Abstract
Recent studies have identified synaptic proteins that undergo synapse-to-nucleus translocation in response to neuronal activity that modulate protein synthesis. One such translational regulatory protein of the postsynaptic density (PSD) is AIDA-1d, which binds to PSD-95 via its C-terminus. Activation of synaptic NMDA receptors induces the cleavage of AIDA-1d, and the N-terminus is then shuttled to nuclear Cajal bodies where it plays a role in regulating global protein synthesis. Chronic ethanol exposure has been shown to increase the synaptic clustering of NMDA receptors and PSD-95. Here, we tested the hypotheses that AIDA-1d regulates chronic ethanol-induced increases in synaptic NMDA receptor expression. As reported, we found that AIDA-1 was highly enriched in dendritic spines and co-localized with PSD-95. Acute NMDA treatment increased AIDA-1 colocalization with p80 coilin, a marker of Cajal bodies. Chronic treatment (4 day) of cultures with ethanol (25 – 100 mM) or with the NMDA receptor antagonist AP-V (50 µM) enhanced the clustering of AIDA-1 at synaptic sites. However, chronic ethanol treatment (50 mM) in the presence of the NMDA receptor agonist NMDA (2.5 µM) prevented this increase. Surprisingly, PSD-95 did not seem to play a role in the synaptic distribution of AIDA-1 as this distribution was not affected by declustering PSD-95 from synapses in response to inhibition of palmitoylation. We found that lentiviral knockdown of AIDA-1d did not affect protein expression levels of NMDA receptor subunits GluN1, GluN1 C2’, or GluN2B. The results of this study demonstrate that synaptic AIDA-1 expression is enhanced by chronic ethanol exposure that can be prevented by concurrent stimulation of NMDA receptors. In addition, we found that the association of AIDA-1 with PSD-95 is not required for its localization to the PSD. Moreover, we found that AIDA-1 does not regulate protein expression levels or alternative splicing of the GluN1 subunit of NMDA receptors.
Keywords: AIDA-1, chronic ethanol, synapse-to-nucleus signaling; NMDA receptors; Cajal bodies; PSD-95
Introduction
Chronic alcohol is well-documented to induce homeostatic neuroadaptations in glutamatergic synapses in key brain regions involved in alcohol dependence (Lovinger and Roberto, 2012). Indeed, chronic treatment of primary hippocampal or cortical neurons with ethanol increases the NMDA receptor expression at synaptic sites (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004; Clapp et al., 2010; Qiang et al., 2007). This synaptic targeting of NMDA receptors by chronic ethanol is associated with an upregulation of the postsynaptic density (PSD) scaffolding protein PSD-95 and an enlargement of dendritic spines (Carpenter-Hyland and Chandler, 2006; Qiang et al., 2007). These findings suggest that chronic ethanol exposure results in an expanded synaptic scaffolding platform for the recruitment and activation of signaling molecules that regulate spine actin dynamics, synaptic plasticity, and protein translation (Carpenter-Hyland and Chandler, 2006, 2007; Mulholland and Chandler, 2007).
Chronic ethanol exposure also alters post-translational modifications and expression of splice variants of NMDA receptor subunits. Previous studies have shown that ethanol increases the expression of the GluN1 subunit variant containing the C2’ cassette in hippocampus (Clapp et al., 2010) and elevates tyrosine phosphorylation of the GluN2B subunit in dorsolateral striatum (Wang et al., 2007; Wang et al., 2010). Neuronal activity has been shown to regulate the alternative splicing of GluN1 (NR1) mRNA alternative splicing (Mu et al., 2003). Taken together, these previous studies as well as our own work suggest that chronic ethanol-dependent inhibition of NMDA receptor function results in homeostatic adaptations at neuronal synapses. How chronic ethanol and neuronal activity control nuclear functions, such as the alternative splicing of GluN1 (NR1) receptors, is unclear. However, a novel class of synaptic proteins that shuttle to the nucleus may enact this regulation (Jordan and Kreutz, 2009).
Synapto-nuclear signaling protein messengers such as AIDA-1, contain a nuclear localization sequences and can shuttle into the nucleus in response to neuronal activity (Dieterich et al., 2008; Jordan et al., 2007; Proepper et al., 2007). Jordan and colleagues have recently shown that AIDA-1d is highly enriched in the PSD and binds to PSD-95 via its C-terminus (Jordan et al., 2007). Activation of synaptic NMDA receptors results in calcium-independent cleavage of AIDA-1d, and the N-terminus of AIDA-1d is then shuttled to nuclear Cajal bodies where it plays a possible role in ribosomal RNA splicing (Jordan et al., 2007). Cajal bodies are subnuculear structures that participate in RNA metabolism and nucleolar formation (Zatsepina et al., 2003). Jordan and colleagues (2007) demonstrated that prolonged NMDA receptor activity produces an AIDA-1d–dependent increase in nucleolar number and global protein synthesis. The following studies were designed to test the hypothesis that chronic ethanol regulate synaptic and nuclear AIDA-1 expression and that this in turn contributes to the enhanced targeting of synaptic NMDA receptors.
Materials and methods
Materials
Sprague-Dawley® rat pups from an established breeding colony were used in these studies. Breeders and dams were originally supplied by Harlan (Indianapolis, IN). All culture medium supplies were obtained from Gibco BRL (Gaithersburg, MD) with the exception of platelet-poor horse serum (PPHS) that was purchased from Sigma-Aldrich, Co. (St. Louis, MO). Unless otherwise noted, all compounds were purchased from Sigma-Aldrich Co. Absolute ethanol was purchased from AAPER Alcohol and Chemical Co. (Shelbyville, KY). Protein determination kit was purchased from Pierce Biotechnology, Inc. (Rockford, IL). Horseradish peroxidase conjugated goat anti-mouse and anti-rabbit secondary antibodies were purchased from Southern Biotechnology Associates, Inc. (Birmingham, AL). Alexa conjugated goat anti-mouse and anti-rabbit secondary antibodies were purchased from Invitrogen (Carlsbad, CA). Enhanced chemiluminescence detection kit was purchased from Amersham Biosciences (Arlington Heights, IL). dl-2-Amino-5-phosphonopentanoic acid (APV) was purchased from Tocris (Minneapolis, MN).
Preparation of primary hippocampal neurons
Procedures for the care and use of animals conformed to National Institutes of Health guidelines and were approved by the Medical University of South Carolina Institutional Animal Care and Use Committee. Newborn rat pups (< 24 hr old) were used to prepare hippocampal neuronal cultures as previously described (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004). In brief, pups were cold anaesthetized and brains were removed and placed in isotonic saline (pH 7.4) containing 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml amphotericin B. Pooled hippocampi were digested in 2 ml of 0.25% trypsin (w/v) in isotonic saline, treated with 20 µg/ml DNase I and dissociated by trituration through a fire-polished Pasteur pipette. The volume of the dissociated cell suspension was increased to 50 ml with Dulbecco’s Modified Eagle’s Medium (DMEM) containing PPHS and centrifuged for 10 min at 600 × g. The resulting pellet was re-suspended in DMEM/PPHS and plated at a density of either 135,000 (low-density) or 750,000 cells (high-density) in poly-L-lysine-precoated 35 mm culture dishes. Three hours after plating, the DMEM/PPHS media was replaced with Neurobasal media that contained B-27 supplement, penicillin G/streptomycin and 0.5 mM GlutaMax. The Neurobasal media was astroglia conditioned (without B27) by incubation for 48 hr in astroglia-confluent flask (24 ml/155 mm flask) prior to the addition of hippocampal cultures. Three days after plating, 5 µM β-cytosine arabinoside was added to each dish. Cultures were maintained in an incubator at 37°C in 7.5% CO2/92.5% air for 16–17 days in vitro (DIV) with a ¼ media change performed every 7 days. Low-density hippocampal cultures were used for immunocytochemistry, whereas high-density cultures were used in immunoblot experiments.
Acute and chronic treatment of hippocampal neurons
Hippocampal neurons were acutely and chronically treated as previously described, with minor modifications as described below (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004). In brief, a concentrated ethanol solution was prepared using culture media and added to cultures to produce 25, 50, and 100 mM ethanol in the culture media. Cultures were then placed in Tupperware containers along with a Nalgene (Naperville, IL) jar that contained 400 ml of an ethanol/water solution, 10% higher in concentration than the cultures. The water in the jar had been temperature and CO2 equilibrated before the addition of ethanol. The vapor chambers were then filled with 8% balanced CO2 and returned to the CO2 incubators for 4 days. Control dishes were treated in the same manner without ethanol. At 20–21 days in vitro, hippocampal neurons were treated with either NMDA (10 µM; 10 min) to induce AIDA-1 translocation to the nucleus or 2-bromo-palmitate (2-Pal; 10 µM; 24 hr) to inhibit palmitoylation and decluster PSD-95 from synaptic sites. 2-Pal is a potent palmitoylation protein acyl-transfer inhibitor (Webb et al., 2000). Additional cultures were exposed to ethanol (25 – 100 mM) or the NMDA receptor antagonist APV (50 µM) for 4 days starting at 16–17 days in vitro. Finally, an additional set of cultures was treated with 50 mM ethanol and a non-toxic concentration of NMDA (2.5 µM) for 4 days (Carpenter-Hyland et al., 2004). The ethanol concentration in the culture media was measured at the end of the chronic exposure period as previously described (Carpenter-Hyland et al., 2004). Analysis indicated that the ethanol concentration in the media was maintained at the appropriate concentration. For 50 mM ethanol treatments, the ethanol media concentration was found to be 52.6 ± 1.7 mM (n = 5 dishes). All experiments were completed in triplicate with 3–4 dishes in each treatment group. Following treatment, neurons were prepared for immunocytochemistry and Western blotting, as described below.
AIDA-1 immunocytochemistry
Immunostaining of AIDA-1 clustering was performed using low-density hippocampal cultures following previously described methods (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004). Briefly, neurons were rinsed twice in ice-cold phosphate-buffered saline (PBS) (in mM: NaCl, 136; KCl, 2.6; KH2HPO4, 1.8; Na2HPO4, 10; pH 7.4) and fixed for 15 min in 4% paraformaldehyde/4% sucrose (PFA-S). Neurons were then permeabilized for 5 min in PBS with 0.2% Triton X-100. Following incubation in 5% normal donkey serum (NDS) for 1 hr, neurons were incubated overnight with rabbit anti-AIDA-1 (1:500; Zymed) and mouse anti-PSD-95 (1:500; NeuroMab) or mouse anti-p80 coilin (1:1000; BD Biosciences, San Jose, CA) at 4°C. After overnight incubation using CoverWell incubation chambers (Invitrogen), the chambers were removed and the dishes were incubated with Alexa 488 goat anti-rabbit (1:000) and Alexa 594 goat anti-mouse secondary antibodies at room temperature for 1 hr. Dishes were then mounted with 1,4-daizabicyclo(2,2,2)octane-containing mounting media and stored at 4°C until imaging. For some experiments, cultures were stained with 5 µM DRAQ5 (nuclear stain) for 10 min prior to mounting. Imaging of hippocampal neurons was performed on a Zeiss LSM 510 confocal microscope using a 63× objective (numerical aperture = 1.4) and a threefold zoom setting. Laser and detector settings were retained for all images collected within a set of experimental dishes. Control and treatment groups were always run in parallel within the same immunocytochemical procedure.
AIDA-1 cluster analysis
To increase the accuracy of identification of AIDA-1 clusters, images were first deconvolved to reduce point-spread functions using the adaptive blind 3D deconvolution method (AutoQuant X, Version X2.1.3, Media Cybernetics, Inc., Bethesda, MD) prior to analysis. We quantified the size and density of AIDA-1 clusters using the Surfaces module of Imaris 3D image analysis software (Version x64 7.2; Bitplane AG, Zurich, Switzerland) and a minimum diameter threshold of 200 nm, following previously described methods (Gorelova et al., 2011). The length of the dendrite was determined using the Measurements module. Data for AIDA-1 cluster volume and number were exported to Excel, and the density (# of clusters/µm) of AIDA-1 clusters was calculated for each dendritic segment. All statistical analyses were performed using GraphPad Prism software (version 4.03; La Jolla, CA).
AIDA-1 colocalization analysis
We sought to determine changes in the subcellular localization of AIDA-1 following chronic ethanol or prolonged inhibition or activation of NMDA receptors. Studies using electron microscopy have reported that the length of the postsynaptic density of asymmetric synapses in hippocampal neurons averages between 180 and 260 nm (Moretti et al., 2006; Takumi et al., 1999). PSD-95 was selected as a marker for asymmetric synapses because ~90% of PSD-95 is expressed in postsynaptic membranes (El-Husseini et al., 2000) and AIDA-1 co-immunoprecipitates with PSD-95 in hippocampus (Jordan et al., 2007). We first identified puncta of AIDA-1 and PSD-95 in dendrites of hippocampal neurons using the ImarisXT interface coupled with the MatLab Spots colocalization extension. The Spots extension was selected for this analysis because it provides a procedure to automatically detect and sort distances between sets of punctate clusters. A minimum diameter filter of 0.27 µm was used to identify punctate clusters of AIDA-1 and PSD-95. The density of AIDA-1 clusters that were < 200 nm (synaptic), between 200 and 600 nm (perisynaptic), or > 600 nm (extrasynaptic) away from a PSD-95 cluster was then calculated.
We next quantified the intensity of AIDA-1 staining in nuclear Cajal bodies stained with p80. First, we identified Cajal bodies using the Surfaces module, and these surfaces were used to mask AIDA-1 staining that was localized inside of the Cajal bodies. Following this, surfaces were created for AIDA-1 staining within Cajal bodies and the mean fluorescence intensity for each of these surfaces was determined. We have previously published using similar methods to quantify staining for proteins using masking technology (Gorelova et al., 2011).
Lentiviral knockdown of AIDA-1
We used pTRIP vectors to generate lentiviral shRNA vectors for knockdown following the methods described (Janas et al., 2006). pTRIP contains an H1 promoter derived from pSUPER vectors to drive shRNA expression, and a GFP construct driven by an elongation factor 1α (EF1α) promoter to identify transduced neurons. The shRNA sequence from rat-derived AIDA-1 was selected using the SFOLD online software prediction algorithms (Wadsworth center) and tested. We selected the sequence at base-pair position 121: 5’ GTAGGACAATGGTTGGAAA 3’. Viruses were generated by triple transfection of pTRIP-shRNA and pCMV-dR8.2 and pMD2.G into 293FT cells (Invitrogen). Medium from the transfected 293FT cells was used for cell transduction. On DIV7 neurons were transduced with a lentivirus that expresses an shRNA targeted against the synaptically-enriched AIDA-1d variant. Additional neurons were transduced with a lentivirus that expresses an shRNA targeted against firefly luciferase to serve as controls. The media was reduced to 1 ml for the first 16 hr of exposure to increase the number of transduced neurons. Neurons were exposed to the lentivirus for 7 days, and we determined the extent of AIDA-1 knockdown using Western blotting and immunocytochemistry.
Western blotting
To confirm the knockdown of AIDA-1, we performed Western blot analysis on high-density hippocampal cultures transduced with lentivirus. Following a 7 day exposure period, neurons were scraped into 100 µl of ice-cold homogenization buffer (50 mM Tris-HCl, 50 mM NaCl, 10 mM EGTA, 5 mM EDTA; 2 mM sodium pyrophosphate, 1 mM activated sodium orthovanadate, 0.2 mM AEBSF, 1 µg/ml aprotinin, 1 mM benzamide, 10 µg/ml leupeptin, 10 µg/ml pepstatin, pH 7.5). Homogenates from three dishes were combined, probe sonicated for ~ 5 sec, and centrifuged at 23,100 × g for 30 min at 4°C. The resulting supernatant was removed and the pellet was resuspended in 2% lithium dodecyl sulfate (LDS) and probe sonicated for ~ 5 sec. An aliquot was taken for determination of protein concentration by the bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, IL). The remaining pellet was stored at −80°C until immunoblot analysis.
An aliquot of each sample was diluted with NuPAGE 4X LDS sample loading buffer (Invitrogen Corp., Carlsbad, CA; pH 8.5) containing 50 mM dithiothreitol, and samples were denatured for 10 min at 70° C. Five µg of each sample was separa ted using the Bis-Tris (375 mM resolving buffer and 125 mM stacking buffer, pH 6.4; 7.5% acrylamide) discontinuous buffer system with MOPS electrophoresis buffer (50 mM MOPS, 50 mM Tris, 0.1% SDS, 1 mM EDTA, pH 7.7). Protein was then transferred to Immobilon-P PVDF membranes (Millipore, Bedford, MA) using a semi-dry transfer apparatus (Bio-Rad Laboratories, Hercules, CA). After transfer, blots were washed with phosphate-buffered saline containing 0.1% Tween 20 (PBST) and then blocked with PBST containing 5% nonfat dried milk (NFDM) for 1 hr at room temperature with agitation. The membranes were then incubated overnight at 4°C with primary antibodies diluted in PBST containing 0.5% NFDM and washed in PBST prior to 1 hr incubation at room temperature with horseradish peroxidase conjugated secondary antibodies diluted 1:2000 in PBST. Membranes received a final wash in PBST and the antigen-antibody complex was detected by enhanced chemiluminescence. Film autoradiograms were quantified by computer-assisted densitometry using Image J 1.42q (National Institutes of Health, MD). Antibodies used in these studies were AIDA-1 (1:1000; Zymed), GluN1 (1:4000; BD Biosciences, Franklin Lakes, NJ), GluN1 C2’ (1:1000; Millipore, Billerica, MA), and GluN2B (1:1000; NeuroMab, Antibodies, Inc. & UC Davis, Davis, CA).
Results
AIDA-1 Subcellular Localization
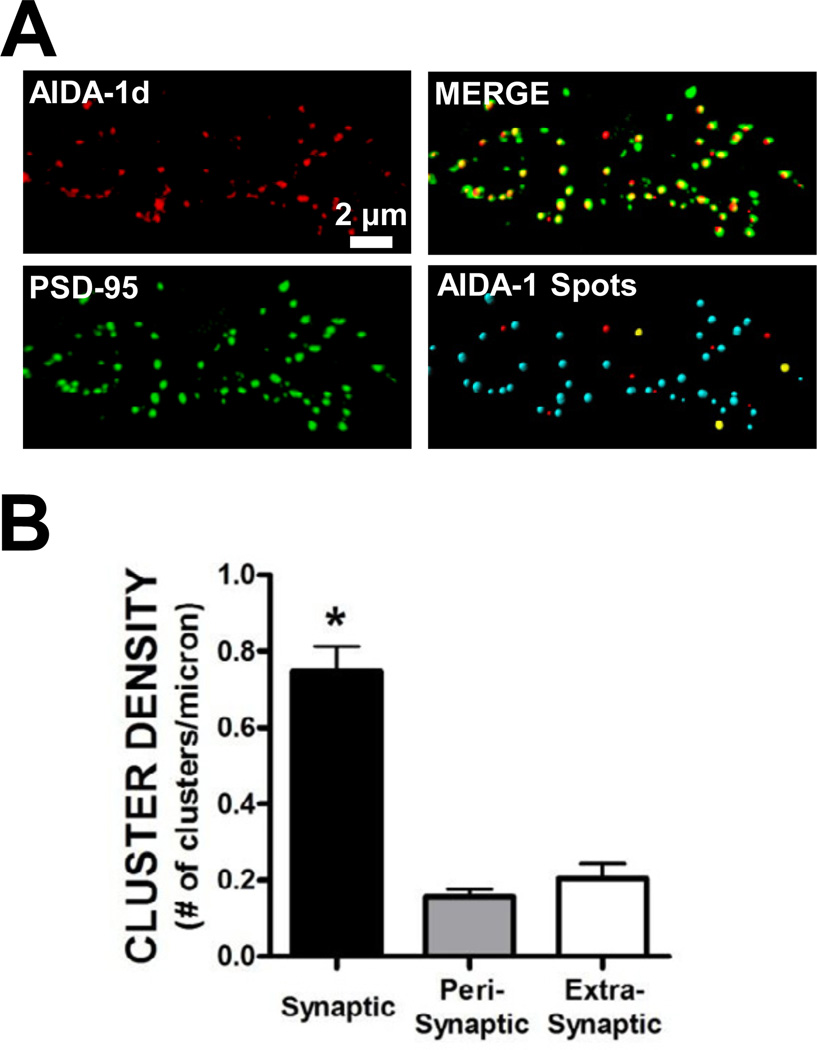
It has previously been reported that AIDA-1 is highly enriched in the PSD at glutamatergic synapses in hippocampal neurons (Jacob et al., 2010; Jordan et al., 2004; Jordan et al., 2007). As expected, we observed extensive colocalization of AIDA-1 with the PSD scaffolding protein PSD-95 in rat primary hippocampal cultures at DIV 21 (Fig. 1A). We then further characterized the subcellular localization of AIDA-1 at synaptic (< 200 nm from a PSD-95 cluster), perisynaptic (between 200 and 600 nm), and extrasynaptic (> 600 nm) sites. Supporting previous evidence (Jordan et al., 2007), the density of AIDA-1 clusters was significantly enriched at synaptic sites under basal conditions (Fig. 1B). The majority (~68%) of AIDA-1 dendritic clusters colocalize with PSD-95 at synaptic sites, while ~14% and ~18% of AIDA-1 clusters were found at perisynaptic and extrasynaptic sites, respectively (Fig 1B).
Fig. 1.
Subcellular localization of AIDA-1 at synaptic, perisynaptic, and extrasynaptic sites in hippocampal neurons. (A) The majority of punctate clusters of AIDA-1 were colocalized with the postsynaptic density scaffolding protein PSD-95. We also observed that a smaller portion of AIDA-1 clusters did not colocalize with PSD-95. Bottom right panel – AIDA-1 Spots: Using the Spots extension in ImarisXT software, AIDA-1 clusters were colocalized within 0.2 µm of a PSD-95 cluster (synaptic, shown as a blue spot) or colocalized within 0.2 and 0.6 µm (perisynaptic, shown as a yellow spot). A subset of AIDA-1 clusters did not colocalize within 0.6 µm (extrasynaptic, shown as a red spot) of a PSD-95 cluster. (B) Significantly more AIDA-1 clusters colocalized with PSD-95 at synaptic sites when compared with AIDA-1 clusters at peri- or extrasynaptic sites. (one-way ANOVA, F[2,63] = 54.06, P < .0001, Student Newman-Keuls (SNK) post-hoc, *P < .001 vs. peri- and extrasynaptic AIDA-1 cluster density, n = 22 dendrites/group).
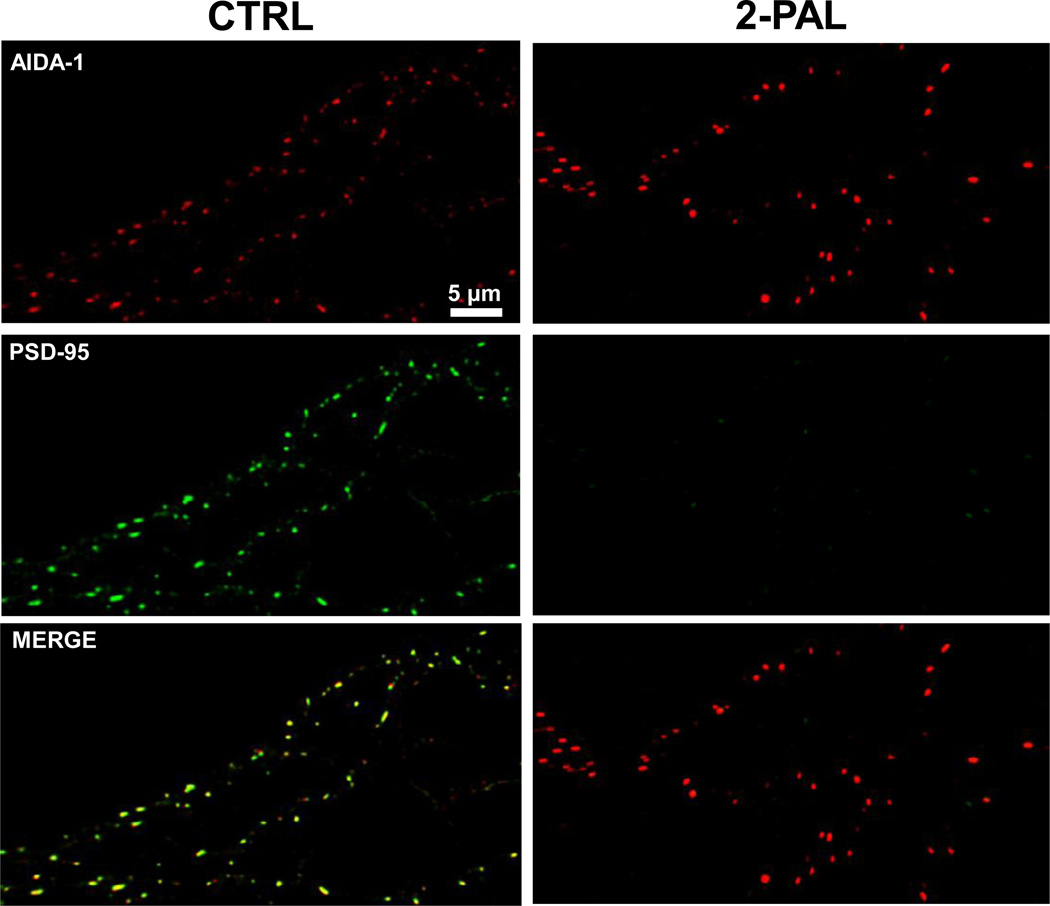
We next explored mechanisms for anchoring AIDA-1 to synaptic sites. AIDA-1d binds to PSD-95 via a C-terminal PDZ ligand (Jordan et al., 2007), suggesting that PSD-95 represents a synaptic anchor for AIDA-1d. The stability of PSD-95 at synaptic sites is regulated by palmitoylation of Cys residues near the N-terminus (El-Husseini et al., 2000; El-Husseini Ael et al., 2002). To directly test the hypothesis that the synaptic localization of AIDA-1 requires PSD-95, we treated neurons with 2-Pal to inhibit palmitoylation and decluster PSD-95. A 24 hr treatment with 10 µM 2-Pal led to a complete loss of PSD-95 clusters (Fig. 2). However, inhibition of palmitoylation for 24 (Fig. 2) or 48 (data not shown) hours did not affect AIDA-1 clustering. These findings suggest that under basal conditions PSD-95 is not necessary for the synaptic localization of AIDA-1.
Fig. 2.
The postsynaptic density scaffolding protein PSD-95 is not necessary for AIDA-1 clustering. Depalmitylation (10 µM 2-Pal, 24 hr) drastically reduced PSD-95 (green) clustering in hippocampal neurons without altering AIDA-1 (red) clustering.
Chronic Ethanol and AIDA-1 Clustering
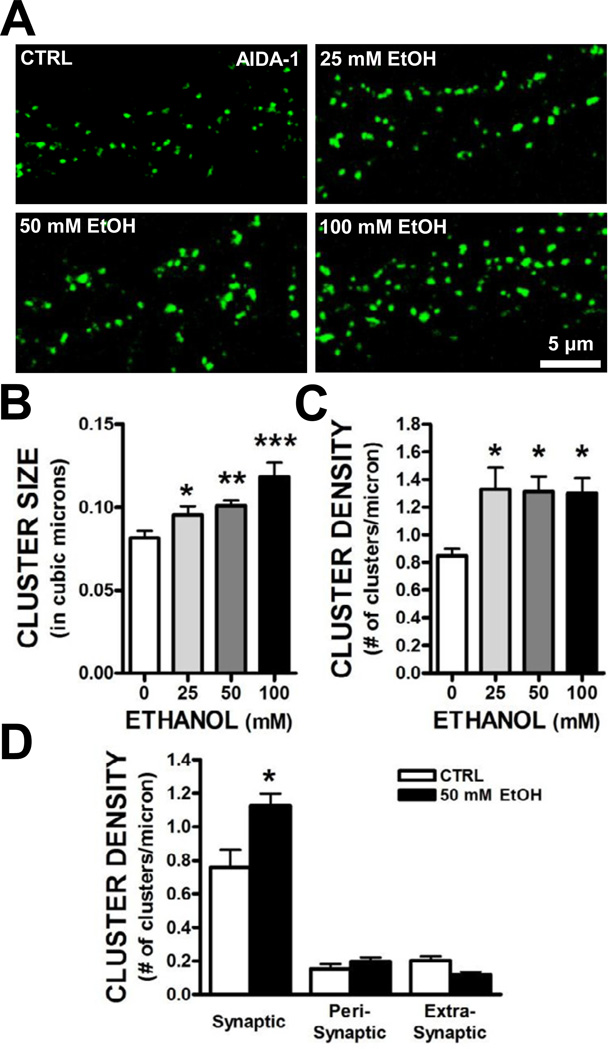
Chronic treatment of primary hippocampal neurons with ethanol increases the targeting of NMDA receptors to synapses (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004). This synaptic targeting was associated with an upregulation of PSD-95 and an increase in dendritic spine size and NMDA receptor function. We observed a concentration-dependent increase in AIDA-1 cluster size in hippocampal neurons treated with 25 – 100 mM ethanol for 4 days (Fig. 3A,B). In addition, all concentrations of ethanol used in this study produced an increase in AIDA-1 cluster density (Fig. 3A,C). We next determined the subcellular localization of the enhanced clustering of AIDA-1 by chronic ethanol using PSD-95 as a marker of glutamatergic synapses. We found that there was a significant ethanol-induced increase in the density of AIDA-1 clusters that were colocalized with PSD-95 without any change in density at peri- or extrasynaptic sites (Fig. 3D).
Fig. 3.
Chronic ethanol exposure increased synaptic clustering of AIDA-1. (A–C) Exposure of hippocampal neurons for 4 days with ethanol (25 – 100 mM) increased AIDA-1 cluster size and density. (Size: one-way ANOVA, F[3,40] = 7.67, P < .001, SNK post-hoc, *P < .05 vs. 0, **P < .01 vs. 0, ***P < .001 vs. 0; Density: one-way ANOVA, F[3,37] = 5.63, P < .01, SNK post-hoc, *P< .05 vs. 0; n = 8–9 images/group). Analysis of AIDA-1 subcellular localization revealed that chronic ethanol significantly increased AIDA-1 cluster density at synaptic sites. (two-way ANOVA, F[2,24] = 7.20, P < .01, SNK post-hoc, *P < .001 vs. CTRL, n = 16–20 dendrites/group).
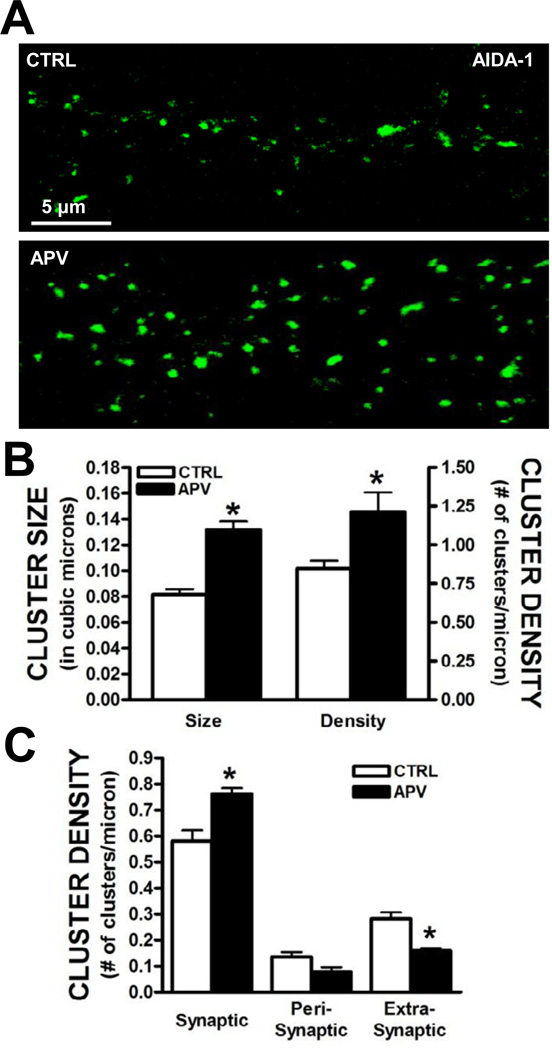
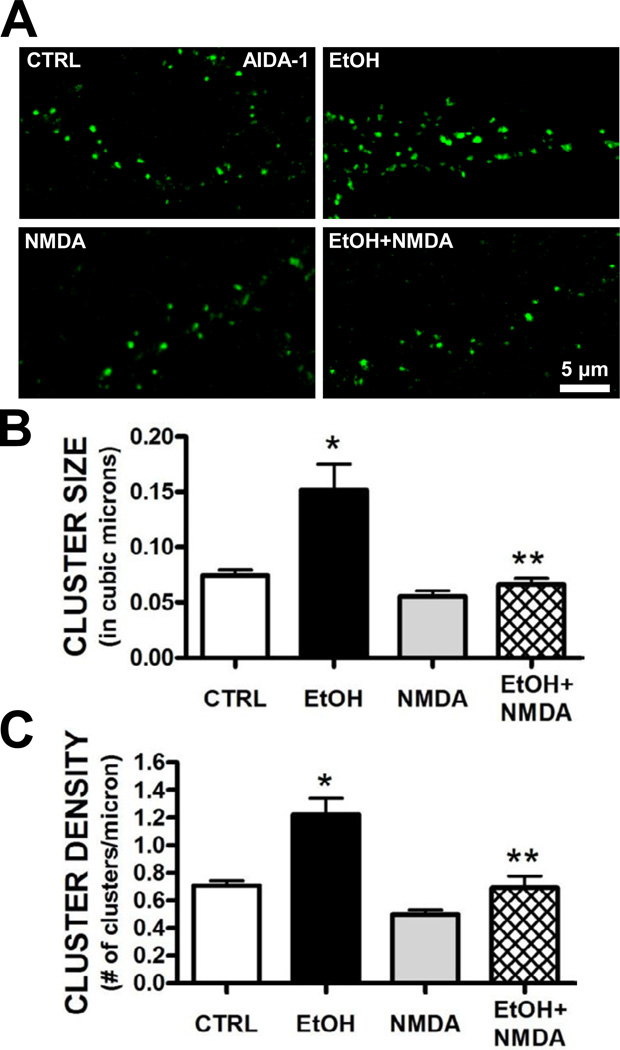
Since ethanol can inhibit NMDA receptor activity, we hypothesized that the loss of NMDA receptor function played a role in the increased clustering of AIDA-1 at synapses. We therefore repeated the experiments described above in the presence of the selective NMDA receptor antagonist APV (50 µM). Similar to our observation with chronic ethanol, 4 day treatment with APV significantly increased dendritic AIDA-1 cluster size and density (Fig. 4A,B). APV treatment also significantly enhanced cluster density at synaptic sites (i.e., clusters colocalized with PSD-95) while reducing the density of AIDA-1 clusters at extrasynaptic localizations (Fig. 4C). We next determined if activation of NMDA receptors would prevent chronic the ethanol-associated upregulation of AIDA-1 at synapses. Treatment of neurons with a non-toxic concentration of NMDA (2.5 µM) blocked chronic ethanol-induced changes in AIDA-1 cluster size and density (Fig. 5). Together, these data suggest that NMDA receptor activity regulates the subcellular localization of AIDA-1.
Fig. 4.
Prolonged inhibition of NMDA receptor activity enhances AIDA-1 clustering. (A,B) Four day treatment of neurons with APV (10 µM) significantly increased AIDA-1 cluster size and density. (Size: two-tailed t-test, t[19] = 3.29, *P < .01; Density: two-tailed t-test, t[21] = 6.41, *P < .001 vs. CTRL; n = 9–11 dendrites/group). (C) Analysis of the subcellular localization revealed that 4 day APV treatment significantly increased AIDA-1 cluster density at synaptic sites and reduced cluster density at extrasynaptic sites. (two-way ANOVA, F[2,24] = 17.04, P < .001, SNK post-hoc, *P < .01 vs. CTRL, n = 16–18 dendrites/group).
Fig. 5.
Chronic activation of NMDA receptors prevents ethanol-induced increases in AIDA-1 clustering. While a non-toxic concentration of NMDA (2.5 µM) did not affect AIDA-1 cluster size or density in ethanol-naive neurons, co-exposure of NMDA significantly blocked the enhanced clustering by 50 mM ethanol. (Size: one-way ANOVA, F[3,15] = 14.30, P < .001, SNK post-hoc, *P < .001 vs. CTRL, **P < .001 vs. EtOH, ***P < .001 vs. 0; Density: one-way ANOVA, F[3,15] = 16.55, P < .001, SNK post-hoc, *P < .05 vs. CTRL, **P < .01 vs. EtOH; n = 12–15 images/group).
Knockdown of AIDA-1
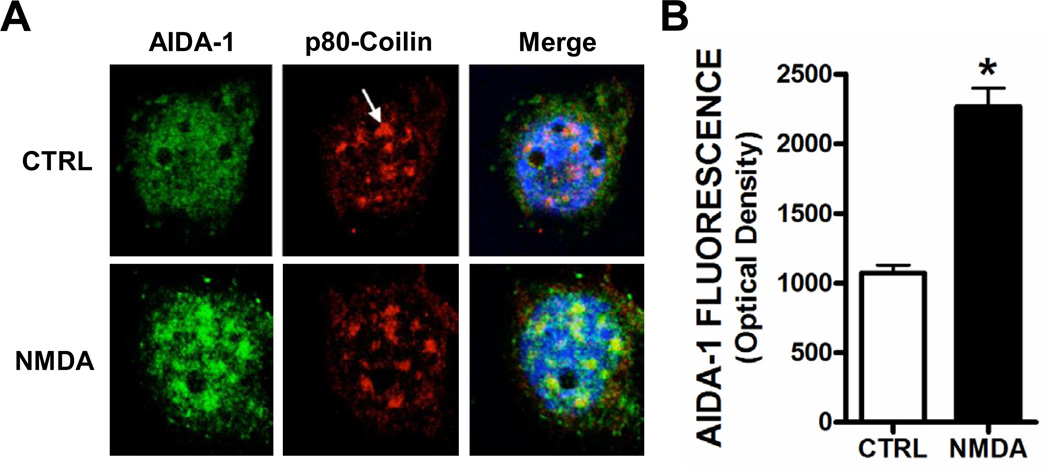
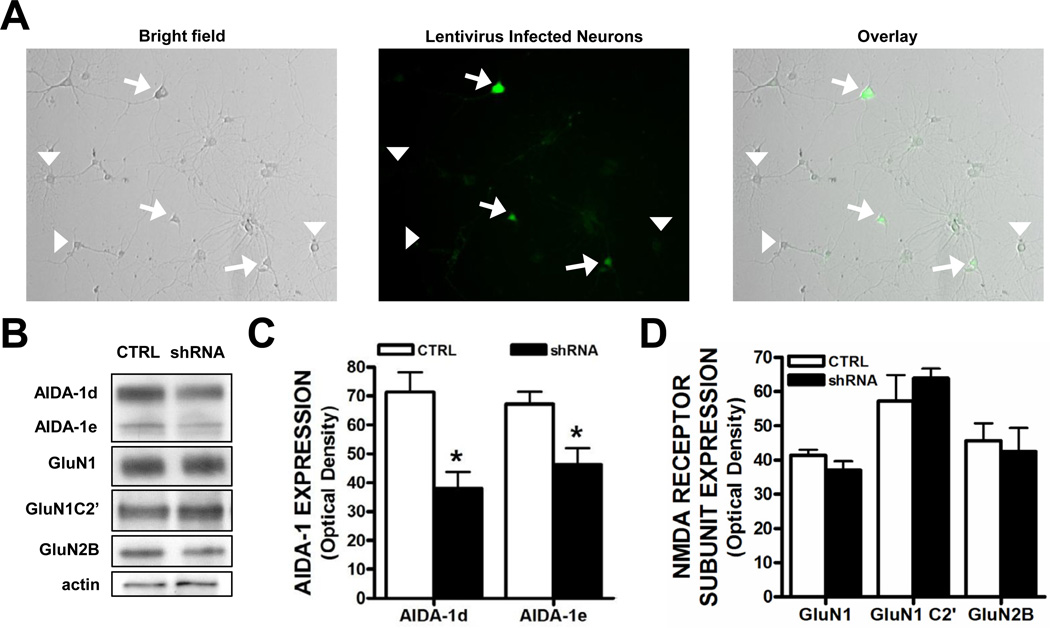
The translocation of AIDA-1d to nuclear Cajal bodies has been proposed to regulate ribosome biogenesis and global protein translation (Jordan et al., 2007; Jordan and Kreutz, 2009). Knockdown of AIDA-1 partially prevented an activity-dependent increase in protein synthesis (Jordan et al., 2007). Consistent with a previous report, we found that 10 min application of NMDA (10 µM) significantly increased the colocalization of AIDA-1 with p80 coilin, a marker of nuclear Cajal bodies (Fig. 6). To determine if the nuclear translocation of AIDA-1 played a role in activity-dependent regulation of NMDA receptor splicing or localization, we knocked down AIDA-1 protein and measured the levels of NMDA receptor subunit expression. Primary hippocampal neurons were transduced on DIV 7 with a lentivirus that expresses an shRNA targeting either firefly luciferase (control) or AIDA-1, and we determined the extent of AIDA-1d knockdown 7 days after transduction. We transduced neurons at a multiplicity of infection level of ~0.5 as approximately half (51.1 ± 5.6%) of the neurons in each dish were positive for GFP (Fig. 7A). We observed a significant reduction in AIDA-1d and AIDA-1e protein levels in neurons transduced with an AIDA-1 specific shRNA lentivirus (Fig. 7B,C). Confirming this knockdown, AIDA-1 cluster size in GFP-positive neurons was markedly smaller or absent when compared with non-transduced neurons (data not shown). However, contrary to our hypothesis, AIDA-1 knockdown did not significantly alter the expression levels of GluN1 or the GluN2B subunits of the NMDA receptor (Fig. 7D). Moreover AIDA-1d knockdown did not affect expression of the alternatively spliced GluN1 C2’ variant (Fig. 7D). These results suggest that surface expression of NMDA receptors remained unchanged and that the nuclear trafficking of AIDA-1 did not regulate alternative splicing.
Fig. 6.
Activation of NMDA receptors increased AIDA-1 in nuclear Cajal bodies. Acute treatment (10 min) of hippocampal neurons with 10 µM NMDA enhanced AIDA-1 fluorescence (green) in Cajal bodies stained with p80 Coilin (red). Nuclei were counterstained with DRAQ-5 (blue). (two-tailed t-test, t[7] = 14.98, *P < .001, n = 8–9 neurons/group).
Fig. 7.
Knockdown of AIDA-1d does not affect NMDA receptor expression. (A) Bright field and GFP expression in transduced hippocampal neurons. (Arrows, GFP-positive neurons; Arrowheads, GFP-negative neurons). (B) Representative Western blots for AIDA-1 variants, NMDA receptor subunits, and actin in neurons transduced with a lentivirus that expresses an shRNA targeting either firefly luciferase (CTRL) or AIDA-1. (C) Quantitation of lentiviral knockdown of AIDA-1 variants (two-tailed t-tests, *P < .05 vs. CTRL, n = 7–9 dishes/group). (D) Quantitation of NMDA receptor subunits following lentiviral knockdown of AIDA-1d (two-tailed t-tests, P > .05, n = 7–9 dishes/group).
Discussion
The principal finding from this study is that chronic ethanol exposure results in homeostatic adaptations that include changes in the synaptic levels of the synpatonuclear signaling protein AIDA-1. Consistent with previous findings from Jordan and colleagues (2007), we demonstrated that AIDA-1 expression was highly enriched at synaptic sites and colocalized with the PSD scaffolding protein PSD-95. Moreover, acute activation of NMDA receptors in ethanol-naïve cultures induced translocation of AIDA-1 to nuclear Cajal bodies. Chronic ethanol exposure of hippocampal neurons increased expression of AIDA-1 at synaptic sites, a process that was dependent upon NMDA receptor activity. These findings raised the possibility that increased AIDA-1 synaptonuclear signaling plays a role in previous findings showing that chronic ethanol can upregulate NMDA receptors at synapses. However, here we show that AIDA-1 does not appear to regulate NMDA receptor subunit expression or the alternative splicing of NMDA receptors. Therefore, AIDA-1 does not appear to play a role in the homeostatic regulation of NMDA receptors in response to chronic alcohol exposure.
To study the effects of ethanol on AIDA-1 subcellular localization, we used a well-characterized model that produces enhanced targeting of NMDA receptors to neuronal synapses and increased dendritic spine size (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004; Clapp et al., 2010). We first characterized the subcellular expression patterns of AIDA-1 using data from electron microscopy studies that reported the length of PSDs from asymmetric synapses in hippocampal neurons, averages between 180 and 260 nm (Moretti et al., 2006; Takumi et al., 1999). We classified punctate clusters of AIDA-1 as synaptic (< 200 nm from a PSD-95 cluster), perisynaptic (between 200 and 600 nm from a PSD-95 cluster), or extrasynaptic (> 600 nm from a PSD-95 cluster). As expected, the majority (~70%) of AIDA-1 clusters were colocalized with PSD-95, suggesting a high level of AIDA-1 expression at glutamatergic synapses. To a much lesser extent, clusters of AIDA-1 were observed at perisynaptic and extrasynaptic localizations. We found that 4 day treatment of hippocampal neurons with ethanol increased AIDA-1 cluster size and density in a concentration-dependent manner. This enhanced clustering was only observed at synapses, as we did not observe ethanol-induced changes in peri- or extrasynaptic AIDA-1 clusters. We previously reported that chronic ethanol treatment of rat primary hippocampal neurons increased expression of NMDA receptors at synaptic, but not extrasynaptic sites (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004). In addition, we demonstrated that chronic ethanol exposure increased GluN2B-PSD-95 complexes at synaptic sites, suggesting an expanded scaffolding platform for recruitment of additional proteins and signaling molecules (Carpenter-Hyland and Chandler, 2006). Jordan and colleagues (2007) recently demonstrated that the C-terminus of AIDA-1d binds to the first two PDZ domains of PSD-95, and that mutating this region markedly reduced its punctate staining in dendritic spines. Together, these data would suggest that AIDA-1 is recruited to the synapse as part of chronic ethanol-induced increases in PSD-95 expression and expansion of the PSD.
Similar to the effects of chronic ethanol on NMDA receptors (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004), the upregulation of AIDA-1 at synapses appears to be dependent upon ethanol-induced inhibition of NMDA receptor activity. This contention is supported by data from two sets of experiments. First, prolonged inhibition of NMDA receptors with the selective antagonist APV produced an increase in the expression of AIDA-1 at synapses that was similar to that observed upon chronic ethanol exposure. Treatment of hippocampal neurons with APV increased AIDA-1 clusters size and density at synaptic localizations and decreased the density of clusters at extrasynaptic localizations. This suggests that chronic inhibition of NMDA receptor activity may increase forward trafficking of AIDA-1 from extrasynaptic sites to the PSD. Alternatively, the increase in AIDA-1 by chronic ethanol may be an increase in AIDA-1 protein synthesis at synaptic sites. We also demonstrated that enhancing NMDA receptor activity completely blocked ethanol-induced adaptations in AIDA-1 clustering. Thus, it appears that NMDA receptor activity can bidirectionally regulate the expression of AIDA-1 at synapses. These data provide further evidence that homeostatic adaptive plasticity contributes to remodeling of glutamatergic synapses by chronic ethanol exposure.
Cajal bodies are nuclear structures prominent in transcriptionally-active neurons and are enriched in proteins and factors involved in assembly and processing of small nuclear and nucleolar ribonucleoproteins (RNPs) and histone gene transcription (Mao et al., 2011). Specific RNPs have been implicated in pre-mRNA splicing and therefore influence alternative splicing (Ule, 2008; Will and Luhrmann, 2011). Our data are consistent with previous findings demonstrating an increase in AIDA-1 colocalization with p80 coilin, a marker of Cajal bodies, in response to acute NMDA receptor activity. Because chronic ethanol increases synaptic AIDA-1 expression, we reasoned that ethanol-induced increases in AIDA-1 may enhance synaptonuclear AIDA-1 shuttling leading to altered expression or mRNA alternative splicing of NMDA receptors. Alternative splicing of the GluN1 C2’ domain accelerates membrane insertion of functional NMDA receptors (Mu et al., 2003; Okabe et al., 1999), and chronic ethanol increases GluN1 C2’ surface expression (Clapp et al., 2010). However, AIDA-1 knockdown did not alter expression of GluN1 C2’. AIDA-1 knockdown also did not alter the expression of GluN1 or GluN2B subunits of NMDA receptors. Together, these data suggest that AIDA-1 does not regulate the expression or alternative splicing of NMDA receptors under basal conditions.
Jordan et al. suggest that PSD-95 provides the necessary scaffolding machinery for maintenance of AIDA-1 at synaptic sites. Dual palmitoylation of Cys residues on the N-terminus (a synaptic targeting motif within the C-terminal domain) and PDZ binding domains are necessary for PSD-95 clustering at asymmetric synapses (Craven et al., 1999; El-Husseini et al., 2000; El-Husseini Ael et al., 2002). Palmitoylation of PSD-95 regulates clustering of the GluR1 subunit of AMPA receptors and Kv1.4 channels, but not NMDA receptor clustering (Carpenter-Hyland and Chandler, 2006; El-Husseini et al., 2000; El-Husseini Ael et al., 2002). To test the hypothesis that PSD-95 represents the AIDA-1 synaptic anchor, we treated neurons with an inhibitor of palmitoylation and found that this led to a complete loss of PSD-95 clustering (Carpenter-Hyland and Chandler, 2006; El-Husseini Ael et al., 2002). However, AIDA-1 clustering was not affected by 2-Pal treatment. Notably, 2-Pal also declusters the scaffolding protein PSD-93 (El-Husseini Ael et al., 2002). Thus, even though AIDA-1 complexes with PSD-95, our data suggest that palmitoylation is not necessary for AIDA-1 clustering and that PSD-95 (and perhaps PSD-93) is not required for maintenance of AIDA-1 at synaptic sites.
To our knowledge, these are the first data to demonstrate chronic ethanol-induced changes in the synaptic levels of a retrograde synaptonuclear shuttling protein. While our data suggest that AIDA-1 does not regulate NMDA receptor expression, the changes we observed in AIDA-1 at the synapse may contribute to homeostatic plasticity associated with chronic ethanol exposure. Thus, future work is necessary to determine which proteins are regulated by AIDA-1 signaling and how AIDA-1 influences ethanol-induced changes in these proteins. In addition, it will be interesting to determine how signaling from the synapse to the nucleus is disrupted by ethanol.
Acknowledgements
The authors would like to acknowledge the supported of NIH Grants AA010983 and AA017922.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Carpenter-Hyland EP, Chandler LJ. Homeostatic plasticity during alcohol exposure promotes enlargement of dendritic spines. Eur. J. Neurosci. 2006;24:3496–3506. doi: 10.1111/j.1460-9568.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- Carpenter-Hyland EP, Chandler LJ. Adaptive plasticity of NMDA receptors and dendritic spines: implications for enhanced vulnerability of the adolescent brain to alcohol addiction. Pharmacol. Biochem. Behav. 2007;86:200–208. doi: 10.1016/j.pbb.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter-Hyland EP, Woodward JJ, Chandler LJ. Chronic ethanol induces synaptic but not extrasynaptic targeting of NMDA receptors. J. Neurosci. 2004;24:7859–7868. doi: 10.1523/JNEUROSCI.1902-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp P, Gibson ES, Dell'acqua ML, Hoffman PL. Phosphorylation regulates removal of synaptic N-methyl-d-aspartate receptors after withdrawal from chronic ethanol exposure. J. Pharmacol. Exp. Ther. 2010;332:720–729. doi: 10.1124/jpet.109.158741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven SE, El-Husseini AE, Bredt DS. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron. 1999;22:497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, et al. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6:e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Craven SE, Chetkovich DM, Firestein BL, Schnell E, Aoki C, Bredt DS. Dual palmitoylation of PSD-95 mediates its vesiculotubular sorting, postsynaptic targeting, and ion channel clustering. J. Cell Biol. 2000;148:159–172. doi: 10.1083/jcb.148.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Gorelova N, Mulholland PJ, Chandler LJ, Seamans JK. The Glutamatergic Component of the Mesocortical Pathway Emanating from Different Subregions of the Ventral Midbrain. Cereb. Cortex. 2011 doi: 10.1093/cercor/bhr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AL, Jordan BA, Weinberg RJ. Organization of amyloid-beta protein precursor intracellular domain-associated protein-1 in the rat brain. J. Comp. Neurol. 2010;518:3221–3236. doi: 10.1002/cne.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas J, Skowronski J, Van Aelst L. Lentiviral delivery of RNAi in hippocampal neurons. Methods Enzymol. 2006;406:593–605. doi: 10.1016/S0076-6879(06)06046-0. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol. Cell. Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat. Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Kreutz MR. Nucleocytoplasmic protein shuttling: the direct route in synapse-to-nucleus signaling. Trends Neurosci. 2009;32:392–401. doi: 10.1016/j.tins.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Roberto M, editors. Synaptic effects induced by alcohol. Springer; 2012. [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Mulholland PJ, Chandler LJ. The thorny side of addiction: adaptive plasticity and dendritic spines. Scientific World J. 2007;7:9–21. doi: 10.1100/tsw.2007.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H. Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J. Neurosci. 1999;19:7781–7792. doi: 10.1523/JNEUROSCI.19-18-07781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proepper C, Johannsen S, Liebau S, Dahl J, Vaida B, Bockmann J, Kreutz MR, Gundelfinger ED, Boeckers TM. Abelson interacting protein 1 (Abi-1) is essential for dendrite morphogenesis and synapse formation. Embo. J. 2007;26:1397–1409. doi: 10.1038/sj.emboj.7601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Denny AD, Ticku MK. Chronic intermittent ethanol treatment selectively alters N-methyl-D-aspartate receptor subunit surface expression in cultured cortical neurons. Mol. Pharmacol. 2007;72:95–102. doi: 10.1124/mol.106.033043. [DOI] [PubMed] [Google Scholar]
- Takumi Y, Ramirez-Leon V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat. Neurosci. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- Ule J. Ribonucleoprotein complexes in neurologic diseases. Curr. Opin. Neurobiol. 2008;18:516–523. doi: 10.1016/j.conb.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Wang J, Carnicella S, Phamluong K, Jeanblanc J, Ronesi JA, Chaudhri N, Janak PH, Lovinger DM, Ron D. Ethanol induces long-term facilitation of NR2B-NMDA receptor activity in the dorsal striatum: implications for alcohol drinking behavior. J. Neurosci. 2007;27:3593–3602. doi: 10.1523/JNEUROSCI.4749-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lanfranco MF, Gibb SL, Yowell QV, Carnicella S, Ron D. Long-lasting adaptations of the NR2B-containing NMDA receptors in the dorsomedial striatum play a crucial role in alcohol consumption and relapse. J. Neurosci. 2010;30:10187–10198. doi: 10.1523/JNEUROSCI.2268-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011:3. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina O, Baly C, Chebrout M, Debey P. The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the Cajal (coiled) body. Dev. Biol. 2003;253:66–83. doi: 10.1006/dbio.2002.0865. [DOI] [PubMed] [Google Scholar]