST surface) or Fc3 (BSA surface) was used. The choice of reference did not affect the results. On the analysis flow cell (Fc2 or Fc4), each cycle of analyte binding was carried out in the following sequence: 1) GST conjugate capture 2) binding (cycloaddition reaction) and 3) surface regeneration. The parameters were set in the following manner: i) capture: GST-TCO (or GST-Tz, 10–20 µg/mL) injection, contact time 120 seconds at 5 µL/min (capture levels ranged from 400–1000 RU) ii) cycloaddition reaction: sample injection Tz-BnNH2, (or TCO-OH) contact time 180 seconds at 30 µL/min (flow rates of 5, 10, 30, or 60 µL/min had negligible effects on association rates, ka) followed by dissociation time 60 seconds (kd = 0 for covalent binding). iii) regeneration: surface regeneration with 2 × 30 second pulses of regeneration solution (10 mM glycine-HCl pH 2.0) at 30 µL/min. Cycloaddition rates were characterized by measuring association at varying concentrations raging from 0.3 to 10 µM during step ii above using three different buffer conditions, PBS-P, RPMI media with 10% FBS and 100% FBS. The rates of cycloaddition of MNP-TCO and MNP-Tz particles to TCO or Tz derivatized surfaces were measured in exactly the same manner as the free small molecules TCO-OH and Tz-BnNH2 above. Cycloaddition rates were characterized by measuring ka at varying concentrations of nanoparticle conjugate raging from 7 to 224 nM in PBS-P and 100% FBS. The fitted sensorgrams and residual scatter plots are displayed in the ESI. Kinetic rates, ka and chi2 (χ2) values are summarized in Table S1 (ESI).
General procedure for the preparation of FKBP12 binding ligands
The syntheses of precursors, carboxylic acids 1a–b35–37,57,58 and Tz-BnNH218 to generate compounds 2a–b, have been reported elsewhere. A 500 µL solution of carboxylic acid 1 (30 mM) in CH2Cl2 was cooled in an ice/water bath, and then 5 µL of Et3N (36 µmol) and 3 µL of pentafluorophenyl trifluoroacetate (17 µmol) were added. After approximately 30 min of stirring, TLC analysis showed complete formation of the corresponding pentafluorophenol ester. The reaction mixture was then applied to a pad of silica, eluted with 2:1 hexanes:ethyl acetate and the solvent was concentrated under reduced pressure. The residue was dissolved in 500 µL CH3CN with 6 µL of Et3N (43 µmol) and while vigorously stirring 300 µL of a 50 mM solution of Tz-BnNH2 (15 µmol) in 2:1 CH3CN:H2O was added. After stirring an additional 10 min, product formation was verified by TLC and LC/MS analysis. The product was then purified by preparatory HPLC. Analytical LC/MS chromatograms and spectra were generated from the purified samples (see ESI). UV-vis absorption in the range of 515–530 nm is characteristic of the benzylamino tetrazine functional group.
The binding of small-molecules 1 to immobilized FKBP12
For binding studies where FKBP12 was immobilized, FKBP12-GST fusion protein was captured on Fc2 (1878 RU) and recombinant GST (2340RU) on Fc1 (reference surface). PBS-P with 2% DMSO was used as the running and dilution buffer. Binding of compounds of type 1 was measured at 25°C. Concentration series in 1:4 dilutions from 2 to 2000 nM in duplicate were assessed for binding. A 40-second association phase was followed by a 60-second dissociation phase at 30 µL/min. Fitted sensorgrams, residual and steady state plots are presented in the ESI.
The binding of FKBP12 to immobilized small molecules 2
Reversed orientation binding studies employed FKBP12 as the analyte and compound of type 2 as immobilized ligands. The general method for this assay was set-up as follows; GST-TCO (20 µg/mL, 250 sec contact time, 5 µL/min) was captured on Fc2 (500–600 RU) followed by cycloaddition of compound 2a (10 µM PBS-P 2%DMSO, 250 sec contact time, 5 µL/min). Flow path was changed to Fc2, Fc1 and FKBP12 binding was assessed in a concentration series in 1:2 dilutions ranging from 0.020 to 5 µg/mL. A 60-second association phase was followed by a 200-second dissociation phase at 30 µL/min. After the series was completed the surface was regenerated (10 mM glycine-HCl pH 2.0) and the same process was repeated using compound 2b. Binding was measured at 25°C using PBS-P as the running and dilution buffer. Fitted sensorgrams, residual and steady state plots are presented in the ESI.
On-Chip nanoparticle derivatization and binding of FKBP12
The general method for nanoparticle derivatization and binding assay was set-up as follows, i. Flow path was set to Fc2, GST-Tz (20 µg/mL) was injected for 60 sec at 5 µL/min (typical capture levels 580–600 RU) ii. MNP-TCO (100 µgFe/mL) was injected for 90 sec at 5 µL/min. iii. compound 2a (10 µM) was injected for 180 sec at 5 µL/min iv. Flow path was changed to Fc2, Fc1 and FKBP12 binding was assessed in a concentration series in 1:2 dilutions ranging from 0.020 to 5 µg/mL. A 60-second association phase was followed by a 200-second dissociation phase at 30 µL/min. After the series was completed the surface was regenerated (10 mM glycine-HCl pH 2.0) and the same process was repeated using compound 2b. Binding was measured at 25°C using PBS-P as the running and dilution buffer. Fitted sensorgrams, residual and steady state plots are presented in the ESI.
Supplementary Material
Acknowledgements
We thank Nikolay Sergeyev for providing the cross-linked magnetic nanoparticles, Mark Karver and Jenna A. Klubnick for reagents, and Paul Belcher (GE Healthcare) for SPR technical support. We acknowledge funding from NIH (NHLBI Contract No. HHSN268201000044C to R.W., S.H. and S.Y.S.).
Footnotes
† Electronic Supplementary Information (ESI) available: [Figures and tables]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Sletten EM, Bertozzi CR. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becer CR, Hoogenboom R, Schubert US. Angew Chem Int Ed Engl. 2009;48:4900–4908. doi: 10.1002/anie.200900755. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 4.Blackman ML, Royzen M, Fox JM. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debets MF, van Berkel SS, Dommerholt J, Dirks AT, Rutjes FP, van Delft FL. Acc. Chem. Res. 2011;44:805–815. doi: 10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 6.Hao Z, Hong S, Chen X, Chen PR. Acc. Chem. Res. 2011;44:742–751. doi: 10.1021/ar200067r. [DOI] [PubMed] [Google Scholar]
- 7.Devaraj NK, Weissleder R. Acc. Chem. Res. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Angew Chem Int Ed Engl. 2002;41:1053–1057. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 10.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dommerholt J, Schmidt S, Temming R, Hendriks LJ, Rutjes FP, van Hest JC, Lefeber DJ, Friedl P, van Delft FL. Angew Chem Int Ed Engl. 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haun JB, Devaraj NK, Marinelli BS, Lee H, Weissleder R. ACS Nano. 2011;5:3204–3213. doi: 10.1021/nn200333m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budin G, Yang KS, Reiner T, Weissleder R. Angew. Chem. Int. Ed. Engl. 2011;50:9378–9381. doi: 10.1002/anie.201103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Nat Nanotechnol. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liong M, Fernandez-Suarez M, Issadore D, Min C, Tassa C, Reiner T, Fortune SM, Toner M, Lee H, Weissleder R. Bioconjug Chem. 2011;22:2390–2394. doi: 10.1021/bc200490r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devaraj NK, Keliher EJ, Thurber GM, Nahrendorf M, Weissleder R. Bioconjug Chem. 2009;20:397–401. doi: 10.1021/bc8004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahrendorf M, Keliher E, Marinelli B, Waterman P, Feruglio PF, Fexon L, Pivovarov M, Swirski FK, Pittet MJ, Vinegoni C, Weissleder R. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7910–7915. doi: 10.1073/pnas.0915163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjug Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA, Weissleder R. Angew Chem Int Ed Engl. 2009;48:7013–7016. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 21.Yuan J, Oliver R, Aguilar MI, Wu Y. Anal. Chem. 2008;80:8329–8333. doi: 10.1021/ac801301p. [DOI] [PubMed] [Google Scholar]
- 22.Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S. Angew Chem Int Ed Engl. 2011;50:11769–11772. doi: 10.1002/anie.201105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keelan JA. Nat Nanotechnol. 2011;6:263–264. doi: 10.1038/nnano.2011.65. [DOI] [PubMed] [Google Scholar]
- 24.Healthcare, G. E. Biacore Sensor Surface Handbook [Google Scholar]
- 25.Adamczyk M, Chen YY, Gebler JC, Johnson DD, Mattingly PG, Moore JA, Reddy RE, Wu J, Yu Z. Steroids. 2000;65:295–303. doi: 10.1016/s0039-128x(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell JS, Wu Y, Cook CJ, Main L. Anal. Biochem. 2005;343:125–135. doi: 10.1016/j.ab.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Douat-Casassus C, Chassaing S, Di Primo C, Quideau S. ChemBioChem. 2009;10:2321–2324. doi: 10.1002/cbic.200900287. [DOI] [PubMed] [Google Scholar]
- 28.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 29.Taylor MT, Blackman ML, Dmitrenko O, Fox JM. J. Am. Chem. Soc. 2011;133:9646–9649. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karver MR, Weissleder R, Hilderbrand SA. Bioconjug Chem. 2011;22:2263–2270. doi: 10.1021/bc200295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sletten EM, Bertozzi CR. J. Am. Chem. Soc. 2011;133:17570–17573. doi: 10.1021/ja2072934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutsell SQ, Kimple RJ, Siderovski DP, Willard FS, Kimple AJ. Methods Mol Biol. 2010;627:75–90. doi: 10.1007/978-1-60761-670-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rufer AC, Thiebach L, Baer K, Klein HW, Hennig M. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:263–265. doi: 10.1107/S1744309105004823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tassa C, Shaw SY, Weissleder R. Acc. Chem. Res. 2011;44:842–852. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 36.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tassa C, Duffner JL, Lewis TA, Weissleder R, Schreiber SL, Koehler AN, Shaw SY. Bioconjug Chem. 2010;21:14–19. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Shannessy DJ, Brigham-Burke M, Soneson KK, Hensley P, Brooks I. Anal Biochem. 1993;212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- 39.Kelly KA, Shaw SY, Nahrendorf M, Kristoff K, Aikawa E, Schreiber SL, Clemons PA, Weissleder R. Integr Biol (Camb) 2009;1:311–317. doi: 10.1039/b821775k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun EY, Josephson L, Kelly KA, Weissleder R. Bioconjug Chem. 2006;17:109–113. doi: 10.1021/bc050290e. [DOI] [PubMed] [Google Scholar]
- 41.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 43.Liong M, Tassa C, Shaw SY, Lee H, Weissleder R. Adv Mater. 2011;23:H254–H257. doi: 10.1002/adma.201101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agasti SS, Liong M, Tassa C, Chung HJ, Shaw SY, Lee H, Weissleder R. Angew Chem Int Ed Engl. 2011 Nov 24; doi: 10.1002/anie.201105670. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkin MR, Wells JA. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell J. Sensors. 2010;10:7323–7346. doi: 10.3390/s100807323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myszka DG. Curr. Opin. Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 48.Mammen M, Choi S-K, Whitesides GM. Angew Chem Int Ed Engl. 1998;37:2754–2899. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 49.Kausaite-Minkstimiene A, Ramanaviciene A, Kirlyte J, Ramanavicius A. Anal. Chem. 2010;82:6401–6408. doi: 10.1021/ac100468k. [DOI] [PubMed] [Google Scholar]
- 50.Giannetti AM, Koch BD, Browner MF. J. Med. Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 51.Kimple AJ, Willard FS, Giguere PM, Johnston CA, Mocanu V, Siderovski DP. Biochim. Biophys. Acta. 2007;1774:1213–1220. doi: 10.1016/j.bbapap.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnamoorthy G, Carlen ET, Bomer JG, Wijnperle D, deBoer HL, van den Berg A, Schasfoort RB. Lab Chip. 2010;10:986–990. doi: 10.1039/c000705f. [DOI] [PubMed] [Google Scholar]
- 53.Kanoh N, Kyo M, Inamori K, Ando A, Asami A, Nakao A, Osada H. Anal. Chem. 2006;78:2226–2230. doi: 10.1021/ac051777j. [DOI] [PubMed] [Google Scholar]
- 54.Keliher EJ, Reiner T, Turetsky A, Hilderbrand SA, Weissleder R. ChemMedChem. 2011;6:424–427. doi: 10.1002/cmdc.201000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Josephson L, Tung CH, Moore A, Weissleder R. Bioconjug Chem. 1999;10:186. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 56.Zhao M, Kircher MF, Josephson L, Weissleder R. Bioconjug Chem. 2002;13:840. doi: 10.1021/bc0255236. [DOI] [PubMed] [Google Scholar]
- 57.Keenan T, Yaeger DR, Courage NL, Rollins CT, Pavone ME, Rivera VM, Yang W, Guo T, Amara JF, Clackson T, Gilman M, Holt DA. Bioorganic & Medicinal Chemistry. 1998;6:1309. doi: 10.1016/s0968-0896(98)00125-4. [DOI] [PubMed] [Google Scholar]
- 58.Yang W, Rozamus LW, Narula S, Rollins CT, Yuan R, Andrade LJ, Ram MK, Phillips TB, van Schravendijk MR, Dalgarno D, Clackson T, Holt DA. J. Med. Chem. 2000;43:1135. doi: 10.1021/jm9904396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.