Abstract
In metazoans, alternative splicing of genes is essential for regulating gene expression and contributing to functional complexity. Computational predictions, comparative genomics, and transcriptome profiling of normal and diseased tissues indicate an unexpectedly high fraction of diseases are caused by mutations that alter splicing. Mutations in cis elements cause mis-splicing of genes that alter gene function and contribute to disease pathology. Mutations of core spliceosomal factors are associated with hematolymphoid neoplasias, retinitis pigmentosa, and microcephalic osteodysplastic primordial dwarfism type 1 (MOPD1). Mutations in the trans regulatory factors that control alternative splicing are associated with autism spectrum disorder, amyotrophic lateral sclerosis (ALS), and various cancers. In addition to discussing the disorders caused by these mutations, this review summarizes therapeutic approaches that have emerged to correct splicing of individual genes or target the splicing machinery.
Keywords: Alternative splicing, Cis mutations, Trans splicing factors, Spliceosome, Splicing related human diseases, Antisense oligonucleotide therapy
Splicing and regulation of gene expression
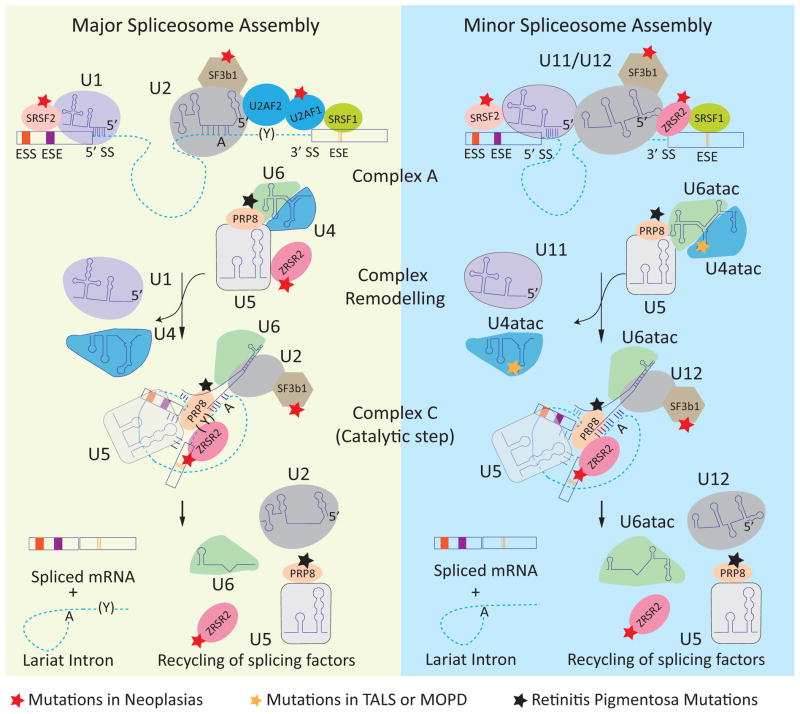
The removal of introns from pre-mRNAs is a precise process required for expression of the majority of human genes. Pre-mRNA splicing and its regulation require a complex array of cis elements (the splicing code) embedded in pre-mRNAs and trans factors that bind to these elements. The core of the splicing code for the majority of introns is comprised of short sequences located at the intron-exon junctions including the 5′ and 3′ splice sites and the branch point. These elements are recognized by the spliceosome, which is composed of five small nuclear ribonucleoproteins (snRNPs) – U1, U2, U4, U5, and U6 – and approximately 150 proteins. The precision of splicing is achieved in part by step-wise assembly of snRNPs on individual introns resulting in two sequential trans-esterification reactions that join the exons while spliceosome components are recycled for additional rounds of splicing (Figure 1) [1].
Figure 1.
Spliceosome assembly and disease-associated mutations in spliceosome components. The dashed lines and rectangles represent introns and exons, respectively. The left panel shows the assembly of the major (U2 type) spliceosome. U1 and U2 snRNPs are recruited to the consensus 5′ splice site (5′ SS) and branch point (A), respectively. The U2-auxiliary factor heterodimer (U2AF2/U2AF1) interacts with the polypyrimidine track (Y) and 3′ splice site (3′ SS), forming complex A. The U4/6 and U5 snRNPs join the assembling spliceosome followed by remodeling of the complex leading to removal of the U1 and U4 snRNP and formation of the catalytic complex (complex C). Two trans-esterification reactions join the exons and release an intron lariat that is subsequently degraded and the spliceosome components are recycled for subsequent rounds of splicing. The right panel shows the assembly of the minor (U12 type) spliceosome, in which U1, U2, U4, and U6 are replaced by homologous U11, U12, U4atac, and U6atac snRNPs, respectively. The red star indicates the components that are mutated in neoplasias. The black star indicates the components that are mutated in retinitis pigmentosa. The orange star indicates the mutation in U4atac that is associated with MOPD1. Abbreviations: ESE, exonic splicing enhancers; ESS, exonic splicing silencers.
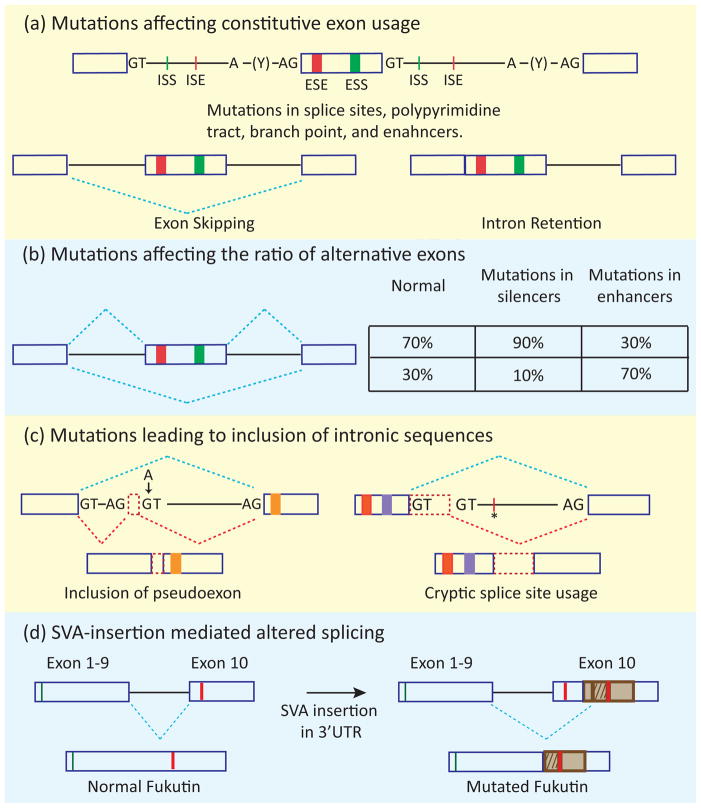
In addition to the core cis-acting elements, other sequences within exons (exonic splicing enhancers, ESEs, and exonic splicing silencers, ESSs) and introns (intronic splicing enhancers, ISEs, and intronic splicing silencers, ISSs) are critical for correct recognition of exons and modulation of splicing outcomes (Figure 2a). The cross-exon communication between the factors that bind to these elements allow the exon to be recognized as a unit (exon definition) prior to intron removal [2].
Figure 2.
Four mechanistic categories of altered gene function by splicing mutations. (a) The basic cis elements of the splicing code are indicated: 5′ and 3′ splice sites (represented by GT and AG), polypyrimidine tract (Y), branch point (A), and exonic and intronic enhancers (ESE and ISE) and silencers (ESS and ISS). Mutations affecting splice sites, the polypyrimidine tract, the branch point, or splicing enhancers lead to exon skipping or intron retention. (b) Mutations in enhancer or silencer elements can change the ratio of isoforms containing alternative exons. (c) Mutations within introns can lead to inclusion of intronic sequences (indicated by red dashed rectangles) by creating a splice site/pseudoexon (indicated by arrow) and/or by creating an enhancer element (indicated by asterisk), allowing recognition of a cryptic splice site. The blue dashed lines indicate the normal splicing pattern, whereas the red dashed lines indicate the splicing pattern caused by the mutation. (d) Insertion of transposable elements (SVAs, represented by a brown rectangle) in the 3′ UTR of the fukutin gene leads to the alternate use of splice sites producing a protein with a different carboxy-terminal sequence (patterned brown rectangle). The green lines indicate the start codon of both the normal and mutated fukutin whereas red lines indicate stop codons.
Although the majority of human introns are of the U2 type, which have the consensus sequences GURAGU and YAG at the 5′ and 3′ ends, respectively, approximately 800 so-called minor or U12 type introns contain different consensus 5′ and 3′ splice sites. U12 introns are spliced by the same mechanism as U2 introns but use a different and homologous set of snRNPs (U11, U12, U4atac, and U6atac that replace U1, U2, U4, and U6, respectively) [3]. By contrast, U5 snRNP and many auxiliary factors are shared between major (U2-type) and minor (U12-type) spliceosomes ([4] and Figure 1).
Alternative splicing allows expression of multiple mRNA isoforms from an individual gene, which often encode protein isoforms of different functionality [5]. A growing number of studies demonstrate that alternative splicing is utilized to adjust gene output in accordance with physiological requirements, and more than 90% of human intron-containing genes undergo alternative splicing, likely affecting all aspects of biology [6]. In this review, we discuss the different outcomes of mutations in splicing cis elements and the splicing machinery that underlie human diseases. We also discuss therapeutic approaches that target splicing to reverse or circumvent pathological processes.
Cis-acting mutations affecting splicing of individual genes
Current estimates of disease causing single nucleotide polymorphisms (SNPs) from the Human Genome Mutation Database (HGMD) predict that 15% of mutations are located within splice sites and more than 20% of missense mutations lie within predicted splicing elements, such that more than one third of the disease-causing SNPs have the potential to disrupt splicing [7, 8]. These estimates do not include mutations that are present in the intronic cis elements separate from the consensus splice sites that can also affect splicing efficiency, particularly the ratio of alternative splicing patterns. A comprehensive list of splicing associated diseases can be found in the databases of aberrantly spliced 3′ and 5′ splices sites (http://www.dbass.org.uk/) [9].
Mutations that cause altered splicing of disease-associated genes
Depending on the location of the disease-associated splicing mutation, various outcomes can be predicted. A large number of reviews illustrate the roles of cis acting mutations [10–12], and a summary of four mechanistic categories is presented below and in Figure 2.
Constitutive exon skipping or intron retention
Mutations that interfere with exon definition by reducing 5′ or 3′ splice site strength or disrupting enhancer elements most commonly result in exon skipping (Figure 2a)[11]. Less frequently, splice site mutations result in intron retention (Figure 2a) [13]. Both effects most often result in gene loss of function either by generating a non-functional protein or by introducing a premature termination codon targeting the mRNA for degradation by non-sense mediated decay (NMD) [14].
Altered inclusion:exclusion ratio of alternative exons
Mutations in silencers (ISS or ESS) or enhancer (ESE or ISE) elements will lead to increased or decreased exon inclusion, respectively (Figure 2b) [15]. Unlike constitutive exons, alternative exons are included or skipped to produce ratios of natural mRNA isoforms that are often regulated in a cell-specific manner. Mutations that affect either the basal level of alternative exon recognition or the binding site of a specific RNA binding protein that regulates exon usage will alter the normal ratio of alternatively spliced RNAs, potentially producing an aberrant biological effect. A well-known example is Frontotemporal Dementia and Parkinsonism linked to chromosome 17 (FTDP-17) in which mutations in exon 10 of the MAPT gene disrupt the ratio of tau protein isoforms [16].
Activation of cryptic splice sites or pseudoexons
Mutations that activate a cryptic splice site (a sequence not normally used for splicing) or induce splicing of an intronic segment, referred to as a pseudoexon, result in inclusion of additional sequence in a spliced mRNA (Figure 2c) [17]. The result is often disruption of the reading frame. Examples of many such disease-causing mutations have been reviewed previously [17, 18].
SVA retrotransposon mediated aberrant splicing
Hominid-specific transposable elements include SVA sequences: composite noncoding SINE (short interspersed sequence)–VNTR (variable number of tandem repeat)–Alu sequences. SVA-mediated inclusion of intronic sequences or pathogenic exonization via activation of cryptic splice site of the host gene has been associated with various diseases [19]. One well characterized example is Fukuyama muscular dystrophy (FCMD) in which an SVA insertion in the 3′ untranslated region (UTR) of the fukutin gene results in inclusion of a new exon from the SVA sequence encoding an altered C-terminus and a nonfunctional protein because of mislocalization (Figure 2d) [20].
Trans-acting mutations affecting the splicing machinery
While there are a large and growing number of examples of cis-acting splicing mutations that cause disease, the examples of trans-acting splicing mutations have been relatively limited. The paucity of trans-acting mutations suggested that mutations within the basal splicing machinery are lethal during embryonic development, if not at the level of individual cells [6]. However, there has been a recent increase in reports of disease-causing mutations within the splicing trans-acting machinery including components of the spliceosome as well as non-spliceosomal components required for regulating alternative splicing.
Mutations in the major and minor core spliceosome components
Hypomorphic mutations in the core spliceosomal components PRPF3, PRPF8, PRPF31, and SNRNP200 are associated with autosomal dominant retinitis pigmentosa, a disease characterized by progressive retinal degeneration. These factors are important for assembling the U4/U6.U5 tri-snRNP of the major spliceosome (Figure 1). PRPF8 and PRPF31 are also components of minor spliceosomes (Figure 1). The specificity of the pathological effects indicates a unique functional requirement in the retina [16, 21], but the disrupted splicing event(s) that are directly responsible for disease remain to be definitively identified.
Two independent studies recently identified several mutations in the gene encoding the U4atac snRNA as a cause of microcephalic osteodysplastic primordial dwarfism type I (MOPD1), also known as Taybi-Linder syndrome (TALS) [22, 23]. U4atac snRNA is the RNA component of the minor spliceosome snRNP (Figure 1). MOPD1 is a multisystemic disease with general growth retardation, abnormal bone morphology, severe brain malformation, and mortality by three years of age. Most of the U4atac mutations affect a conserved stem-loop structure important for association of the 15.5K protein that nucleates binding of proteins required for catalysis of the splicing trans-esterification reactions [24]. Most of the minor, but none of the major, spliceosome introns tested in MOPD1 fibroblasts showed reduced splicing efficiency and increased intron retention [23]. These results are consistent with the predicted loss of function specifically of the minor spliceosome.
Whole genome or exome sequencing of tumor and matched normal tissues in myeloid neoplastic syndrome (MDS), acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), and chronic lymphocytic leukemia (CLL) patients, along with targeted sequencing of candidate genes, identified a high percentage of recurrent somatic mutations in genes encoding spliceosome components [25–30]. Multiple independent somatic mutations were identified in the genes encoding U2AF1, ZRSR2, SF3B1, and SRSF2. U2AF1 is exclusively required for splicing of U2 introns, whereas ZRSR2 is required for splicing of both U2 and U12 introns [31, 32]. SF3B1 and SRSF2 are expected to participate in the splicing of both U2 and U12 introns (Figure 1). SF3B1 mutations strongly correlate with refractory anemia with ring sideroblasts (RARS), a distinct type of low-risk MDS, however studies differ regarding the prognostic relevance of these mutations in this subtype [25, 33]. Similarly, the relevance of mutations in U2AF1, SRSF2, and ZRSR2 in predicting disease progression or treatment outcomes for various neoplasias needs further studies. Nevertheless, spliceosomal mutations in these neoplasms may provide valuable prognostic markers as well as bring new insight into the complex biology of cancer initiation and progression.
Misregulation of trans factors by microsatellite expansion mutations
Myotonic dystrophy (DM) is the best characterized example of a disease in which RNA transcribed from a microsatellite expansion causes disease. DM type 1 (DM1) is caused by expanded CTG repeats in the 3′ UTR of the dystrophia myotonica-protein kinase (DMPK) gene, and DM type 2 (DM2) is caused by expanded CCTG repeats within intron 1 of the zinc finger protein 9 (ZNF9) gene. In both diseases, pathogenesis is due to expression of toxic RNA containing the expanded repeats, resulting in an RNA gain of function [34]. The expanded CUG or CCUG RNAs accumulate in nuclear foci and form double-stranded RNA structures that sequester muscleblind-like (MBNL) splicing regulators, resulting in a loss of MBNL function. RNA containing expanded CUG repeats also increases the stability of CUG-BP and ETR-3-like factor 1 (CELF1) because of increased phosphorylation resulting from activation of protein kinase C [35]. Alterations of these trans factors manifest in various ways in different DM tissues. Altered splicing of chloride channel protein (Clcn1), Bin1, ryanodine receptor 1 (RyR1), myotubularin-related 1 gene (Mtmr1), sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA) 1 or 2, and insulin rector (IR) in skeletal muscle are proposed to contribute to myotonia, muscle weakness, impaired calcium homeostasis, and insulin resistance [36–43]. Altered splicing of cardiac troponin T in heart and tau in brain is thought to contribute to cardiac and brain abnormalities, respectively [44, 45]. CELF and MBNL function in other aspects of mRNA processing and translation, such that their altered function is likely to contribute to disease pathology by multiple mechanisms. For example, increased levels of CELF1 in DM1 is associated with translation of a dominant negative isoform of a transcription factor CCAAT/enhancer-binding protein β (C/EBP β)[46]. An RNA gain of function mechanism has also been proposed to explain pathogenic effects in other expansion diseases such as spinocerebellar ataxia type 8 (SCA8), SCA10, SCA12, fragile X tremor ataxia syndrome (FXTAS), and more recently in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [47–52]. A GGGGCC hexanucleotide expansion in the noncoding region of the C9ORF72 gene accounts for 23–50% of familial ALS, 21% of sporadic ALS, and 11% of familial FTD [51, 52]. RNA containing the GGGGCC expansion was detected in nuclear foci in neurons from patients carrying the mutation, suggesting an RNA gain-of-function mechanism in ALS-FTD [52].
RBFOX1 is associated with autism spectrum disorder (ASD)
The approximately fifty RNA binding proteins shown to regulate alternative splicing are a small fraction of the more than 650 RNA binding proteins encoded by the human genome [5, 53], and a growing number of alternative splicing regulators has been linked with various human diseases. Cytogenetic and association studies identified the conserved splicing regulator Rbfox1 as an autism-associated gene [54, 55]. Expression of the three Rbfox genes is enriched in brain, heart, and skeletal muscle, and RNA-Seq analysis of brain cortex tissue from three individuals with ASD showed significantly reduced expression (~6 fold) of Rbfox1 compared with three control samples [56]. RNA-Seq also identified 36 alternative splice events that are altered in ASD samples and which are predicted to be Rbfox targets based on enrichment of the Rbfox binding motif near the alternatively spliced exons. Four of these events were previously shown to be directly regulated by Rbfox1 (ATP5C1, ATP2B1, GRIN1, and MEF2C), and also changed in ASD patients consistent with loss of Rbfox1 function [56, 57]. The genes that exhibited aberrant alternative splicing in ASD brains are associated with synaptic functions, synaptogenesis, and actin cytoskeletal organization, consistent with a role in neuronal connectivity in the cortex [56].
TDP-43 in ALS
The most common cellular feature of ALS and frontotemporal lobar degeneration (FTLD) is ubiquitin positive cytoplasmic inclusion bodies in neurons and astroglia, with TAR DNA binding protein-43 (TDP-43) as one of the main components [58]. More than 40 mutations in TDP-43 have been identified, accounting for about 5% of inherited cases of ALS [59]. Genome-wide identification of in vivo binding sites of TDP-43 using in vivo crosslinking and immunoprecipitation (CLIP) identified hundreds of intronic binding sites, and exons proximal to these binding sites underwent splicing changes in TDP-43 knockdown in mouse brain striatum and the human neuroblastoma cell line SH-SY5Y [60, 61]. These splicing events include genes involved in neuronal function and neurodegenerative disease such as MEF2D, sortilin, tau, BIM, AP2, parkin, huntingtin, and ataxin; each of which could contribute to disease pathology [60, 61]. Splicing was also altered for BIM in a postmortem brain from FTLD patients, encoding the proapoptotic isoform that is consistent with neuronal apoptosis and neuronal degeneration [60]. However, whether these events directly contribute to disease due to altered regulation of TDP-43 remains to be demonstrated. In addition to TDP-43, mutations in the structurally similar RNA-binding proteins FUS and TAF15 have been identified as a genetic cause of ALS, suggesting a misregulated RNA processing pathway and altered gene splicing may be a common feature in these neuropathies [59, 62, 63]. Identification of in vivo binding sites and direct RNA targets of TDP-43, FUS, and TAF15 in disease relevant model systems have implications not only in ALS but also for other neurodegenerative diseases where TDP-43 inclusions are also found, such as Alzheimer’s and Huntington’s Disease [58].
Splicing trans regulatory factors in cancer
Initiation, progression, and metastasis of cancer require a complex interplay of cell–cell signaling, epigenetics, transcriptional and post-transcriptional mechanisms, and environmental factors [64–66]. Altered expression or localization of several RNA binding proteins such as SRSF1, hnRNPA1, hnRNP H, TIA-1/TIAR, Sam68, RBM5, HuR, SON, and RBM10 have been linked with various cancers [67, 68]. Here, we focus on specific aspects of tumor biology that are explained by misregulation of trans-acting splicing regulators. Additional aspects of splicing in cancer have been reviewed elsewhere [68–70].
A metabolic switch from oxidative phosphorylation to aerobic glycolysis is beneficial for the pathological proliferation of cancerous cells, and is known as the Warburg effect [71]. A key molecular change associated with the metabolic switch is generation of the pyruvate kinase M2 isoform in tumors that is determined by alternative splicing of the PKM gene [72]. Transcriptional upregulation of splicing regulators hnRNPA1, hnRNPA2, and PTB1 by the oncogenic transcriptional factor c-myc coordinates the alternative splicing of the pyruvate kinase gene to generate PKM2 in human gliomas [73]. High expression of the PKM2 isoform is almost universal in cancer in contrast to the lower fraction that overexpress c-myc; the mechanism of PKM2 expression in cancers that do not overexpress c-myc will be of particular interest.
SRSF1, a member of the SR family of proteins, is upregulated in multiple cancers and has been demonstrated to be an oncogene based on overexpression studies [74, 75]. SRSF1 target genes are involved in tumor initiation, survival, and tumor progression. Overexpression of SRSF1 promotes production of the Bin1 isoform lacking the domain required for tumor suppressor activity, whereas the isoforms of the MNK2 and S6K1 kinases produced in SRSF1 overexpressing cells have proliferative and antiapoptotic effects [74]. Overexpression of SRSF1 also induces alternative splicing of the RON tyrosine kinase receptor to produce a constitutively active isoform that imparts enhanced cell mobility and cell invasion properties characteristic of aggressive tumor cells [76]. In addition, a global siRNA screen to identify splicing regulators of the apoptosis regulatory genes Bcl-x and Mcl1 identified SRSF1 as a critical driver for coordinated regulation of the Bcl-2 family, suggesting a role for SRSF1 in cancer cell survival [77].
Increased expression of hnRNPH in glioblastoma multiforme (GBM) is thought to impart tumor survival by promoting the antiapoptotic, rather than the apoptotic, isoform (IG20) of the MADD gene [78]. Increased expression of hnRNPH is also associated with generation of the constitutively active isoform of RON in GBM potentially enhancing cell migration [78].
Overall, these examples emphasize the importance of moderating alternative splicing events by tight and coordinated control of trans regulatory factors.
Therapeutic approaches in splicing diseases
The recent realization that a large fraction of disease-causing mutations affects splicing has driven several therapeutic approaches to correct aberrant splicing [21, 79–81]. Two particularly robust approaches are the use of modified antisense oligos (ASOs) targeted to specific RNA sequences to redirect splicing and small molecules identified by large-scale screens or strategic chemical design.
Splicing redirection using ASOs
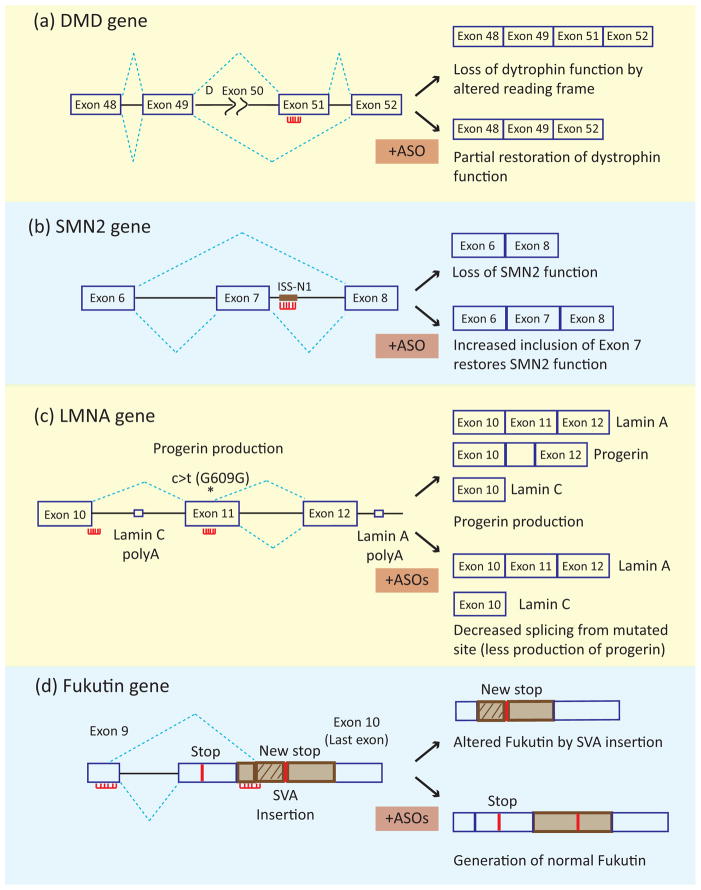
To redirect splicing, an ASO is targeted to bind specific complementary 15–20 nucleotide regions of the pre-mRNA and sterically blocking interactions of the splicing machinery (Figure 3). ASOs directed to splice sites or enhancer elements will induce exon skipping, whereas ASOs directed to splicing silencer elements will enhance exon inclusion. The effectiveness of ASOs has been demonstrated in clinical trials for Duchenne muscular dystrophy (DMD), which is caused by partial or complete loss of dystrophin function. A large fraction of DMD mutations are genomic deletions such that splicing of the remaining exons produce out of frame mRNAs. ASOs are used to induce skipping of specific exons to restore the reading frame or remove exons containing mutations (Figure 3a) [82, 83]. In two independent phase II clinical trials, systemic administration of different ASOs targeting exon 51 restored the dystrophin reading frame from the mutant gene and increased the number of dystrophin-positive fibers [84, 85]. While no significant improvement in muscle function was observed, these initial results are encouraging as both ASOs were well tolerated without significant side effects, supporting the efficacy and safety of ASO as splicing redirecting therapy.
Figure 3.
Use of antisense oligonucleotides for splicing correction therapy. The splicing patterns of the mutated gene in the absence and presence of ASOs are indicated in the top and bottom panels, respectively. The red lines with projections indicate the ASOs. (a) In DMD a deletion of exon 50 (indicated by Δ exon 50) leads to disruption of the reading frame resulting in loss of dystrophin function. ASO-induced skipping of exon 51 restores the reading frame and partially rescues dystrophin function. (b) In SMA, loss of SMN protein from the mutated SMN1 gene can be rescued by inducing inclusion of exon 7 of the SMN2 gene. The use of an ASO targeting an intronic splicing silencers leads to enhanced inclusion of SMN2 exon 7. (c) In Hutchinson-Gilford progeria syndrome, a silent mutation in exon 11, indicated by asterisk labeled c>t (G609G), leads to activation of a cryptic 5′ splice site resulting in the production of a toxic protein, progerin. Combined administration of two ASOs to block the 3′ splice site of exon 10 and the cryptic splice site inhibit progerin production. (d) In FCMD, insertion of an SVA leads to activation of a cryptic 5′ splice site in exon 9 and use of a 3′ splice site in the SVA segment. Combined administration of three ASOs to block the aberrant 3′ splice site and a nearby ESE (shown together), and an ISE near the aberrant 5′ splice site promote use of correct 5′ and 3′ splice sites producing fukutin protein.
Spinal Muscular Atrophy (SMA) is a motor neuron disease caused by loss of function of the survival motor neuron gene 1 (SMN1). A paralogue in humans, SMN2, contains a silent mutation in exon 7 that leads to exon skipping to produce a truncated, unstable SMN protein that does not rescue the loss of SMN1. Residual expression of full length SMN2 to various levels results in different classes of SMA severity. In general, SMN1 deficiency results in progressive loss of motor neurons leading to paralysis, skeletal muscle atrophy, and mortality. A major treatment strategy for SMA has focused on increasing expression of full-length SMN protein using a variety of approaches, including use of an ASO targeting an ISS to promote inclusion of SMN2 exon 7. An extensive systematic analysis of tiled ASOs targeting the intron adjacent to SMN2 exon 7 identified specific ASOs that dramatically promoted exon 7 inclusion (Figure 3b) [86, 87]. Using a mouse model for SMA, a single intracerebroventricular (ICV) injection of one ASO (ASO-10-27) targeting this region showed dose-dependent increased exon 7 inclusion, SMN protein synthesis, and number of motor neurons as well as improved muscle physiology, motor function, and increased survival [86]. Importantly, systemic delivery of ASO-10-27 in neonatal SMA mice increased SMN levels in peripheral tissues (liver, heart, muscle, and kidney) as well as in the central nervous system (spinal cord and brain) and resulted in increased skeletal muscle function and a remarkable increase in survival from 10 days to more than 500 days [88]. The restoration of SMN protein in CNS and the dramatic increase in survival as a result of systemic ASO delivery indicates the importance of peripheral restoration of SMN [88]. In addition, the incomplete blood–brain barrier in neonates along with retrograde transport of ASO is thought to provide sufficient ASO for partial rescue of SMN expression in brain [88, 89]. The effectiveness of ASO-10-27 (ISIS-SMNRx) in mouse models has led to FDA approval for its use in clinical trials in human patients which are currently in progress (ClinicalTrial.gov Identifier: NCT01494701).
The use of a cryptic 5′ splice site in exon 11 of the LMNA1 gene due to a silent mutation leads to production of a mutant protein, progerin, and is the primary cause of Hutchinson-Gilford progeria syndrome (HGPS) [90]. Mice containing a knock-in of the analogous mutation recapitulate misplicing, production of progerin as well as many associated molecular and clinical features of HGPS including shortened life-span, reduced body weight, abnormally shaped nuclei, and bone and cardiovascular abnormalities [91]. Systemic administration of a combination of ASOs targeting the exon 10 5′ splice site and the cryptic 5′ splice site in exon 11 reduced use of the cryptic splice site and produced significant improvement in life-span (155 to 190 days) and body-weight consistent with the observed reduced expression of progerin in most tissues (Figure 3c) [91].
SVA insertion-mediated altered splicing of the fukutin gene is responsible for production of altered fukutin protein product associated with FCMD. Transfection of three ASOs targeting the aberrant 3′ splice site, a predicted ESE within the SVA transposon sequence, and a predicted ISE near the 5′ splice site of exon 10 in cell lines derived from FCMD patients restored normal fukutin production and function (Figure 3d) [20]. Furthermore, intravenous delivery as well as local injection of this cocktail in skeletal muscle of the knock-in mouse model restored normal fukutin function supporting the use of an ASO cocktail as a therapeutic strategy for FCMD [20].
Alternative splice site usage within exon 23 of signal transducer and activator of transcription 3 (STAT3) generates an isoform (STAT3β) that lacks the C-terminal transactivation domain and is proposed to have a dominant negative effect associated with induction of apoptosis and cell cycle arrest in tumors [92, 93]. In a xenograft mouse model using implantation of MDA-MD-435s cells, intratumor injection of ASOs targeting the 3′ splice site and an ESE that causes exon 23 skipping showed tumor regression consistent with switching from normal STAT3α to antitumorigenic STAT3β isoform [94]. These results support the idea that splicing redirection of an endogenous key molecule to induce apoptosis or cell cycle arrest can be used for personalized cancer therapy combined with traditional cancer therapy. A database of such alternative splicing events with antitumorigenic effects on specific tumor type will be of tremendous value for such approaches.
Disruption of RNA-protein interaction in myotonic dystrophy
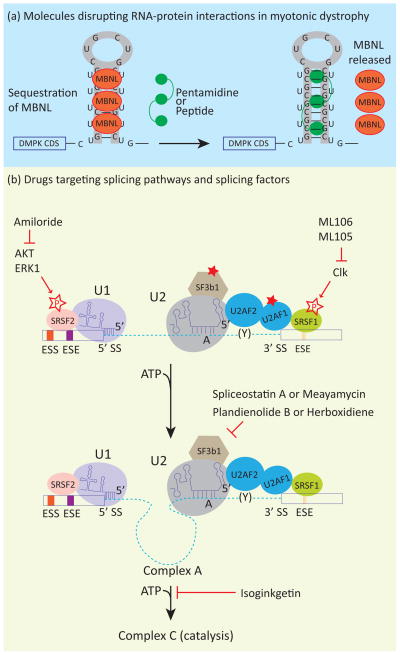
One of the primary pathogenic effects of the expanded repeat RNA in myotonic dystrophy is a trans-dominant effect on RNA binding proteins that regulate alternative splicing. Strategies using ASOs, small molecules, and small peptides have been devised to disrupt interactions between repeat-containing RNA and MBNL and restore MBNL protein function (Figure 4a) [95–97]. Similar strategies have been applied for other diseases caused by repeat-containing RNA including Huntington’s disease and spinocerebellar ataxias [98, 99].
Figure 4.
(a) Small molecules disrupt aberrant RNA-protein interactions in myotonic dystrophy. Small molecules and peptides have been used to disrupt the interaction between CUG repeat RNA and MBNL. This results in rescue of MBNL splicing functions and reversal of some features of myotonic dystrophy. (b) Several compounds inhibit splicing at various steps of splicing assembly and have potential as antitumorigenic drugs. Meayamycin, a compound derived from spliceostatin A, as well as Pladienolid B and Herboxidiene, bind to the SF3b complex of the U2 snRNP and inhibit the formation of complex A. Isoginkgetin prevents the formation of complex B. Amiloride inhibits Akt1 and Erk1 kinases which phosphorylate SR proteins that are important for regulation of constitutive and alternative splicing. Similarly, ML105 and ML106 were identified in a screen for correction of splicing of the LMNA minigene. These compounds were found to inhibit cdc2-like kinases (Clk) that also phosphorylate SR proteins.
One particularly effective approach is an ASO targeted to the expanded CUG repeats expressed in DM1 cells to block binding of MBNL1. A 25 nucleotide CAG repeat morpholino oligonucleotide (CAG25) was shown to prevent binding of MBNL1 to expanded CUG repeat RNA in vitro, prevent MBNL1 sequestration in vivo, correct misregulated splicing and increase translocation of repeat-containing RNA to the cytoplasm in a DM1 mouse model [100]. A second ASO containing seven CAG repeats (CAG)7 with 2′-OMe chemistry was shown to reduce CUG repeat containing RNA in cultured immortalized myoblasts from a DM1 mouse model as well as in skeletal muscle from a second mouse model [101]. In a third example, ASOs modified to promote RNase H-mediated degradation resulted in the loss of CUG repeat containing RNA in skeletal muscle tissue correlating with mild rescue of misregulated splicing [95]. These results provide a strong foundation for the use of ASOs to reduce the levels of toxic CUG repeat containing RNA below a pathogenic threshold.
A screen for D-amino acid containing hexapeptides that disrupt foci formation in a Drosophila model of DM1 identified a peptide (ABP1) that binds to double stranded CUG repeat RNA and changes its structure to single stranded RNA in turn releasing MBNL. This peptide reduced foci formation and rescued MBNL distribution and splicing changes associated with loss of function of MBNL in both Drosophila and mouse DM1 models [97].
A screen of 25 molecules that bind to structured nucleic acids identified pentamidine and Hoechst 33258 as having the ability to disrupt binding of MBNL1 to expanded CUG repeats [102, 103]. Pentamidine partially rescued splicing of MBNL targets in a DM1 mouse model, however the severe toxicity of pentamidine prevented the use of higher doses to achieve full-rescue [103]. In a separate study, Hoechst 33258 was used to synthesize a pentamer that binds specifically to CUG repeat RNA in nanomolar range and displayed 23-higher affinity to CUG repeat RNA than MBNL1 in vitro [102]. A rational design approach was used to produce a molecule that specifically recognizes the U-U mismatch in a CUG repeat RNA structure and intercalates in double stranded RNA produced ligand 1, a compound that inhibits the MBNL1 CUG RNA interaction in vitro [104]. Similar design principles and further optimization can lead to discovery of small compounds that have the potential to disrupt specific disease related RNA-protein interactions.
Small molecules targeting splicing pathways
The small molecule approaches used to develop therapies for myotonic dystrophy are specifically targeted to disrupt interactions of MBNL with repeat containing RNA, however there are much broader applications of small molecules to reverse aberrant splicing. Given that splicing is an essential step of gene expression, splicing inhibition provides a new mode of action for antitumor drugs. Various antitumor drugs have been reported to inhibit splicing: isoginkgetin, pladienolide B, herboxidiene (GEX1A) and spliceostatin A (SSA) (Figure 4b) [105–108]. All four drugs have antitumorigenic effects on various human cancer cell lines and mouse xenograft models [108–111]. Herboxidiene is structurally similar to pladienolide B and both bind to SF3b components, which are constituents of the U2 snRNP, and likely function by inhibiting U2 snRNP function [108]. Similar to pladienolide B and herboxidiene, the SSA target protein is also a component of the SF3b complex, however, instead of general splicing inhibition, SSA produced alternative splicing changes in cell cycle genes [112]. Pladienolide B and SSA inhibit formation of complex A, and early steps of spliceosome assembly (Figure 1), whereas isoginkgetin inhibits progression of complex A to complex B [105]. The target protein of isoginkgetin is not yet known. The discovery of these drugs has fuelled studies to synthesize more potent derivatives. Meayamycin, a derivative of SSA is reported to be active at picomolar concentrations, 100-fold more potent than the SSA [113]. Additionally, meayamycin treatment resulted in pronounced cell death in the A549 human lung cancer cell line compared to the IMR-90 nontumorigenic lung fibroblast cell line suggesting selectivity of meayamycin for transformed cells [113]. Further studies with these compounds should identify dose-dependent effects on various types of cancer.
A quest for small molecules that inhibited cryptic splicing of the LMNA1 gene to prevent HGPS led to the discovery of compounds that modulate splicing by inhibiting cdc-2-like-kinases (Clk) [114]. The Clk kinases phosphorylate SR proteins, and many of these phosphorylation events are known to be required for SR activity and appropriate nuclear localization [115, 116]. Similarly, a small molecule screen to identify drugs that can modulate alternative splicing of BCL-X, HIPK3, and RON transcripts in a hepatocarcinoma cell line identified amiloride as a potent inducer of splicing changes in these transcripts [117]. Amiloride treated cells showed decreased levels of SRp20, and hypophosphorylation of SR proteins, decreased phosphorylation of AKT, ERK1/2 kinases, and PP1 phosphatase, as well as increased phosphorylation of p38 and JNK kinases (Figure 4b) [117]. Small compounds that affect the activities of specific splicing factors can be useful for disrupting cancer-associated splicing pathways and as potential therapeutic agents.
Concluding remarks and future perspectives
Pre-mRNA splicing is intimately connected with multiple aspects of gene expression. Splicing is regulated by secondary structure of the pre-mRNA, noncoding RNAs, chromatin modifications, RNA polymerase II elongation rate and various signaling pathways [118–120]. Splicing also affects upstream events, such as chromatin modification, transcription kinetics, and mRNA export and translation [119, 121]. The crosstalk between splicing and the continuum of gene expression raises the possibility that disruption of splicing has a determinative effect on other regulatory steps, and thereby causing or modifying a human disease by indirect mechanisms.
A clear understanding of disease causing events is important to develop individual therapies. As diagnostic approaches become individualized through whole genome sequencing as well as transcriptome and epigenetic profiling, it is likely that a large fraction of disease-causing mutations and SNPs that predispose to disease or modulate disease severity will affect splicing. ASOs, for example, provide a highly specific approach for targeting splicing modulation and are a promising approach for individualized treatment. Additional corrective strategies such as trans splicing, ribozymes, and modified snRNA molecules have also proved successful to varying degrees [83]. Currently, ASOs and small molecules show promising results for splicing directed therapy, but with improved delivery systems other strategies can also be applied to increase the therapeutic possibilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 3.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 4.Will CL, Luhrmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nature reviews Genetics. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim KH, et al. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krawczak M, et al. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 9.Buratti E, et al. DBASS3 and DBASS5: databases of aberrant 3′- and 5′-splice sites. Nucleic acids research. 2011;39:D86–91. doi: 10.1093/nar/gkq887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper TA, et al. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Bigas N, et al. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 13.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. Journal of medical genetics. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green RE, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19:I118–I121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 15.Sterne-Weiler T, et al. Loss of exon identity is a common mechanism of human inherited disease. Genome research. 2011;21:1563–1571. doi: 10.1101/gr.118638.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 17.Dhir A, Buratti E. Alternative splicing: role of pseudoexons in human disease and potential therapeutic strategies. Febs J. 2010;277:841–855. doi: 10.1111/j.1742-4658.2009.07520.x. [DOI] [PubMed] [Google Scholar]
- 18.Perez B, et al. Present and future of antisense therapy for splicing modulation in inherited metabolic disease. J Inherit Metab Dis. 2010;33:397–403. doi: 10.1007/s10545-010-9135-1. [DOI] [PubMed] [Google Scholar]
- 19.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Human genetics. 2010;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi-Ikeda M, et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–131. doi: 10.1038/nature10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulos MG, et al. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3:a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edery P, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 23.He H, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nottrott S, et al. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. The EMBO journal. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. The New England journal of medicine. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England journal of medicine. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 29.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nature genetics. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makishima H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H, et al. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes & development. 2010;24:2389–2394. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kielkopf CL, et al. U2AF homology motifs: protein recognition in the RRM world. Genes & development. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirabayashi S, et al. Spliceosomal gene aberrations are rare, coexist with oncogenic mutations, and are unlikely to exert a driver effect in childhood MDS and JMML. Blood. 2012;119:e96–99. doi: 10.1182/blood-2011-12-395087. [DOI] [PubMed] [Google Scholar]
- 34.Ranum LP, Cooper TA. RNA-Mediated Neuromuscular Disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 35.Kuyumcu-Martinez MN, et al. Increased steady state levels of CUGBP1 in Myotonic Dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura T, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 37.Savkur RS, et al. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fugier C, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 39.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 40.Charlet-B N, et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 41.Dansithong W, et al. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 42.Ho TH, et al. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 43.Buj-Bello A, et al. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum Mol Genet. 2002;11:2297–2307. doi: 10.1093/hmg/11.19.2297. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H, et al. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 45.Philips AV, et al. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 46.Timchenko NA, et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 47.Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS genetics. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White M, et al. Transgenic mice with SCA10 pentanucleotide repeats show motor phenotype and susceptibility to seizure: A toxic RNA gain-of-function model. J Neurosci Res. 2011 doi: 10.1002/jnr.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes SE, et al. Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nature genetics. 1999;23:391–392. doi: 10.1038/70493. [DOI] [PubMed] [Google Scholar]
- 50.Iwahashi CK, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 51.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabut M, et al. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192, e191. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Martin CL, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 55.Barnby G, et al. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. American journal of human genetics. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011 doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagier-Tourenne C, et al. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Human molecular genetics. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Current opinion in neurobiology. 2011 doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Couthouis J, et al. Feature Article: A yeast functional screen predicts new candidate ALS disease genes. Proceedings of the National Academy of Sciences of the United States of America; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5 doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandler DS, et al. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66:9502–9508. doi: 10.1158/0008-5472.CAN-05-4271. [DOI] [PubMed] [Google Scholar]
- 65.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 67.Ahn EY, et al. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & development. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 70.Dutertre M, et al. Alternative splicing and breast cancer. RNA biology. 2010;7:403–411. doi: 10.4161/rna.7.4.12152. [DOI] [PubMed] [Google Scholar]
- 71.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 73.David CJ, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ezponda T, et al. The oncoprotein SF2/ASF promotes non-small cell lung cancer survival by enhancing survivin expression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:4113–4125. doi: 10.1158/1078-0432.CCR-10-0076. [DOI] [PubMed] [Google Scholar]
- 76.Ghigna C, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 77.Moore MJ, et al. An alternative splicing network links cell cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lefave CV, et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. The EMBO journal. 2011;30:4084–4097. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia-Blanco MA. Alternative splicing: therapeutic target and tool. Prog Mol Subcell Biol. 2006;44:47–64. doi: 10.1007/978-3-540-34449-0_3. [DOI] [PubMed] [Google Scholar]
- 80.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 81.Wood MJ, et al. RNA-targeted splice-correction therapy for neuromuscular disease. Brain. 2010;133:957–972. doi: 10.1093/brain/awq002. [DOI] [PubMed] [Google Scholar]
- 82.Aartsma-Rus A, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Human mutation. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 83.Hammond SM, Wood MJ. Genetic therapies for RNA mis-splicing diseases. Trends in genetics : TIG. 2011;27:196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 84.Goemans NM, et al. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. The New England journal of medicine. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 85.Cirak S, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Passini MA, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hua Y, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ek CJ, et al. Structural characteristics and barrier properties of the choroid plexuses in developing brain of the opossum (Monodelphis Domestica) J Comp Neurol. 2003;460:451–464. doi: 10.1002/cne.10661. [DOI] [PubMed] [Google Scholar]
- 90.Lopez-Mejia IC, et al. A conserved splicing mechanism of the LMNA gene controls premature aging. Human molecular genetics. 2011;20:4540–4555. doi: 10.1093/hmg/ddr385. [DOI] [PubMed] [Google Scholar]
- 91.Osorio FG, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3:106ra107. doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 92.Caldenhoven E, et al. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. The Journal of biological chemistry. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 93.Xu G, et al. Dominant negative STAT3 suppresses the growth and invasion capability of human lung cancer cells. Mol Med Report. 2009;2:819–824. doi: 10.3892/mmr_00000178. [DOI] [PubMed] [Google Scholar]
- 94.Zammarchi F, et al. Antitumorigenic potential of STAT3 alternative splicing modulation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17779–17784. doi: 10.1073/pnas.1108482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee JE, et al. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1117019109. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foff EP, Mahadevan MS. Therapeutics development in myotonic dystrophy type 1. Muscle & nerve. 2011;44:160–169. doi: 10.1002/mus.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garcia-Lopez A, et al. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Friedlander RM. Using noncoding small RNAs to develop therapies for Huntington’s disease. Gene therapy. 2011;18:1139–1149. doi: 10.1038/gt.2011.170. [DOI] [PubMed] [Google Scholar]
- 99.Krzyzosiak WJ, et al. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic acids research. 2012;40:11–26. doi: 10.1093/nar/gkr729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wheeler TM, et al. Reversal of RNA-dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mulders SA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pushechnikov A, et al. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Warf MB, et al. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0903234106. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arambula JF, et al. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci U S A. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Brien K, et al. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. The Journal of biological chemistry. 2008;283:33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 107.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 108.Hasegawa M, et al. Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS chemical biology. 2011;6:229–233. doi: 10.1021/cb100248e. [DOI] [PubMed] [Google Scholar]
- 109.Yoon SO, et al. Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression. Mol Cancer Ther. 2006;5:2666–2675. doi: 10.1158/1535-7163.MCT-06-0321. [DOI] [PubMed] [Google Scholar]
- 110.Nakajima H, et al. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot (Tokyo) 1996;49:1204–1211. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 111.Mizui Y, et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibiot (Tokyo) 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- 112.Corrionero A, et al. Reduced fidelity of branch point recognition and alternative splicing induced by the antitumor drug spliceostatin A. Genes & development. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Albert BJ, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol Cancer Ther. 2009;8:2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Auld D, et al. A high-throughput screen for pre-mRNA splicing modulators. Probe Reports from the NIH Molecular Libraries Program 2010 [Google Scholar]
- 115.Ngo JC, et al. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Molecular cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 116.Prasad J, Manley JL. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Molecular and cellular biology. 2003;23:4139–4149. doi: 10.1128/MCB.23.12.4139-4149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chang JG, et al. Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS ONE. 2011;6:e18643. doi: 10.1371/journal.pone.0018643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and noncoding RNA in alternative splicing regulation. Current opinion in genetics & development. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. The Journal of biological chemistry. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 121.Kim S, et al. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]