Abstract
Regulated centrosome biogenesis is required for accurate cell division and for maintaining genome integrity1. Centrosomes consist of a centriole pair surrounded by a protein network known as pericentriolar material (PCM)1. PCM assembly is a tightly regulated, critical step that determines a centrosome’s size and capability2–4. Here, we report a role for tubulin in regulating PCM recruitment via the conserved centrosomal protein Sas-4. Tubulin directly binds to Sas-4; together they are components of cytoplasmic complexes of centrosomal proteins5,6. A Sas-4 mutant, which cannot bind tubulin, enhances centrosomal protein complex formation and has abnormally large centrosomes with excessive activity. These suggest that tubulin negatively regulates PCM recruitment. Whereas tubulin-GTP prevents Sas-4 from forming protein complexes, tubulin-GDP promotes it. Thus, tubulin’s regulation of PCM recruitment depends on its GTP/GDP-bound state. These results identify a role for tubulin in regulating PCM recruitment independent of its well-known role as a building block of microtubules7. Based on its guanine bound state, tubulin can act as a molecular switch in PCM recruitment.
Centrosome biogenesis is a multi-step process that begins with centriole formation followed by PCM recruitment to form a functional organelle4. PCM recruitment begins with the formation of cytoplasmic protein complexes and requires Sas-4/CPAP3,5,8,9. Recently, we reported that Sas-4, a protein known to have a role in centriole and PCM formation3,10,11, scaffolds centrosomal protein complexes (S-CAP complexes) which include Cnn, Asl, D-PLP, CP-190, and tubulin (αβ–tubulin dimer), and tethers the S-CAP complexes to centrosomes5. Sas-4 also exists in complexes with γ-tubulin9 and γ-tubulin ring proteins (S-γ-tubulin complexes), suggesting that Sas-4 may also be associated with the assembly intermediates of γ-tubulin ring complexes (Figs. 1). S-γ-tubulin complexes are recruited to developing centrosomes in a Sas-4-dependent manner (Fig. 2). Together, these suggest that Sas-4 regulates PCM recruitment via several protein complex types. Interestingly, it appears that tubulin exists in the multiple Sas-4 complex types (Fig. 1b–d). Since tubulin is significantly more abundant than other centrosomal proteins, Sas-4 likely interacts with free tubulin prior to formation of the centrosomal protein complexes. If the Sas-4-tubulin interaction is a first step, then tubulin may regulate centrosomal complex formation and PCM recruitment.
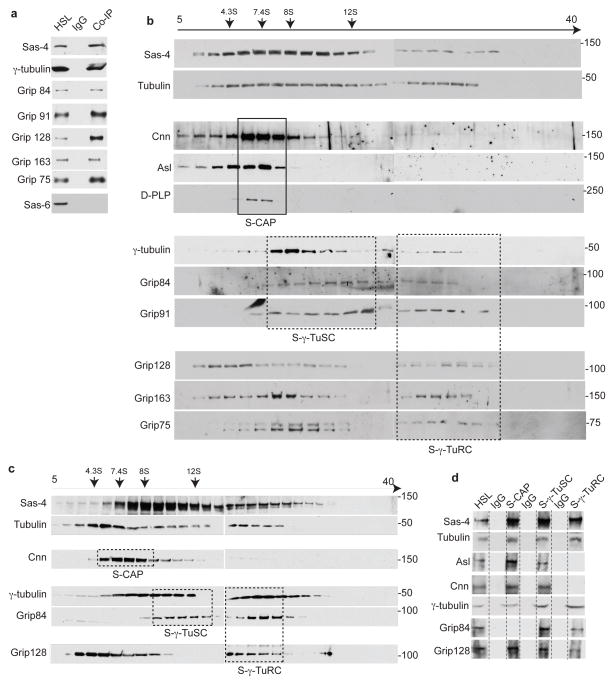
Fig. 1. Tubulin is present in each Sas-4 complex type.
(a) Immuno-purification of Sas-4 complexes from high-speed lysate (HSL) of Drosophila embryonic extracts using anti-Sas-4 antibody revealed associations between Sas-4 with Grip proteins that are components of γ-TuRCs: Grip91, Grip84, Grip163, Grip128, and Grip75, and γ-tubulin small complexes (γ-TuSCs): Grip91 and Grip8436. Embryonic extracts were used as a positive control; mouse IgG beads were used as a negative control. The use of extract depleted of centrosomes and the absence of the centriole-core protein Sas-6 indicate that the purified complexes were not centrosomes.
(b) Immuno-purified Sas-4 complexes fractionate at distinct densities in a 5–40% sucrose gradient. Individual fractions are analyzed by Western blot. Tubulin co-fractionates with Sas-4 across the gradient. Co-fractionation patterns likely represent different complex types: S-CAP (Cnn, Asl, and D-PLP, proteins); dashed boxes, S-γ-TuSC, and S-γ-TuRC; dashed boxes. The fractionation pattern of S-CAP complexes in a narrow range of low-density fractions and γ-tubulin and Grip proteins fractionation at intermediate and high-densities were consistent with the previous reports5, 36. However, the fractionation pattern of γ-TuSC and γ-TuRC proteins complexes do not exhibit clear peaks as previously demonstrated suggesting that Sas-4 interact with assembly intermediates of γ-tubulin ring proteins.
(c–d) The immuno-purified Sas-4 complexes are unlikely to be part of an unstable large complex that destabilizes during immuno-purification. HSL of Drosophila embryonic extract was first fractionated in a 5–40% sucrose gradient (c) and the immuno-purifications of the distinct Sas-4 complex types were performed on distinct fractions (marked by solid boxes and named S-CAP, S-γ-TuSCs and S-γ-TuRCs) (d). Note that Sas-4 and tubulin were detected in all of the complex types. Embryonic extract was used as a positive control; mouse IgG beads were used as a negative control.
In experiments (a–d), HSLs were diluted such that the tubulin concentration was below 0.2μM in order to prevent tubulin polymerization; additionally, Sas-4 complexes were purified at 4°C in the presence of nocodazole (330 nM). In experiments (b and c), inverted arrows mark the corresponding peaks of the sedimentation coefficient standards.
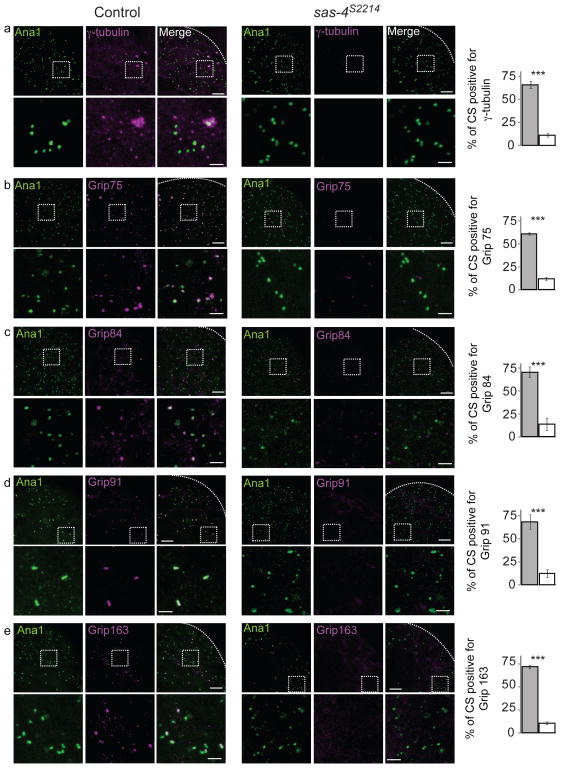
Fig. 2. Sas-4 is essential for recruiting S-γ-tubulin complexes to centrosomes.
Centriolar structures labeled by Ana-1-GFP in control testes but not in sas-4s2214 null mutant testes recruit components of S-γ-tubulin complexes as tested using antibodies specific to γ-tubulin, Grip75, Grip84, Grip91 and Grip163 (a–e). Dashed boxes mark the enlarged areas shown in the lower panels. Charts on the right show the fraction of Ana-1 positive centriolar structures (CS) that are also positive for the respective proteins tested in the control (gray filling) and in sas-4s2214 (white filling). As described previously5, Ana-1-GFP-labeled centriolar structures from each testis were counted within a 20μm2 area that is ~25 μm away from the tip of a testis (dotted lines). The mean ± SEM of three independent testes are shown. (a–e) Scale bar 10 μm; and 1 μm for lower and higher magnification (inset), respectively. *** marks significant difference (P<0.001)
We began testing this hypothesis, by comparing the abilities of Sas-4, which can bind tubulin, to a mutated version of Sas-4, which cannot bind tubulin. For this, we used an N-terminal fragment of Sas-4 (Sas-4-N) that includes Sas-4’s tubulin binding site12,13; we also used a mutated version of Sas-4-N (Sas-4-NΔT), which lacks the two amino acids essential for tubulin binding12,13. As expected, Sas-4-NΔT failed to pull-down tubulin from embryonic high-speed lysates (HSLs). Surprisingly, Sas-4-NΔT pulled-down significantly more Cnn, Asl, D-PLP, γ-tubulin, and Grip128 than was pulled-down by Sas-4-N (Fig. 3a). We then tested the effects of tubulin on the ability of Sas-4-N or Sas-4-NΔT to bind centrosomal proteins. Increasing amounts of tubulin progressively inhibited Sas-4-N’s binding to centrosomal proteins but did not inhibit Sas-4-NΔT’s binding (Fig. 3b–c). These suggest that tubulin can negatively regulate the formation of centrosomal protein complexes.
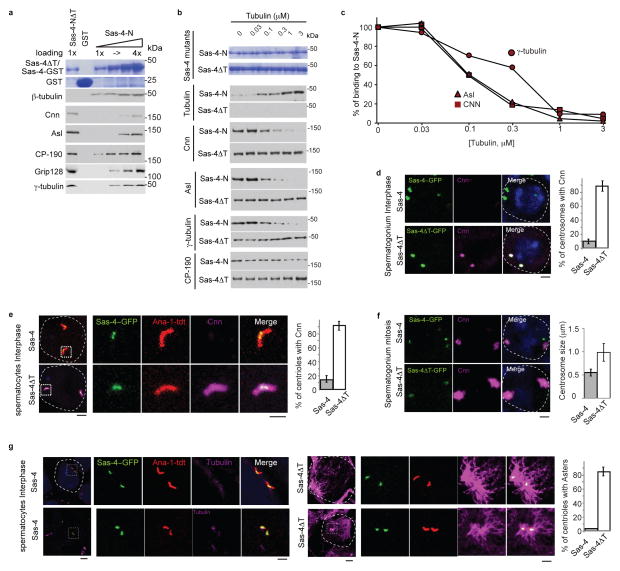
Fig. 3. Tubulin negatively regulates PCM recruitment.
(a) Comparison of the ability of Sas-4-NΔT and Sas-4-N to interact with centrosomal proteins in embryonic extracts. Increasing loading amounts of Sas-4-N (1 to 4-fold) pulls down Sas-4 interacting proteins from embryonic HSL. Sas-4-NΔT does not pull down tubulin, but pulls down approximately three times more Cnn, Asl, γ-tubulin, and Grip128 than Sas-4-N. Purified recombinant proteins are shown in Coomassie stained gels
(b–c) Addition of increasing amounts of free tubulin to Sas-4-N pull down experiments from HSL proportionally inhibits Sas-4-N binding to its interacting partners (a) with an IC50 of 0.1–0.3 μM (b). There is no significant change in the ability of Sas-4-NΔT to bind Cnn and Asl in the presence of tubulin. CP-190 binding to Sas-4N did not change significantly; suggesting that tubulin specifically interferes with the binding of Cnn, Asl and γ-tubulin. The purified recombinant proteins used are shown in Coomassie stained gels.
(d) In Sas-4::sas-4s2214 testes Cnn immunoreactivity (Magenta) is not detected in interphase centrosomes. In contrast, sas-4ΔT::sas-4s2214 (sas-4ΔT) interphase centrosomes contain Cnn. Dotted lines mark a cell boundary. Charts show the percentage of centrosomes positive for Cnn. The mean±SEM of six independent testes are shown. p<0.001. Scale bar, 2 μm..
(e) Unlike Sas-4::sas-4s2214, sas-4ΔT::sas-4s2214 interphase spermatocyte centrosomes contain Cnn. Centrosomes are marked by Ana-1-tdT (Red) and Sas-4-GFP (Green). Boxes mark the magnified areas. Charts show the percentage of centrosomes positive for Cnn. The mean±SEM of six independent testes are shown, p<0.001. Scale bar, 2 μm.
(f) sas-4ΔT::sas-4s2214 mitotic centrosomes have increased Cnn immunoreactivity. The chart shows centrosome size for Sas-4::sas-4s2214 and sas-4ΔT::sas-4s2214, as measured by Cnn immunolabeling. The mean±SEM of six independent testes are shown. p<0.001. Scale bar, 2μm.
(g) sas-4ΔT::sas-4s2214 interphase spermatocyte centrosomes emanate microtubule asters. Microtubules are stained by α-tubulin (Magenta). Boxes mark the magnified areas. The chart shows the percentage of centrosomes emanating microtubule asters in sas-4::sas-4s2214 (grey) and sas-4ΔT::sas-4s2214 (white). The mean±SEM of six independent testes are shown. p<0.001. Scale bar, 2 μm.
To test this hypothesis in vivo, we generated transgenic Drosophila that express full-length Sas-4ΔT in the sas-4s2214 null (sas-4ΔT::sas-4s2214). Sas-4ΔT failed to fully rescue the sas-4s2214 phenotype of uncoordination14: sas-4ΔT::sas-4s2214 flies stood but could barely walk (Supplementary Movie). Furthermore, sas-4ΔT::sas-4s2214 sperm axonemes were abnormal (Supplementary Fig. S1a). These phenotypes suggest defects in centrosome biogenesis14,15 and that tubulin binding to Sas-4 is essential for centrosome function and proper cilia formation. However, sas-4ΔT::sas-4s2214 flies had correct numbers of centrosomes, indicating that Sas-4ΔT rescued this aspect of the sas-4s2214 phenotype14 (Supplementary Fig. S1b). Thus, the Sas-4-tubulin interaction is not essential for maintaining centrosome number. Yet, sas-4ΔT::sas-4s2214 spermatocyte centrioles were slightly shorter (Supplementary Fig. S1c). This is consistent with reports that Sas-4, and in particular, the tubulin-Sas-4 interaction is required for centriole elongation13,16–18.
In addition to its well-known role in centriole formation, Sas-4 plays an important role in PCM formation and in regulating centrosome size3,10,11. Achieving proper centrosome size and capability requires Sas-4 and Cnn2,3. Since Sas-4 scaffolds centrosomal complexes that include Cnn, regulation of Sas-4 complex formation may indirectly control centrosome size. Indeed, although Cnn is normally detected only in mitotic or meiotic centrosomes, interphase spermatogonium and spermatocytes centrosomes of sas-4ΔT::sas-4s2214 contained Cnn (Fig. 3d–e)19. Moreover, mitotic and meiotic centrosomes of sas-4ΔT::sas-4s2214 contained twice the Cnn immunolabelling as control centrosomes (Fig. 3f and Supplementary Fig. S2a). Thus, tubulin can negatively regulate the timing, distribution and quantity of protein recruitment to centrosomes, via Sas-4.
In Drosophila, interphase centrosomes do not nucleate microtubules20. Since sas-4ΔT::sas-4s2214 centrosomes prematurely contain Cnn and Cnn’s human ortholog stimulates microtubule nucleation21, we tested whether the sas-4ΔT mutation affects microtubule nucleation. Interphase sas-4ΔT::sas-4s2214 centrosomes had premature microtubule nucleation (Fig. 3g) and their meiotic centrosomes had massive microtubule asters, which could fill a significant fraction of a cell (Supplementary Fig. S2b–c). Similarly, in cultured cells, Sas-4ΔT produced massive asters (Supplementary Fig. S2d–e). These results suggest that the tubulin present in wild-type Sas-4 complexes is not a building block of microtubule asters, but instead appears to be essential in the regulation of PCM recruitment.
To gain insight into how disruption of the Sas-4-tubulin interaction affects meiosis and mitosis, we analyzed spermatids and larval brain cells. We found that over 95% of sas-4ΔT::sas-4s2214 round spermatids exhibit normal morphology, suggesting that meiotic cell division can conclude normally (Supplementary Fig. S2f). In larval brain cells, unlike control cells, which recruit significant amounts of Cnn and form robust asters only during mitosis22, some sas-4ΔT::sas-4s2214 cells recruited Cnn and formed asters before entry into mitosis (Supplementary Fig. S3a–c). During mitosis, control larval brain cells have Cnn enrichment in only one centrosome22, 23, but in sas-4ΔT::sas-4s2214 cells, Cnn was distributed more evenly to both centrosomes (Supplementary Fig. S3d). Finally, spindle orientation relative to Bazooka’s crescent (a polarity establishment marker), were abnormal in sas-4ΔT::sas-4s2214, suggesting that these centrosomes have difficulty properly aligning their spindles (Supplementary Fig. S3e–f). Taken together, these results suggest that the interaction of tubulin with Sas-4 is essential for normal PCM recruitment and centrosome function in larval brain cells.
To better understand how tubulin operates in the regulation of PCM recruitment, we focused on the biochemical properties of the Sas-4-tubulin interaction. Tubulin is a guanine binding protein having GTPase activity, which hydrolyzes tubulin-GTP into tubulin-GDP7. Tubulin has a different conformation when present as tubulin-GTP versus tubulin-GDP and tubulin’s confirmation acts as a molecular switch that regulates microtubule dynamics7. Therefore, we speculated that tubulin’s confirmation might also regulate the formation of Sas-4 complexes. For this, we analyzed tubulin’s binding to Sas-4-N in the presence of GDP or GMPCPP (a non-hydrolyzable GTP analog)24. Sas-4-N, which includes Sas-4’s tubulin binding site, prevents microtubule polymerization when present in excess6,12,25. Tubulin-GMPCPP at 0.5μM (which is below the concentration necessary for microtubule polymerization26) had four-fold less binding to Sas-4-N than tubulin-GDP at the same concentration (Fig. 4a). Similar results were obtained by isothermal titration calorimetry experiments, indicating that tubulin-GDP has a higher affinity for Sas-4 than tubulin-GMPCPP has (Supplementary Fig. S4). However, since the affinity of tubulin to Sas-4 appears to be high (Fig. 3b–c and Supplementary Fig. S4) relative to the cytoplasmic concentration of free tubulin (~10 μM27), it is likely that cytoplasmic Sas-4 is bound to either tubulin-GDP or tubulin-GTP. Therefore, it is possible that conformation of this bound tubulin (depending on which guanine is present) regulates the formation of Sas-4-containing complexes.
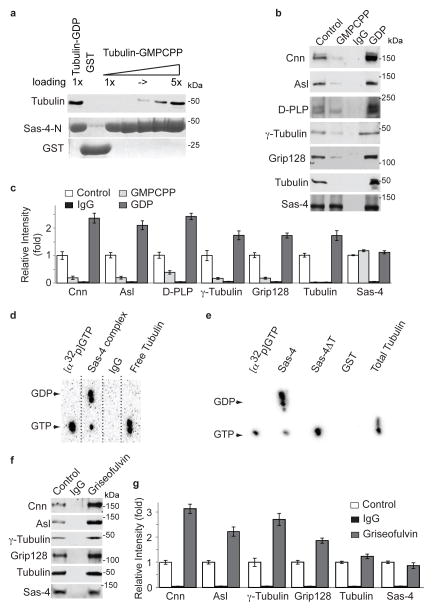
Fig. 4. Tubulin regulates Sas-4 complex formation.
(a) Sas-4-N GST (which includes the first 190 amino acids of Sas-4 and contains Sas-4’s tubulin binding site, 0.5 μM) binds over four times as much tubulin-GDP as tubulin-GMPCPP. Purified recombinant proteins (Sas-4-N and GST) are shown in Coomassie stained gels. Note that increasing loading amounts of Sas-4-N-GST (1- to 5-fold) are shown, and that Sas-4N-GST binds tubulin as much as five-fold Sas-4-N-GST. To prevent microtubule polymerization, this experiment contained nocodazole, the tubulin concentration was below the concentration necessary for microtubule polymerization, and the Sas-4-N concentration used prevents microtubule polymerization6, 12, 25.
(b–c) Immuno-purification of Sas-4 complexes from Drosophila embryonic HSL in the absence of additional nucleotides (control), or presence of additional GMPCPP, or GDP (2 mM). GDP enhances Sas-4’s complex association, whereas GMPCPP reduces assembly of the Sas-4 complex (b). The amount of Sas-4 was unchanged among the different experiments, however GDP enhanced the association of PCM components. Quantification of signal intensity (n=3). (c) The mean±SEM of three independent experiments are shown.
(d–e) Tubulin in complex with Sas-4 contains GDP. Embryonic HSL was supplemented with [α32p]GTP. (d) Complexes purified using anti-Sas-4 antibody (Sas-4 complex) but not mock IgG (IgG) contain [α32p]GDP. In contrast, tubulin purified using an anti-tubulin antibody (tubulin) after depletion of the Sas-4 complexes, bound [α32p]GTP. (e) Pull down assay from HSL using Sas-4N-GST, but not Sas-4NΔT-GST, contained GDP. The detected GTP in the Sas-4 complex may be from γ-tubulin, which is a GTP binding protein. The relative increase in GTP detected in Sas-4NΔT may reflect the increase inγ-tubulin binding (see Fig. 1a). Standards of GDP and GTP were run in parallel and arrowheads indicate their position.
(f–g) Griseofulvin enhances Sas-4 complex formation. Immuno-purification of Sas-4 complexes fro Drosophila embryonic HSL treated with 250 μM Griseofulvin. The amount of Sas-4 is unchanged among experiments. Quantification of signal intensity (n=3) (g). The mean±SEM of three independent experiments are shown.
To test this, we purified and analyzed Sas-4 complexes from HSLs exposed to GDP or GMPCPP. Although the quantity of Sas-4 present in the purified complexes was unaffected by GDP or GMPCPP exposure, the amounts of other centrosomal proteins in the Sas-4 complex were affected. More specifically, HSLs exposed to GDP had 6 to 12 fold-increases in the amounts of particular centrosomal proteins relative to HSLs exposed to GMPCPP (Fig. 4b–c). Therefore, when bound to tubulin-GDP, Sas-4 acts similar Sas-4ΔT in that it accumulatesexcess centrosomal proteins in its complexes. Perhaps, tubulin-GTP’s binding to Sas-4 sterically hinders Sas-4’s binding to other centrosomal proteins and tubulin-GDP reverses the steric hindrance, allowing Sas-4 to bind the centrosomal proteins. Together, it appears that Sas-4’s binding to tubulin-GDP (but not tubulin-GTP) favors formation of centrosomal protein complexes.
To confirm this, we first tested whether Sas-4 complexes preferentially contain GDP. We immuno-purified Sas-4 complexes from embryonic HSLs treated with [α32p]GTP and analyzed the complexes using thin-layer chromatography. Tubulin that was not bound to Sas-4 contained [α32p]GTP, whereas purified Sas-4 complexes instead contained mostly [α32p]GDP, which is the hydrolyzed product of [α32p]GTP (Fig. 4d). Sas-4N, but not Sas-4NΔT, was able to pull-down GDP (Fig. 4e). Accordingly, when in a Sas-4 complex, tubulin binds GDP.
Second, we tested the effects on the composition of Sas-4 complexes of treatments with Griseofulvin, a compound that changes tubulin’s conformation and induces hydrolysis of tubulin’s bound GTP into GDP28. Griseofulvin increased the quantity of centrosomal proteins in purified Sas-4 complexes (Fig. 4f–g); this is consistent with our data of HSLs exposed to GDP (Fig. 4b–c). Together, these suggest that tubulin’s conformation can regulate the formation of cytoplasmic Sas-4 complexes.
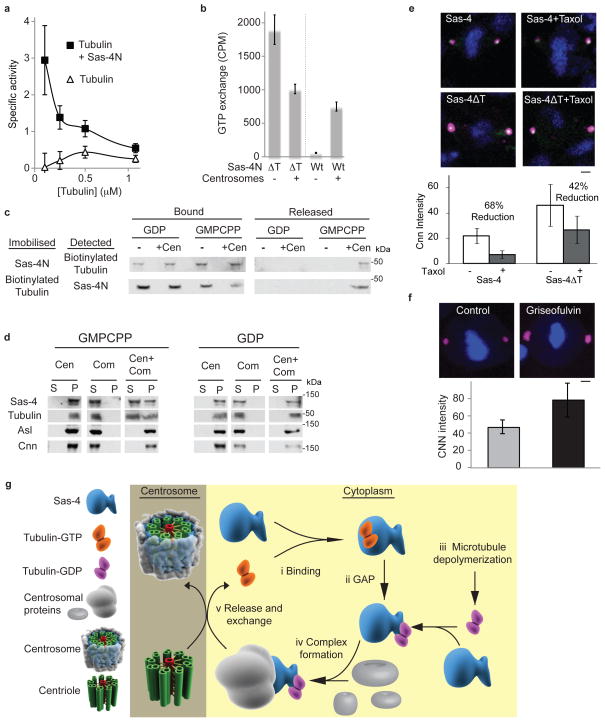
We then studied how tubulin modulates PCM recruitment. Typical GTP-binding proteins (G-proteins), i.e., heterotrimeric G-proteins and the small GTPases belonging to the Ras superfamily, act as molecular switches whose function depends on its GTP- or GDP-bound state. G-proteins have both low intrinsic GTPase and guanine exchange activities and require GTPase activating proteins (GAPs) and guanine exchange factors (GEFs) as catalysts29, 30. Accordingly, we tested whether tubulin can exhibit the characteristics of a typical G-protein during PCM recruitment by acting as a molecular switch.
It is known that free tubulin has low intrinsic GTPase activity and exists as tubulin-GTP7. Therefore, if a tubulin switch is involved in PCM recruitment, it is expected that a GAP exists which induces tubulin to hydrolyze its bound GTP into GDP. We tested whether Sas-4 functions as a tubulin GAP and found that Sas-4-N enhanced the intrinsic GTPase activity of tubulin, as measured by the release of inorganic phosphate (Fig. 5a). This suggests that Sas-4 can function as a tubulin GAP.
Fig. 5. GAP and guanine exchange activities in PCM recruitment.
(a) Sas-4-N functions as a tubulin GAP. Specific activity of tubulin GTPase as determined by [Pi] release (μM, min-1,μM tubulin-1). Since the GTPase activity is greater at low tubulin concentrations, the observed increase in GTPase activity is unlikely to be mediated by tubulin-tubulin interactions occurring during microtubule polymerization6,12,25.
(b) Centrosomes induce guanine exchange of the tubulin-Sas-4 complex. [α32p]GTP was added to biotinylated-tubulin bound to Sas-4 (Sas-4-N) or not bound to Sas-4 (Sas-4-NΔT). Scintillation counting shows that the inclusion of centrosomes increases the GTP exchange of the Sas-4-tubulin complex but not free tubulin.
(a–b) The mean±SEM of three independent experiments are shown.
(c) Centrosomes disrupt the Sas-4-tubulin-GDP interaction. When GMPCPP or GDP is added to the Sas-4-N-biotinylated-tubulin-GDP complex immobilized to resin via Sas-4-N, biotinylated-tubulin remains bound to tubulin-GDP (upper row); likewise, when GMPCPP or GDP is added to the Sas-4-N-biotinylated-tubulin-GDP complex immobilized to resin via biotinylated-tubulin, Sas-4-N remains bound to tubulin-GDP (lower row). However, when centrosomes (+Cen) and GMPCPP are added together, the interaction between tubulin and Sas-4 is weakened, releasing the partner that is not immobilized to the resin; this is not observed when centrosomes and GDP are added together.
(d) Centrosomes can induce Sas-4 complex disassembly allowing Sas-4 interacting proteins to remain in the centrosome while Sas-4 is released into the cytoplasm. Isolated centrosomes (Cen) were mixed with Sas-4 complexes (Com) in the presence of GMPCPP or GDP, and subjected to velocity sedimentation. Proteins bound to the centrosome are found in the pellet (P), while proteins not bound to the centrosome are found in the supernatant (S).
(e) Taxol treated (1 μm) mitotic centrosomes of S2 cells have a reduced amount of Cnn (magenta). S2 cells transfected with Sas-4ΔT but not with Sas-4 are less sensitive to taxol treatment. Scale bar, 2 μm.
(f) Griseofulvin treated (250 μM) mitotic centrosomes of S2 cells have an increased amount of Cnn. Scale bar 2 μm.
(e–f) Signal intensity with mean±SEM of ten cells is shown.
(g) Model for tubulin in regulating PCM recruitment
Tubulin is known to have high guanine exchange activity and readily exchanges its GDP with GTP31. On the other hand, although tubulin-GTP disfavors the formation of centrosomal protein complexes (Fig. 4b), Sas-4 complexes are quite stable regardless of whether they are exposed to GDP or GMPCPP (Supplementary Fig. S5a–c). Therefore, for a Sas-4 complex to remain stable, tubulin’s guanine exchange activity must remain low. To assay the effects of Sas-4 on tubulin’s guanine exchange activity, we added [α32p]GTP to tubulin bound to Sas-4 (Sas-4-N) or not bound to Sas-4 (Sas-4-NΔT). The amount of exchanged [α32p]GTP was then determined by scintillation counter and thin-layer chromatography (Fig. 5b; Supplementary Fig. S5d). Consistent with previous reports, tubulin had a high rate of guanine exchange in the absence of bound Sas-431. In contrast, GTP exchange was not observed in the presence of Sas-4. These suggest that Sas-4 inhibits tubulin’s guanine exchange activity and can stabilize the Sas-4-tubulin complex.
Eventually, Sas-4 complexes are recruited to centrosomes. So, we tested how centrosomes affect guanine exchange and the stability of Sas-4 complexes. In the presence of centrosomes, tubulin did not have an increase in guanine exchange when tubulin is unbound to Sas-4 (Fig. 5b; Supplementary Fig. S5d). This indicates that centrosomes cannot increase the intrinsic guanine exchange activity of tubulin. In contrast, in the presence of centrosomes, tubulin’s guanine exchange was significantly increased when tubulin is bound to Sas-4. Therefore, centrosomes appear to undo Sas-4’s inhibition of tubulin’s guanine exchange activity. Consistently, centrosomes also destabilize the Sas-4-tubulin interaction (Fig. 5c). In the absence of centrosomes or in the presence of centrosomes exposed to GDP, Sas-4-tubulin complexes remained stable; however, in the presence of centrosomes exposed to GMPCPP, Sas-4-tubulin complexes were destabilized and dissociated, potentially allowing Sas-4 to be released into the cytoplasm (Fig. 5c).
To further test this, we mixed isolated centrosomes, purified Sas-4 complexes, and either GMPCPP or GDP. The reaction mixture was subjected to velocity sedimentation, which pelleted centrosomes along with their bound proteins5. When exposed to GDP, Sas-4-complex proteins, Sas-4, and tubulin were in the pellet (Fig. 5d), indicating that the Sas-4 complexes were bound to centrosomes. However, when exposed to GMPCPP, the Sas-4-complex proteins were in the pellet, yet some Sas-4 and tubulin were released into the supernatant. This indicates that centrosomes have guanine exchange activity that releases Sas-4 and tubulin from Sas-4 complexes, whereas other complex proteins remain in the centrosome (Fig. 5d). This is consistent with the observation that Sas-4 traffics between centrosomes and cytoplasm8.
PCM recruitment is tightly coupled to the cell cycle32. Mathematical models33 and analyses of global cytoskeleton remodeling34 predict that microtubule breakdown releases tubulin-GDP causing tubulin-GDP’s concentration to increase when cells enter mitosis. Given our above observations that tubulin-GDP promotes complex formation, this increase in tubulin-GDP concentration may promote PCM recruitment. Currently, there are no tools to determine this directly. Therefore, we analyzed centrosomes of cells treated with taxol, a compound that stabilizes microtubules and reduces tubulin-GDP release into the cytoplasm35. As expected, taxol-treated mitotic centrosomes of Sas-4-GFP transfected cells had significantly less Cnn, whereas, mitotic centrosomes of Sas-4ΔT transfected cells were less sensitive to taxol (Fig. 5e). Although taxol may affect centrosomes via multiple mechanisms, our data suggests that cytoskeleton remodeling regulates recruitment of Sas-4 complexes to centrosomes. Furthermore, treating cells with Griseofulvin, which enhances Sas-4 complex formation, increased Cnn incorporation into the centrosome (Fig. 5f). Together, the taxol and Griseofulvin experiments show that modulating tubulin in cells affects PCM formation.
Our findings reveal a previously unknown function of tubulin. We show that tubulin can negatively control Sas-4 complex formation and, thereby regulate PCM recruitment. Tubulin is a molecular switch that can regulate the formation of Sas-4 complexes and the recruitment of centrosomal proteins to a developing centrosome. The data described above was used to formulate a model whereby tubulin coordinates normal PCM recruitment (Fig. 5g). In the cytoplasm, tubulin-GTP binds Sas-4, which prevents Sas-4 from forming complexes with centrosomal proteins (Fig. 5gi). When Sas-4 activates tubulin’s GTPase, hydrolysis of tubulin-GTP into tubulin-GDP takes place; tubulin-GDP can initiate Sas-4 complex formation (Fig. 5gii). Additionally, Sas-4 complex formation may be enhanced when the tubulin-GDP concentration in the cytoplasm is increased due to microtubule depolymerization (Fig. 5giii). Sas-4 binding to tubulin-GDP stabilizes the Sas-4-tubulin complex by blocking the exchange of GDP with GTP. Sas-4-tubulin-GDP then interacts with other centrosomal proteins to form one of the various types of Sas-4-containing complexes (Fig. 5giv). When a Sas-4 complex tethers to a centrosome, tubulin’s guanine exchange activity is induced by the centrosome, causing the release of tubulin and Sas-4 and allowing the recruitment of centrosomal proteins to the centrosome (Fig. 5gv).
Typical G-proteins act as molecular switches whose function depends on their GTP- or GDP-bound state. Here, during PCM recruitment, we show that tubulin acts as a molecular switch whose function depends on its GTP- or GDP-bound state. Therefore, in PCM recruitment, tubulin acts like a typical G-protein. By manipulating this switch, it may be possible to target cancerous cells, which are known to have abnormal centrosomes and PCM1.
Supplementary Material
Acknowledgments
We would like to thank Dr. J. Iwasa for scientific illustrations; Drs. T. Mitchision A. Johnson, I. Cheeseman and J. Malicki for their scientific discussions; Dr. T. Kaufman, Dr. J. Raff, and Dr. B. Raynaud-Messina, Dr. T. K. Tang for reagents; Drs. Eric, Hari, Rodriguez for technical help with biophysical experiments, Dr. E. Koundakjian scientific editing and discussions, and EM facility staff at HMS for help with EM analyses. This work was supported by a grant (R01GM098394) from National Institute of General Medical Sciences.
Footnotes
Author Contributions J.G. and T.A.R. conceived the project. J.G. performed most of the experiments described herein. T.A.R. supervised the project. Y.F.C. performed phase and electron microscopy analyses. A.H. performed biochemical complex analyses. M.L.B. generated constructs and took part in the biochemical purification of recombinant proteins. D.A.L. and N.M.R. advised and discussed larval brain analyses.
Author Information: The authors declare that they have no competing financial interests.
Bibliography
- 1.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 2.Conduit PT, et al. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 4.Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalakrishnan J, et al. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun. 2011;2:359. doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung LY, Chen HL, Chang CW, Li BR, Tang TK. Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Mol Biol Cell. 2004;15:2697–2706. doi: 10.1091/mbc.E04-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 8.Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung LY, Tang CJ, Tang TK. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol. 2000;20:7813–7825. doi: 10.1128/mcb.20.20.7813-7825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 11.Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 12.Hsu WB, et al. Functional characterization of the microtubule-binding and -destabilizing domains of CPAP and d-SAS-4. Exp Cell Res. 2008;314:2591–2602. doi: 10.1016/j.yexcr.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated proteinthat controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 14.Basto R, et al. Flies without Centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Blachon S, et al. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blachon S, et al. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182:133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohlmaier G, et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Li K, et al. Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J Cell Biol. 1998;141:455–467. doi: 10.1083/jcb.141.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusan N, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. The Journal of cell biology. 2007;177:13–33. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giansanti M, Gatti M, Bonaccorsi S. The role of centrosomes astral microtubules during asymmetric division of Drosophila neuroblasts. Development (Cambridge, England) 2001;128:1137–1182. doi: 10.1242/dev.128.7.1137. [DOI] [PubMed] [Google Scholar]
- 24.Sandoval IV, Jameson JL, Niedel J, MacDonald E, Cuatrecasas P. Role of nucleotides in tubulin polymerization: effect of guanosine 5′-methylene diphosphonate. Proc Natl Acad Sci US A. 1978;75:3178–3182. doi: 10.1073/pnas.75.7.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cormier A, et al. The PN2–3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J Biol Chem. 2009;284:6909–6917. doi: 10.1074/jbc.M808249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozziconacci J, Sandblad L, Wachsmuth M, Brunner D, Karsenti E. Tubulin dimers oligomerize before their incorporation into microtubules. PLoS One. 2008;3:e3821. doi: 10.1371/journal.pone.0003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiller G, Weber K. Radioimmunoassay for tubulin: a quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 28.David-Pfeuty T, Simon C, Pantaloni D. Effect of antimitotic drugs on tubulin GTPase activity and self-assembly. J Biol Chem. 1979;254:11696–11702. [PubMed] [Google Scholar]
- 29.Cabrera-Vera TM, et al. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 30.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 31.Melki R, Carlier MF, Pantaloni D, Timasheff SN. Cold depolymerization of microtubules to double rings: geometric stabilization of assemblies. Biochemistry. 1989;28:9143–9152. doi: 10.1021/bi00449a028. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janulevicius A, van Pelt J, van Ooyen A. Compartment volume influences microtubule dynamic instability: a model study. Biophys J. 2006;90:788–798. doi: 10.1529/biophysj.105.059410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci U S A. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oegema K, et al. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR. 2001;20:11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- 38.Moritz M, et al. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gopalakrishnan J, et al. Self-assembling SAS-6 multimer is a core centriole building block. JBiol Chem. 2010;285:8759–8770. doi: 10.1074/jbc.M109.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.