Abstract
A panel of 133 allergens derived from 28 different sources, including fungi, trees, grasses, weeds and indoor allergens, was surveyed utilizing prediction of HLA class II binding peptides and ELISPOT assays with PBMC from allergic donors, resulting in the identification of 257 T cell epitopes. More than 90% of the epitopes were novel, and for 14 allergen sources were the first ever identified. The epitopes identified in the different allergen sources summed up to a variable fraction of the total extract response. In cases of allergens where the identified T cell epitopes accounted for a minor fraction of the extract response, fewer known protein sequences were available, suggesting that for “low epitope coverage” allergen sources, additional allergen proteins remain to be identified. IL-5 and IFN-γresponses were measured as prototype Th2 and Th1 responses, respectively. While in some cases (e.g., Orchard Grass, Alternaria, Cypress, and Russian Thistle) IL-5 production greatly exceeded IFN-γ, in others (e.g., Aspergillus, Penicillum, and Alder) the production of IFN-γ exceeded IL-5. Thus, different allergen sources are associated with variable polarization of the responding T cells. The present study represents the most comprehensive survey to date of human allergen derived T cell epitopes. These epitopes might be used to characterize T cell phenotype/T cell plasticity as a function of seasonality, or as a result of SIT treatment or varying disease severity (asthma or rhinitis).
INTRODUCTION
Allergic reactions to common environmental allergens are associated with serious clinical manifestations, such as rhinitis and asthma, translating into high morbidity and societal costs. Furthermore, the incidence and prevalence of allergic disease is constantly rising. Current treatments for allergic disease are not fully satisfactory and, coincidentally, our understanding of the fundamental aspects of allergic disease remains incomplete (1, 2).
It is well appreciated that the initiation and maintenance of allergic disease is due to a complex series of molecular events, including both innate and adaptive immunity (3-5). In terms of adaptive immunity, IgE is of particular importance (6). Its cross-linking by antigen is a main cause of the release of histamine and other mediators, and subsequent clinical manifestations of an allergic response (7). In fact, IgE reactivity is utilized to define the particular allergens contained in a given allergen source (8). However, in addition to IgE responses, T cells also vitally contribute to allergic disease (9). This contribution may be indirect, by promoting the production of IgE and the differentiation of eosinophils, or direct, through the release of various pro-inflammatory cytokines, such as IL-5, which promotes eosinophilic inflammation. A possible beneficial role of Tregs, resulting in suppression of allergic reactions, has also been indicated by several studies (10). Our knowledge of the role of antigen specific T cells in allergic reactions in humans is limited, and detailed studies are hampered by a relative paucity of information relating to the actual epitopes recognized by allergen-specific T cells.
Several studies have identified epitopes recognized by human T cells, but for many allergen systems the information is either fragmentary or lacking altogether. Responses to complex allergens in humans are very heterogeneous and involve recognition of a large number of epitopes (11-26). At the same time, the most dominant and prevalent responses encompass an appreciable fraction, and in some cases the majority, of the response, and these dominant epitopes can be predicted on the basis of their capacity to bind, and be recognized in the context of, multiple HLA DR, DP and DQ allelic variants (11) (Oseroff et al., in press).
Here, we took advantage of this approach to investigate a large panel of allergen proteins derived from 28 common allergen sources, which included fungi (Alternaria, Aspergillus, Cladosporuim and Penicillium), trees (Alder, Ash, Birch, Black Walnut, Cypress, Juniper, Oak and Palm), grasses (Bermuda, Canary, Kentucky Blue, Orchard, Rye and Sweet Vernal), weeds (English Plantain, Giant Ragweed, Mugwort, Russian Thistle, and Western Ragweed) and various indoor allergens (American Cockroach, Cat dander, Dog dander and Dust Mites). We identified over 250 different antigenic regions, most of which have not been previously described, and provide the first actual epitope data for several allergen sources. We find that T cell responses to the allergens previously defined on the basis of IgE reactivity account for a variable, and in many cases a surprisingly small, fraction of T cell responses, suggesting that several antigens involved in T cell recognition are yet to be described. We further find that different allergen sources are differentially polarized in terms of their capacity to recall production of Th1 versus Th2 associated lymphokines.
MATERIALS AND METHODS
Patient donor population
Patient recruitment for this study was performed at the University of California, San Diego (UCSD), and at National Jewish Health (NJH) in Denver, CO, under each local IRB approved protocol, as well as LIAI Institutional Review Board IRB approved protocol VD1-059-0311 (Federal Wide Assurance #00000032). Informed consent, study ID numbers, clinical case histories and other information were collected and recorded by clinical investigators. Immediate hypersensitivity skin test reactivity to a panel of extracts from 28 common allergens, as well as positive (histamine), and negative controls (diluent), was determined by standard methods. Both wheal (mm) and flare (mm) were measured at 15 minutes. All volunteers were asked to provide a 5 ml serum sample and a unit of peripheral blood.
An allergic donor was defined based on a history of allergic rhinitis and a positive skin test (a wheal of at least 3 mm in diameter greater than the diluent negative control) to one or more of the allergens tested. A total of 87 allergic donors were investigated. The donor cohort included 45 females and 42 males, and ranged between 20-63 years of age. Of the 87 donors, 52 had rhinitis and another 29 were categorized as having rhinitis and asthma (6 were not classified).
Our main goal was to identify a majority of the epitopes that are frequently recognized in the human population. By testing at least ten donors for each allergen of interest, we have a sufficient sample size such that 80% of all epitopes that are recognized in 15% or more allergic individuals in the general population are expected to give a positive response in one or more of the ten individuals in our study (assuming a binomial distribution). Similarly, 95% of all epitopes recognized in 40% of the allergic population are expected to give responses in two or more individuals in our study. Based on these considerations, we consider studying ten donors per allergen as sufficient to identify targets of frequent responses.
Bioinformatic analyses
Uniprot accession IDs for each of a panel of 28 common allergens were collected from the IUIS Allergen Nomenclature database, then used to retrieve sequences from the Uniprot database (see Supplemental Table 1). For those allergens without Uniprot IDs, we used the sequences provided by IUIS. If an allergen (e.g. Alt a1) had multiple sequences, a representative sequence for each allergen was selected. The representative sequence should have the longest length and greatest sequence coverage (or sequence identity > 90%) compared to other sequences. If the sequence identity between two sequences that belong to the same allergen is < 40%, both sequences were selected. As a result, 169 non-redundant sequences were identified.
Sequences, including isoforms, were then scanned for unique 15-mer peptides overlapping by 10 residues. Each peptide was then predicted for its capacity to bind to a panel of 20 of the most common HLA class II alleles (DPA1*0103/DPB1*0201, DPA1*0201/DPB1*0101, DPA1*0201/DPB1*0501, DPA1*0301/DPB1*0402, DQA1*0101/DQB1*0501, DQA1*0301/DQB1*0302, DQA1*0401/DQB1*0402, DQA1*0501/DQB1*0301, DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0405, DRB1*0701, DRB1*0802, DRB1*1101, DRB1*1302, DRB1*1501, DRB3*0101, DRB4*0101, DRB5*0101) using the consensus prediction described in (27). Peptides with predicted binding scores in the top 20% for a given allele were considered potential binders, and the number of HLA molecules each peptide was predicted to bind was enumerated. All peptides predicted to bind 10 or more HLA molecules were selected for synthesis and further study.
To make sure each allergen is adequately represented in the peptide set, at least 2 peptides were included for each allergen. We further stipulated that these 2 peptides should not be located in the first 20 residues (which likely contain signal sequences), and that the 2 peptides should not directly overlap (e.g. peptide 56-70 and 61-75). These additional requirements could be met for all but 4 proteins (Sec c 1, Sec c 20, Ant o 1, Dac g 2) for which only fragmentary sequence information was available. The complete list of peptides studied is shown in Supplemental Table 1.
Peptide synthesis
Peptides for screening studies were purchased from Mimotopes (Clayton, Victoria, Australia) and/or A and A (San Diego, CA) as crude material on a small (1 mg) scale. Peptides utilized as radiolabeled ligands were synthesized on larger scale, and purified (>95%) by reversed phase HPLC.
HLA binding assays
Assays to quantitatively measure peptide binding to MHC class II molecules, based on the inhibition of binding of a high affinity radiolabeled peptide to purified MHC molecules, have been described in detail elsewhere (28). Briefly, MHC molecules were purified from EBV transformed homozygous cell lines by monoclonal Ab-based affinity chromatography. HLA-DR, DQ and DP molecules were captured by repeated passage of lysates over LB3.1 (anti-HLA-DR), SPV-L3 (anti-HLA-DQ) and B7/21 (anti-HLA-DP) columns.
For inhibition experiments, 0.1-1 nM of radiolabeled peptide was co-incubated at room temperature or 37°C with 1 μM to 1 nM of purified MHC in the presence of a cocktail of protease inhibitors and various amounts of inhibitor peptide. Following a 2 to 4 day incubation, the percent of MHC bound radioactivity was determined by capturing MHC/peptide complexes on LB3.1 (DR), L243 (DR), HB180 (DR/DQ/DP), SPV-L3 (DQ) or B7/21 (DP) Ab coated Optiplates (Packard Instrument Co., Meriden, CT), and bound cpm measured using the TopCount (Packard Instrument Co.) microscintillation counter. Inhibitor peptides were tested in at least three or more independent assays at six different concentrations covering a 100,00-fold dose range. Under the conditions utilized, where [label]<[MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values (29, 30).
PBMC isolation and HLA typing
PBMC were obtained by density gradient centrifugation (Ficoll-Hypaque, Amerhsam Biosciences, Uppsala, Sweden) from one unit of blood (450 ml), according to manufacturer's instructions, and cryo-preserved for further analysis. An aliquot of serum was obtained for RAST IgE and IgG analyses (performed at NJMRC, Denver, CO & Phadia).
HLA typing was performed according to standard methods. Briefly, genomic DNA isolated from PBMC of the study subjects by standard techniques (QIAmp, Qiagen, Valencia, CA) was used for HLA typing. High resolution Luminex-based Sequence-Specific Oligonucleotide (SSO) typing for HLA Class I and Class II was utilized according the manufacturer's instructions (One Lambda, Canoga Park, CA). Where needed, PCR based Sequence-Specific Primer (SSP) typing methods were used to provide high resolution sub-typing (One Lambda, Canoga Park, CA).
In vitro expansion of allergen-specific T cells
PBMCs were cultured in RPMI 1640 (V Scientific, Tarzana, CA) supplemented with 5% human serum (Cellgro, Herndon, VA) at a density of 2×106 cells/ml in 24-well plates (BD Biosciences, San Jose, CA) and stimulated with 2 to 50 μg/ml of allergen extract (Greer, Lenoir, NC) depending on the allergen (see Supplemental Table 2). Cells were kept at 37°C in 5% CO2 and additional IL-2 (10 U/ml; eBioscience, San Diego, CA) was added every 3 days after initial antigenic stimulation. On day 14, cells were harvested and screened for reactivity against the allergen-specific peptide pools or individual peptides. LPS content of the various extracts was measured by Indoor Biotechnologies (Charlottesville, VA) using standard Limulus Amebocyte Lysate (LAL) methodology.
ELISPOT assays
The production of IL-5 and IFN-γ was analyzed in ELISPOT assays. Flat-bottom 96-well nitrocellulose plates (Millipore, Bedford, MA) were prepared according to manufacturer's instructions and coated with 10 μg/ml anti-human IL-5 (Clone TRFK5; Mabtech, Cincinnati, OH) and anti-human IFN-γ (Clone 1-D1K; Mabtech). Cells were then incubated at a density of 1×105/well either with peptide pools or individual peptides (10μg/ml), extract (2-50 μg/ml), PHA (10 μg/ml), or medium containing 0.1% DMSO (corresponding to the percentage of DMSO in the pools/peptides) as a control. After 24 hours, cells were removed, and plates were incubated with either 2 μg/ml biotinylated anti-human IL-5 Ab (Mabtech) and 1:200 HRP-conjugated anti-human IFN-γ Ab (Mabtech) at 37°C. After 2 hours, spots corresponding to the biotinylated Abs (IL-5) were developed by incubation with Alkaline Phosphatase-Complex (Vector Laboratories, Burlingame, CA) followed by incubation with Vector Blue Alkaline Phosphatase Substrate Kit III (Vector Laboratories) according to the manufacturer's instructions. Spots corresponding to the HRP-conjugated Ab (IFN-γ) were developed with 3-amino-9-ethylcarvazole solution (Sigma-Aldrich, St. Louis, MO). Spots were counted by computer-assisted image analysis (Zeiss, KS-ELISPOT reader, Munich, Germany).
Each assay was performed in triplicate. The level of statistical significance was determined with a Student's t-test using the mean of triplicate values of the response against relevant pools or individual peptides versus the response against the DMSO control. Criteria for peptide pool positivity were 100 spot-forming cells (SFCs)/106 PBMC, p ≤ 0.05 and a stimulation index (SI) ≥ 2, while criteria for individual peptide positivity were ≥ 20 SFC/106 PBMC, p ≤ 0.05, and a SI ≥ 2. The SFC/10^6 criteria utilized (in conjunction with also passing a T test with p< 0.05, and a SI>2) have been used in several recent studies from our group (see, e.g., (16-19, 31-36). In particular, in the context of allergen epitope identification, a recent study analyzing responses to Timothy Grass allergens validated much of the methodology applied in the present study. Together, in these studies it was also noted that, in general, epitope pools yielding significant but relatively weaker responses (in the 20 to 100 SFC range) did not lead to the identification of significant and consistent responses at the level of individual peptides. For this reason, and because cells are often limiting, and pool deconvolution is the most demanding step in terms of cell requirement, in those studies, as well as in the present study, only pools yielding 100 SFC/10^6 were deconvoluted.
HLA restriction
To determine the HLA locus restriction of identified epitopes, mAb inhibition assays were performed as described previously (11, 37). Preliminary determinations were made on control T cell clones of known specificity to determine optimal antibody doses leading to complete inhibition of the specific clones, and not associated with inhibition of clones known to be restricted by a different HLA allele or locus. This antibody concentration was then utilized in experiments where a dose response of antigenic peptide was tested in the presence or absence of the specific antibodies. Experimental determinations were performed utilizing ELISPOT assays specific for the particular lymphokine utilized to originally identify the particular epitope mapped.
For each antigenic region/donor combination short-term T cell lines were derived by extract stimulation of triplicate cultures of 2-3 million cells. IL-2 was added 5-8 days following stimulation. After 14 days of stimulation with the corresponding allergen extract (2-50 μg/ml), the HLA locus that restricted the response to the specific lymphokine was determined by measuring the capacity of mAbs specific for HLA-DR, DP or DQ to inhibit (block) the response. For this, PBMCs were incubated with 10 μg/ml of mAbs (Strategic Biosolutions, Windham, ME) against HLA-DR (LB3.1), DP (B7/21) or DQ (SVPL3) 30 minutes prior to addition of 10 μg/ml of peptide. Cytokine production induced by positive peptides was then measured in ELISPOT assays as described above. The pan MHC class I Ab (W6/32) was used as a control. The decrease (inhibition) in cytokine production in the presence of an HLA locus specific mAb, relative to production in the absence of mAb, was determined. A response was considered as restricted by HLA alleles at a specific locus when ≥50% inhibition of the response was observed in the presence of the corresponding mAb.
RESULTS
Selection of a panel of common allergens for investigation
T cell responses to complex allergens in humans are very heterogeneous and involve recognition of a large number of epitopes (11-26). Recent work by our group in the Timothy Grass (11) and German cockroach systems (Oseroff et al., in press) utilized an approach based on in vitro stimulation of PBMC from allergic donors followed by testing of peptide pools, and then further characterization of responses by testing individual peptides from positive pools. Here, we took advantage of this approach to investigate a large panel of allergen proteins derived from 28 different allergen sources. This investigation broadly addressed inhalants and contact allergens, including allergens derived from fungi (Alternaria, Aspergillus, Cladosporuim and Penicillium), trees (Alder, Ash, Birch, Black Walnut, Cypress, Juniper, Oak and Palm), grasses (Bermuda, Canary, Kentucky Blue, Orchard, Rye and Sweet Vernal), weeds (English Plantain, Giant Ragweed, Mugwort, Russian Thistle, and Western Ragweed) and various indoor allergens (American Cockroach, Cat dander, Dog dander and Dust Mites) (Table I). These allergen sources were selected because of their common diagnosis by extract reactivity in allergic patients at the two participating clinical sites, and availability in the IUIS database of at least some allergen protein sequences (for a complete list of these allergen proteins see Supplemental Table 1).
Table I.
Identification of 257 epitope regions from common allergen sources
| Allergen category | Allergen Source | Number of allergens | Number of peptides tested | Unique epitopes | Antigenic regions |
|---|---|---|---|---|---|
| Fungi | Alternaria Rot Fungus | 10 | 88 | 29 | 25 |
| A. fumigatus | 23 | 229 | 11 | 11 | |
| C. herbarum | 6 | 53 | 15 | 15 | |
| P. chrysogenum | 2 | 38 | 18 | 18 | |
| Grasses | Bermuda Grass | 5 | 21 | 3 | 3 |
| Canary Grass | 2 | 20 | 9 | 7 | |
| Kentucky Blue Grass | 2 | 31 | 12 | 9 | |
| Orchard Grass | 4 | 15 | 5 | 4 | |
| Rye Grass | 6 | 70 | 34 | 30 | |
| Sweet Vernal Grass | 5 | 3 | 3 | 2 | |
| Indoor allergens | American Cockroach | 5 | 79 | 3 | 3 |
| Cat epithelia | 4 | 55 | 10 | 8 | |
| D. farinae | 11 | 160 | 23 | 22 | |
| D. pteronyssinus | 14 | 224 | 40 | 35 | |
| Dog epithelia | 4 | 44 | 5 | 5 | |
| Trees | Alder | 2 | 11 | 4 | 3 |
| Ash | 1 | 6 | 3 | 3 | |
| Birch | 6 | 47 | 14 | 13 | |
| Black Walnut | 2 | 22 | 0 | 0 | |
| Cypress | 5 | 65 | 16 | 12 | |
| Date Palm | 1 | 5 | 3 | 3 | |
| Prickly Juniper | 1 | 3 | 3 | 3 | |
| White Oak | 1 | 5 | 4 | 2 | |
| Weeds | English Plantain | 1 | 3 | 2 | 2 |
| Giant Ragweed | 1 | 5 | 4 | 3 | |
| Mugwort | 4 | 22 | 0 | 0 | |
| Russian Thistle | 4 | 83 | 16 | 16 | |
| Western Ragweed | 1 | 4 | 0 | 0 | |
| Total | 133 | 1411 | 289 | 257 | |
In our investigations we utilized in vitro stimulation for a 14-17 day period, as the studies in the Timothy Grass and German Cockroach systems mentioned above indicated these time points to be optimal. As shown in Figure 1, additional data from three individual donors for two different allergen sources (Alternaria and Rye Grass) demonstrate that both IFN-γ and IL-5 responses are optimally detected in the 14-17 day window. The doses of individual extracts used for in vitro stimulation were chosen on the basis of initial dose titration experiments. For each extract the magnitude of responses, as well as cell viability at the end of the in vitro stimulation step, were noted. For certain extracts toxicity (loss of viability) was observed. In all experiments we used the highest dose not associated with noticeable toxicity. The exact dose utilized for each extract source is shown in Supplemental Table 2.
Figure 1. IFN-γ and IL-5 responses are optimally detected on day 14.
IFN-γ and IL-5 responses to Rye Grass or Alternaria extract, in three correspondingly allergic donors each, were measured in ELISPOT assays on days 0, 3, 6, 14 and 17. The doses of individual extracts used for in vitro stimulation were chosen on the basis of initial dose titration experiments, and were as shown in Supplemental Table 2.
Experimental strategy to identify allergen T cell epitopes
The strategy previously validated in the Timothy Grass model system (11) was utilized to predict potential allergen derived T cell epitopes. As described in the Materials and Methods, peptides ranking in the top 20% of predicted affinities for 10 or more of 20 common HLA class II alleles were selected for synthesis and further analysis. In total (see Table I), 133 different proteins were analyzed, and a total of 1411 predicted promiscuous binders were synthesized, corresponding to an average of 10.6 peptides per protein. On average, about 50 peptides were made for each allergen source, with a range of 3 to 229 (a complete list of the peptides synthesized is also provided in Supplemental Table 1).
Short-term lines stimulated with extract, as described above, were tested with pools of 15-20 peptides from the proteins of the corresponding allergen, and then individual epitopes were identified by deconvolution of positive pools (11). The dose of peptide utilized in the experiments was selected based on the fact that for pools of 20 peptides, in which the initial individual peptide stocks are 40 mg/ml in 100% DMSO, the highest final concentration of each individual peptide that can be achieved, without reaching a total pool DMSO concentration that is toxic in the assay (0.25-0.5%), is 4-5 μg/ml. A dose of 10 μg/ml was used for experiments with single peptide stimulations. This is consistent with our experience in other systems (see, e.g., (11, 17, 21, 31, 33, 38)), and is also a dose routinely used in the literature relating to stimulation of human class II restricted T cells.
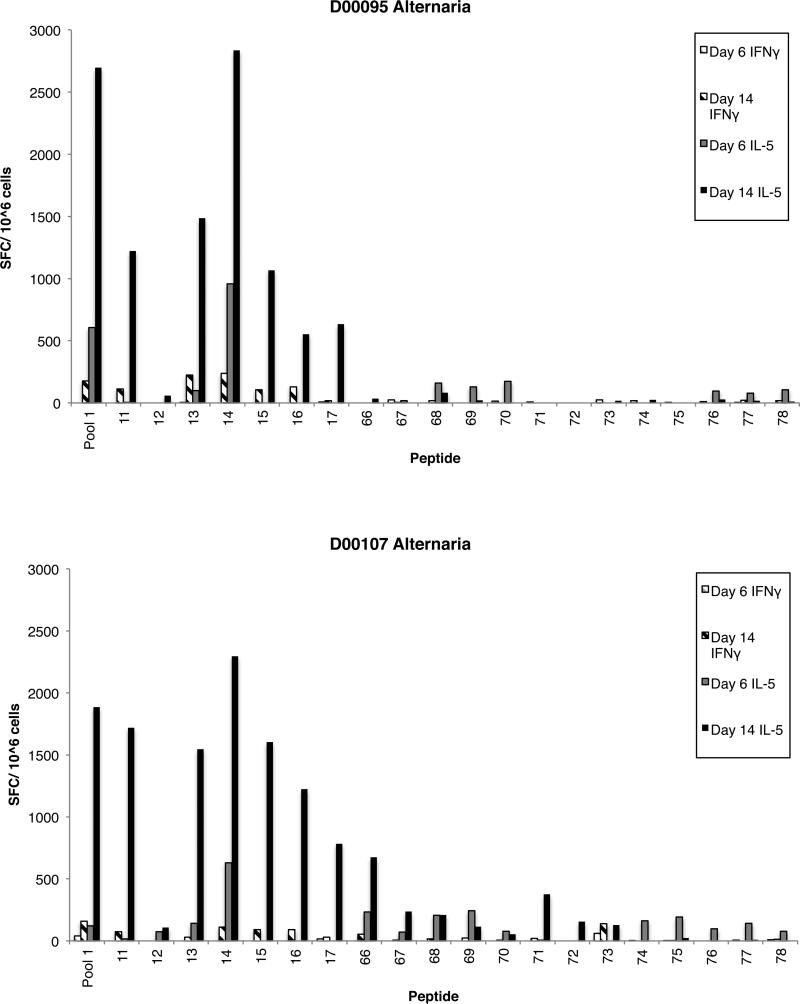
An additional issue to be addressed is whether different epitopes may be recognized on day 6 compared to day 14 of cultures. To address this issue, the pattern of epitopes identified was compared for two representative donors (D00095 and D00107) in the Alternaria system. The data, shown in Figure 2, illustrate how the epitopes identified on day 6 were also identified on day 14. In addition, because of the stronger signal obtained on day 14, additional peptides that would have been missed on day 6 are revealed at the day 14 time-point.
Figure 2. Comparison of day 6 and day 14 epitope repertoires.
The pattern of Alternaria epitopes identified on days 6 and 14 in two representative donors (D00095 and D00107) were compared. IL-5 or IFN-γ ELISPOT assays were performed, and positive responses defined, as described in the Material and Methods. In terms of IL-5 responses in donor D00095, only peptide 14 was recognized on day 6; by day 14, peptides 11, 13, 14, 15, 16 and 17 were all positive. Similarly, in donor D00107 peptides 14 and 66 gave positive responses on days 6 and 14, while peptides 11, 12, 13, 15, 16, and 17 only gave responses on day 14. For IFN-γ, in both donors positive responses were only seen on day 14.
Different lymphokines are differentially produced and regulated (39). However, in our experience, the epitopes that are recognized by IL-5 producing cells are essentially the same ones recognized by IL-4/IL-13 producing cells. Furthermore, IL-5 and IL-4/IL-13 producing cells are largely overlapping. Accordingly, in the present study, we focused on IL-5 as a representative lymphokine of the Th2 lineage. To illustrate our rationale, Figure 3 summarizes an analysis of IL-4, IL-5 and IL-13 producing cells in representative donors in response to Alternaria and Rye Grass peptides. These data demonstrate that, indeed, the epitopes recognized by IL-5 producing cells are the same ones recognized by IL-4/IL-13 producing cells. Furthermore, the data show that IL-5 and IL-4/IL-13 producing cells are largely overlapping and that there is little to gain by measuring all three lymphokines. Thus, while measuring all three lymphokines separately would be of obvious interest, it essentially doubles the number of cells required, and thus would not be easily compatible with the large number of allergens studied, and the relatively high-throughput nature of the assay strategy utilized in the present study.
Figure 3. Comparison of allergen epitope induced IL-4, -5 and -13 responses.
An analysis of IL-4, IL-5 and IL-13 producing cells in representative donors in response to Alternaria and Rye Grass peptides. Responses to IL-4, -5 and -13 were measured in ELISPOT assays on day 14, as described in the Materials and Methods. In donor U00064 Alternaria peptides 11, 13, 14, 15, 16, and 17 induced IL-5, IL-4 and IL-13 responses. Additionally, IL-5 responses were induced by peptides 12, 66, 67, 68 and 70. Together, these data show that all epitopes inducing IL-4 and IL-13 responses also induced IL-5 responses. A similar pattern was noted in donor U00099 in response to Rye Grass peptides, where peptides 1168, 1175, 1181 and 1184 all induced IL-5 responses, and the same peptides encompassed all IL-4 and IL-13 responses.
Identification of 257 different antigenic regions from common allergen sources
PBMC from donors with positive skin tests to the allergen in question were restimulated in vitro with the various corresponding allergen extracts. Allergen extract stimulation was effective in most of the donors (94% overall, and on average across all sources; range 67 to 100% for the different extracts; Supplemental Table 2). These short-term, extract-stimulated, lines were tested with pools of 15-20 peptides from the proteins of the corresponding allergen, and then individual epitopes were identified by deconvolution of positive pools (11). For each allergen source, we tested PBMCs from at least ten different allergic donors (range 10-15, average 11), as determined by skin test to the corresponding allergen. in standard dual ELISPOT assays detecting IFN-γ and IL-5. We acknowledge that this is a rather limited number of donors. This number was chosen to enable identification of epitopes that are most frequently recognized in the donor population, consistent with the promiscuous HLA binding principle used to select the candidate epitopes. According to our power calculations, by studying the response in 10 allergic donors, 80% of all epitopes that are recognized in 15% or more allergic individuals in the general population would be identified (assuming a binomial distribution). Similarly, 95% of all epitopes recognized in 40% of the allergic population are expected to give responses in two or more individuals in our study. This is consistent with the scope of the current investigation, which was to survey a large number of allergen sources, side-by-side, utilizing the same experimental design, and characterizing the most dominant responses.
In total, 322 peptides were positive in at least one donor. ELISPOT results from each individual peptide are presented in Supplemental Table 3. The high rate at which predicted peptides were found to be antigenic (322/1405 = 22.9%) demonstrates the power of the approach based on prediction of promiscuous HLA class II binding. Some epitopes were highly homologous because they were derived from allergen isoforms, or from the same allergen protein and covered largely overlapping regions. After removal of redundancies and consolidation of largely overlapping regions, a total of 289 unique epitopes, corresponding to 257 distinct antigenic regions, were identified (Table I).
Novelty of the allergen epitopes identified
Next, the sequences of the 257 antigenic regions identified by our study were compared to the known human Class II/CD4 epitopes curated in the IEDB (www.iedb.org; (40)). To obtain a data set with characteristics comparable to the one obtained in the screening described herein, the IEDB was queried for all epitopes defined in human hosts, either ex vivo or by utilizing short-term lines, and for which CD4/class II restriction could either be demonstrated or inferred on the basis of the assay methodology. The purpose of this analysis was to determine the extent to which the identified epitopes were either novel or previously cited.
Overall, epitopes were identified for 25 of the 28 allergen sources studied. For 14 of the allergen sources all of the epitopes identified were completely novel, in that no epitope could be found in the IEDB that satisfied the criteria defined above (Table IIA). By contrast, and as expected, in the cases of 11 other allergen sources which have been previously and more extensively studied, many of the epitopes identified mapped to regions reported as being antigenic in allergic patients (Table IIB). Nonetheless, in those cases, the antigenic regions identified in the current study were still, to a great extent novel, as 84% of the epitopes identified had not been previously reported. Conversely, only 38% of the epitopes already curated by the IEDB were re-identified by the present study. This is consistent with the results obtained in the Timothy Grass study, which estimated that the predictive approach would identify the most dominant and prevalently recognized epitopes, corresponding to about 50% of the total T cell response (11). In conclusion, the analysis presented in this section underscores the novel nature of a large number of the epitopes identified in the current study.
Table II.
Novelty of the epitopes identified
| A. Novel sources | |||
|---|---|---|---|
| Regions identified |
|||
| Source | This study | IEDB | Novel |
| Alternaria Rot Fungus | 25 | 0 | 25 |
| P. chrysogenum | 18 | 0 | 18 |
| Russian Thistle | 16 | 0 | 16 |
| C. herbarum | 15 | 0 | 15 |
| Canary Grass | 7 | 0 | 7 |
| Orchard Grass | 4 | 0 | 4 |
| American Cockroach | 3 | 0 | 3 |
| Ash | 3 | 0 | 3 |
| Prickly Juniper | 3 | 0 | 3 |
| Date Palm | 3 | 0 | 3 |
| Giant Ragweed | 3 | 0 | 3 |
| White Oak | 2 | 0 | 2 |
| Sweet Vernal Grass | 2 | 0 | 2 |
| English Plantain | 2 | 0 | 2 |
| Total | 106 | 0 | 106 |
| B. Sources for which epitopes were previously described | ||||||
|---|---|---|---|---|---|---|
| Regions identified |
||||||
| Source | This study | IEDB | Overlap | Novel | %Novel | %IEDB reidentified |
| Alder | 3 | 3 | 2 | 1 | 33.3 | 66.7 |
| D. farinae | 22 | 7 | 2 | 20 | 90.9 | 28.6 |
| A. fumigatus | 11 | 3 | 1 | 10 | 90.9 | 33.3 |
| Bermuda Grass | 3 | 5 | 0 | 3 | 100.0 | 0.0 |
| Birch | 13 | 7 | 3 | 10 | 76.9 | 42.9 |
| Cat epithelia | 8 | 4 | 4 | 4 | 50.0 | 100.0 |
| Dog epithelia | 5 | 5 | 0 | 5 | 100.0 | 0.0 |
| D. pteronyssinus | 35 | 8 | 5 | 30 | 85.7 | 62.5 |
| Japanese Cypress | 12 | 10 | 3 | 9 | 75.0 | 30.0 |
| Kentucky Blue Grass | 9 | 7 | 2 | 7 | 77.8 | 28.6 |
| Rye Grass | 30 | 4 | 2 | 28 | 93.3 | 50.0 |
| Total | 151 | 63 | 24 | 127 | 84.1 | 38.1 |
Multiple allergen sequences are necessary to capture T cell epitope responses
The distribution of epitopes identified was next analyzed as a function of the various allergen sources. As mentioned above, allergen extract stimulation was effective in 95% of the donors. However, in some cases a relatively large number of epitopes were identified, while in other cases the screen revealed no, or only a few, epitopes. To quantitatively express these variations, for each allergen source we calculated the ratio of the total epitope specific IL-5 and IFNγ response to the total response observed against the corresponding crude allergen extract (Table III). In 16 cases the peptide specific responses corresponded to 15% or more of the response detected against the extract, while in 12 cases the peptide responses totaled less than this arbitrary threshold. Indeed, the 16 allergen sources associated with the larger total peptide responses accounted for 230 of the 257 (89%) epitope regions identified, and 74 of the 81 (91%) regions recognized by 2 or more donors (see also Supplemental Table 3).
Table III.
Heterogeneity in epitope coverage of the different allergen sources
| Response (SFC) |
|||||
|---|---|---|---|---|---|
| Allergen Source | Regions | Peptide | Extract | %Peptide /Extract | IUIS Proteins |
| Rye Grass | 30 | 29722 | 19437 | 152.9 | 4 |
| D. pteronyssinus | 35 | 28129 | 22505 | 125.0 | 14 |
| D. farinae | 22 | 9402 | 9707 | 96.9 | 11 |
| Alternaria Rot Fungus | 25 | 31162 | 35207 | 88.5 | 10 |
| Cypress | 12 | 14310 | 18160 | 78.8 | 5 |
| Cat epithelia | 8 | 20068 | 28607 | 70.2 | 5 |
| Birch | 13 | 9690 | 17110 | 56.6 | 6 |
| White Oak | 2 | 8520 | 18067 | 47.2 | 1 |
| Canary Grass | 7 | 8217 | 22054 | 37.3 | 2 |
| P. chrysogenum | 18 | 9723 | 26643 | 36.5 | 2 |
| Russian Thistle | 16 | 11848 | 32723 | 36.2 | 5 |
| C. herbarum | 15 | 9200 | 31020 | 29.7 | 6 |
| Kentucky Blue Grass | 9 | 11007 | 39232 | 28.1 | 7 |
| Alder | 3 | 3877 | 14384 | 27.0 | 2 |
| Orchard Grass | 4 | 5474 | 25990 | 21.1 | 2 |
| A. fumigatus | 11 | 3953 | 23180 | 17.1 | 23 |
| Median | 12.5 | 9707 | 22843 | 42.2 | 5.0 |
| Juniper | 3 | 1033 | 17540 | 5.9 | 1 |
| Sweet Vernal Grass | 2 | 1667 | 35263 | 4.7 | 5 |
| Ash | 3 | 767 | 17073 | 4.5 | 1 |
| Bermuda Grass | 3 | 903 | 21083 | 4.3 | 2 |
| English Plantain | 2 | 688 | 16963 | 4.1 | 1 |
| Giant Ragweed | 3 | 533 | 13960 | 3.8 | 1 |
| American Cockroach | 3 | 690 | 27854 | 2.5 | 5 |
| Dog epithelia | 5 | 353 | 14616 | 2.4 | 4 |
| Date Palm | 3 | 233 | 23417 | 1.0 | 1 |
| Black Walnut | 0 | 0 | 28913 | 0.0 | 2 |
| Mugwort | 0 | 0 | 21793 | 0.0 | 4 |
| Western Ragweed | 0 | 0 | 16733 | 0.0 | 1 |
| Median | 3 | 611 | 19312 | 3.1 | 1.5 |
When the number of known allergenic proteins described in the IUIS database, and utilized for the present analysis, was scrutinized (Table III), a correlation between the fraction of the extract response that could be attributed to the peptides studied, and the number of representative protein sequences available, became apparent. In other words, the sources for which the set of T cell epitopes identified accounted for only a minor fraction of the extract response were represented by fewer protein sequences (median 1.5) as compared to the sources where the epitopes accounted for a larger fraction of the extract response (median 5 protein sequences, RS = 0.49; p = 0.01). These results suggest that, in the cases of the “low epitope coverage” allergen sources, the number of antigenic T cell targets was too limited to allow for capture of the complexity of responses. Thus, it is likely that for these sources additional allergens remain to be identified and described.
Exceptions to this trend were noted, however. For example, in the case of White Oak, only one protein was represented in the IUIS. However, peptides from that one protein accounted for almost 50% of the extract response. Other instances where an appreciable fraction (21-47%) of the extract response was associated with just a few proteins were also noted. Indeed, as shown in Table III, for 5 of the 16 allergens for which >15% of the extract response was identified with antigen specific peptide pools, only one or two proteins were available for study. These data suggest that for these allergen sources the major T cell epitopes are derived from a limited number of proteins. Conversely, it is remarkable that for some organisms, such as Aspergillus, where many proteins were scanned (n=23), only a few (n=1) were found to have epitopes. Finally, in some cases, such as American Cockroach, the paucity of epitopes might be reflective of the weak level of sensitization of the particular patient population investigated in this study, as judged by low skin test reactivity.
Promiscuous restriction of dominant epitopes
In the next series of experiments we focused on 74 epitope regions that were recognized in two or more donors and derived from the 16 allergen sources in which peptide specific responses could account for 15% or more of the responses detected with the extract. These more prominent epitopic regions, listed in Table IV, accounted for 70% of the total SFC response detected against all of the epitope regions identified from these 16 different allergen sources.
Table IV.
HLA restriction of prevalently recognized epitope regions
| Region restriction(s) |
|||||||
|---|---|---|---|---|---|---|---|
| Organism | Source | Donors responding | % responding | Total SFC | DR | DQ | DP |
| Alt. Rot Fungus | Alt a 1 (6-25) | 4 | 33.3 | 5572 | 4 | 0 | 0 |
| Alt a 1 (116-135) | 7 | 58.3 | 7450 | 3 | 3 | 0 | |
| Alt a 1 (141-157) | 5 | 41.7 | 5768 | 5 | 0 | 0 | |
| Alt a 5 (51-65) | 2 | 16.7 | 3450 | 1 | 0 | 0 | |
| Alt a 6 (161-175) | 2 | 16.7 | 1238 | 0 | 0 | 1 | |
| Alt a 7 (6-20) | 2 | 16.7 | 727 | 1 | 0 | 0 | |
| Alt a 7 (190-204) | 2 | 16.7 | 397 | 0 | 0 | 0 | |
| A. fumigatus | Asp f 17 (91-105) | 2 | 18.2 | 813 | 1 | 0 | 0 |
| C. herbarum | Cla h 5 (6-20) | 2 | 20.0 | 247 | 0 | 0 | 0 |
| Cla h 6 (161-175) | 3 | 30.0 | 2082 | 1 | 0 | 1 | |
| Cla h 8 (101-115) | 2 | 20.0 | 317 | 0 | 0 | 0 | |
| Cla h 8 (151-165) | 2 | 20.0 | 470 | 1 | 0 | 0 | |
| Cla h 8 (181-195) | 2 | 20.0 | 817 | 0 | 0 | 0 | |
| Cla h 8 (236-250) | 2 | 20.0 | 4067 | 1 | 1 | 0 | |
| P. chrysogenum | Pen ch 13 (101-115) | 2 | 20.0 | 677 | 1 | 0 | 0 |
| Pen ch 13 (271-285) | 2 | 20.0 | 1463 | 1 | 0 | 0 | |
| Pen ch 18 (226-240) | 2 | 20.0 | 607 | 1 | 0 | 0 | |
| Pen ch 18 (291-305) | 3 | 30.0 | 2480 | 2 | 0 | 0 | |
| Cat epithelia | Fel d 1 (41-60) | 3 | 27.3 | 6065 | 3 | 0 | 0 |
| Fel d 1 (78-92) | 3 | 27.3 | 3453 | 3 | 0 | 0 | |
| Fel d 1 (21-40) | 6 | 54.5 | 7128 | 2 | 0 | 2 | |
| Fel d 1 (36-50) | 2 | 18.2 | 2837 | 1 | 0 | 0 | |
| D. farinae | Der f 1 (131-150) | 2 | 20.0 | 1448 | 2 | 0 | 0 |
| D. pteronyssinus | Der p 1 (131-150) | 2 | 18.2 | 2417 | 2 | 1 | 0 |
| Der p 1 (176-190) | 2 | 18.2 | 890 | 2 | 0 | 0 | |
| Der p 1 (226-240) | 2 | 18.2 | 2783 | 1 | 1 | 0 | |
| Der p 4 (96-120) | 3 | 27.3 | 3877 | 2 | 0 | 0 | |
| Der p 4 (251-265) | 2 | 18.2 | 763 | 0 | 0 | 1 | |
| Der p 4 (321-335) | 2 | 18.2 | 1570 | 0 | 1 | 0 | |
| Der p 4 (381-400) | 4 | 36.4 | 2573 | 2 | 0 | 0 | |
| Der p 4 (411-430) | 2 | 18.2 | 1117 | 0 | 0 | 1 | |
| Der p 4 (482-502) | 2 | 18.2 | 1160 | 1 | 0 | 0 | |
| Alder | Aln g 1 (11-30) | 2 | 16.7 | 2036 | 0 | 1 | 1 |
| Aln g 1 (111-125) | 2 | 16.7 | 1244 | 1 | 1 | 0 | |
| Birch | Bet v 1 (96-110) | 2 | 20.0 | 462 | 0 | 0 | 0 |
| Bet v 1 (111-125) | 3 | 30.0 | 1132 | 2 | 0 | 0 | |
| Bet v 6 (211-230) | 2 | 20.0 | 807 | 0 | 1 | 0 | |
| Cypress | Cha o 1 (86-110) | 3 | 30.0 | 1922 | 1 | 0 | 0 |
| Cha o 1 (211-225) | 2 | 20.0 | 835 | 0 | 0 | 0 | |
| Cup a 1 (71-90) | 3 | 30.0 | 3067 | 1 | 0 | 0 | |
| Cup a 1 (316-330) | 3 | 30.0 | 740 | 1 | 1 | 0 | |
| Cup s 1 (91-115) | 3 | 30.0 | 3627 | 1 | 0 | 0 | |
| Cup s 1 (211-225) | 2 | 20.0 | 1650 | 1 | 0 | 0 | |
| White Oak | Que a 1 (11-35) | 8 | 61.5 | 7183 | 1 | 0 | 3 |
| Que a 1 (141-155) | 4 | 30.8 | 1337 | 1 | 0 | 1 | |
| Orchard Grass | Dac g 1 (181-195) | 4 | 36.4 | 2795 | 2 | 0 | 0 |
| Dac g 2 (11-30) | 2 | 18.2 | 1917 | 1 | 0 | 0 | |
| Dac g 4 (1-15) | 3 | 27.3 | 482 | 0 | 0 | 0 | |
| Rye Grass | Lol p 1 (176-190) | 3 | 27.3 | 1180 | 1 | 0 | 0 |
| Lol p 2 (51-70) | 4 | 36.4 | 1163 | 3 | 0 | 0 | |
| Lol p 2 (76-90) | 3 | 27.3 | 1253 | 0 | 2 | 0 | |
| Lol p 3 (11-25) | 3 | 27.3 | 2070 | 2 | 0 | 0 | |
| Lol p 3 (51-65) | 2 | 18.2 | 1803 | 1 | 0 | 0 | |
| Lol p 3 (76-90) | 2 | 18.2 | 3342 | 0 | 0 | 0 | |
| Lol p 5 (71-85) | 3 | 27.3 | 3325 | 0 | 2 | 1 | |
| Lol p 5 (156-175) | 3 | 27.3 | 3437 | 0 | 2 | 0 | |
| Lol p 5 (236-250) | 2 | 18.2 | 460 | 0 | 0 | 0 | |
| Lol p 5 (306-320) | 2 | 18.2 | 1695 | 0 | 1 | 0 | |
| Lol p 5 (181-195) | 3 | 27.3 | 2260 | 1 | 1 | 1 | |
| Lol p 11 (116-135) | 2 | 18.2 | 2393 | 0 | 0 | 1 | |
| Canary Grass | Pha a 1 (186-200) | 4 | 40.0 | 2095 | 3 | 1 | 0 |
| Pha a 5 (121-140) | 3 | 30.0 | 1680 | 1 | 1 | 0 | |
| Pha a 5 (166-180) | 2 | 20.0 | 2222 | 0 | 1 | 1 | |
| Pha a 5 (181-195) | 2 | 20.0 | 493 | 2 | 0 | 0 | |
| Pha a 5 (201-220) | 4 | 40.0 | 1502 | 3 | 1 | 0 | |
| Ken. Blue Grass | Poa p 1 (121-135) | 3 | 25.0 | 1357 | 1 | 0 | 0 |
| Poa p 5 (51-75) | 7 | 58.3 | 3528 | 0 | 1 | 1 | |
| Poa p 5 (121-135) | 3 | 25.0 | 435 | 1 | 0 | 0 | |
| Poa p 5 (131-145) | 2 | 16.7 | 243 | 1 | 0 | 0 | |
| Poa p 5 (176-190) | 6 | 50.0 | 1862 | 3 | 0 | 1 | |
| Poa p 5 (211-230) | 4 | 33.3 | 1738 | 3 | 0 | 0 | |
| Poa p 5 (221-235) | 2 | 16.7 | 1370 | 1 | 0 | 0 | |
| Russian Thistle | Sal k 1 1 (191-205) | 2 | 20.0 | 2060 | 0 | 1 | 1 |
| Sal k 1 1 (321-335) | 2 | 20.0 | 1947 | 1 | 0 | 1 | |
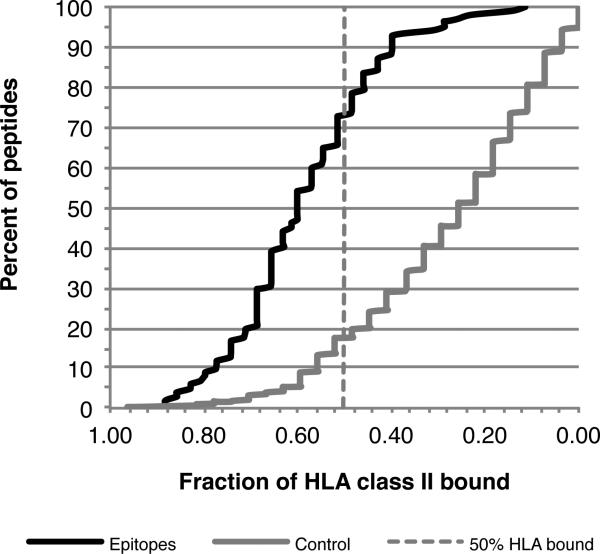
Each of the individual 15-mer peptides associated with these regions was tested for its capacity to bind a panel of 35 different DR, DP and DQ molecules representative of the most common allelic variants worldwide (41-43), including all 20 molecules comprising the prediction panel. It was found that 73% of the epitopes bound 50% or more of the molecules tested with an affinity of 1000 nM, or better (Figure 4; binding data for each peptide is presented in Supplemental Table 4). By contrast, in a control set of a panel of 425 unbiased, non-redundant, peptides representing 15-mers, overlapping by 10 residues, spanning the entire sequences of the P. pratense 1, 2, 3, 4, 5, 6, 7, 11, 12, and 13 pollen antigens, only 17% bound 50% or more of the same molecules. As the allergen-derived peptides tested herein were selected on the basis of predicted promiscuous HLA class II binding capacity, overall, these data support the validity of the predictive approach for identification of promiscuous binding peptides.
Figure 4. Promiscuous HLA binding capacity of dominant epitopes.
Individual 15-mer epitopes (n=96) from the 74 epitope regions derived from the 16 allergen sources where peptide specific responses could account for 15% or more of the responses detected with the extract, and that were recognized in two or more donors, were tested for their capacity to bind a panel of 35 different DR, DP and DQ molecules. The cumulative percent of epitopes (black line) binding various fractions of the class II panel is shown. 73% of the epitopes bound 50% or more of the molecules tested with an affinity of 1000 nM, or better. Also shown is the cumulative percentage of peptides in a control panel of 425 unbiased, non-redundant, peptides binding various fractions of the same class II panel; only 17% of the control peptides bound 50% or more of the molecules tested.
Based on the predictive approach taken and the data presented above, we expected that a diverse set of HLA allelic variants would be restricting the T cell responses to the epitopes identified. To address this issue experimentally, the HLA locus restriction of the more frequently recognized epitopes was determined by inhibition experiments utilizing DR-, DP- and DQ-specific antibodies. Locus restriction could be determined for a total of 65 antigenic regions (Table IV). In the remaining combinations (about a third of the cases), locus restriction could not be determined, either due to a scarcity of cells, low responses, or because 50% inhibition by locus specific mAb could not be achieved, perhaps reflective of promiscuous locus restriction at the level of individual donors.
Multiple restricting loci were observed in about a third of the cases (21/65; 32.3%). Furthermore, in 10 of the 17 (59%) cases where a single locus was indicated by the antibody inhibition experiments as restricting a specific epitope in multiple donors, no single allelic variant capable of binding the epitope in vitro was shared by all of the responding donors, thus implying intra-locus promiscuous restriction as well. Overall, the data presented in this section highlights the promiscuous restriction of the more prevalent and dominant epitopes identified.
T cell responses to different allergen sources are differentially polarized
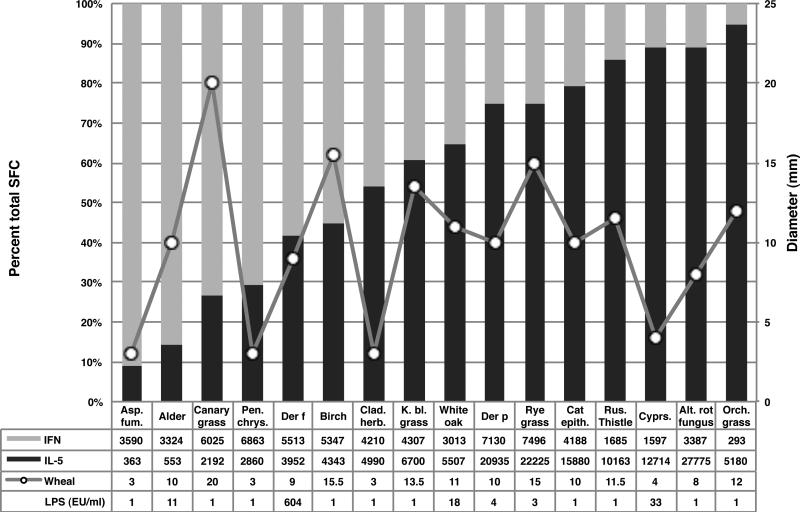
As described above, the epitope specific responses for the various allergen sources were determined by utilizing both IL-5 and IFN-γ ELISPOT assays. These lymphokines were chosen as prototype Th2 and Th1 responses, respectively. For each of the allergen sources, we next inspected the relative proportion of the response attributed to each of these two lymphokines. As expected, overall IL-5 production exceeded IFN-γ production by a ratio of approximately 2:1. Interestingly, however, different patterns were noted for individual allergen sources (Figure 5). In some cases (e.g., Orchard grass, Alternaria, Cypress, and Russian Thistle), IL-5 production exceeded IFN-γ production by 5-10-fold. In other cases (e.g., Aspergillus, Penicillum, and Alder), the converse was noted, as production of IFN-γ exceed IL-5 production by more than 3-10-fold. These results suggest that different allergen sources are associated with different degrees of polarization of the responding T cell subsets. We hypothesized that differential polarization may correlate with the skin test response, such that a late phase skin response may associate more frequently with the Th1-type allergen/epitope. However, examination of the data revealed no discernable trend (Figure 5). Similarly, it was hypothesized that the polarization observed was due to the different allergenic extracts having different levels of LPS content. When the LPS content of the various extracts was measured (see the table embedded in Figure 5) we found no correlation (r=0.14, -0.16 and -0.16 for IFNγ, IL-5 and the IFNγ/IL-5 ratio, respectively) between the LPS content the Th1/Th2 skewing we observed.
Figure 5. T cell responses to different allergen sources are differentially polarized.
Percentage of the total response to the various allergen sources attributable to IL-5 or IFN-γ, representative of Th2 and Th1 responses, respectively, are shown as a bar graph. Median wheal size (mm in diameter) measured in correspondingly allergic donors are plotted on the secondary axis.
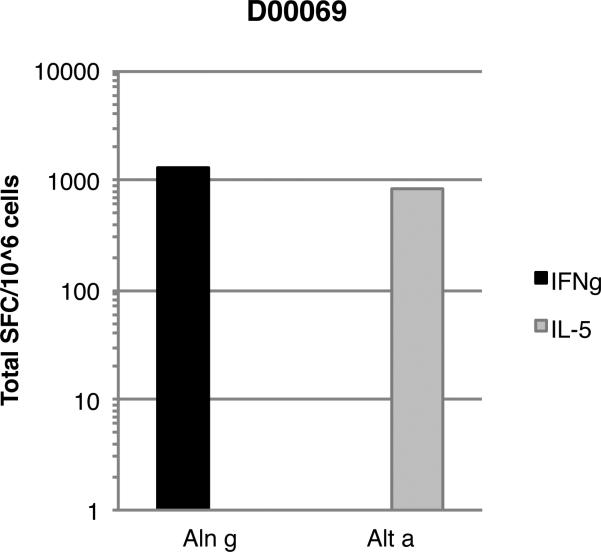
Strikingly, even within an individual donor, responses to different allergen sources could be differentially polarized, with responses to one allergen dominated by Th1 responses, and to a different allergen dominated by Th2 responses. An example of this type of situation is shown in Figure 6, which depicts the T cell responses observed in a donor who responded to epitopes from Alder and Alternaria rot fungus, allergens associated with Th1 and Th2 polarization, respectively. As shown, the response by this donor to Alder (Aln g) was completely Th1, while the response to Alternaria (Alt a) was completely Th2.
Figure 6. Polarized T cell responses to different allergens within an individual donor.
IFNγ (black bars) and IL-5 (gray bars) responses, associated with Th1 and Th2 polarization, respectively, in donor D00069. As described in the text, the T cell response, in terms of total SFC, to Aln g was associated only with only IFN-γ production, while the response to Alt a was only mediated by IL-5, highlighting the polarization of the responses to epitopes derived from Alder (Aln g) and Alternaria Rot Fungus (Alt a).
DISCUSSION
Herein, we report the results of a systematic survey of T cell epitopes derived from common allergens. To the best of our knowledge this is the first such study simultaneously investigating 28 different common allergen sources with the same controlled and uniform technical approach. This approach is designed to allow relatively high throughput analysis, while still capturing a large fraction of the total class II restricted allergen specific T cell response, and was previously validated in the Timothy Grass (11) and German Cockroach (Oseroff et al., in press) systems. The approach is based on prediction of the peptides most likely to bind a panel of HLA molecules chosen to be representative of the most common HLA class II allelic variants at the DR, DP and DQ loci.
Each peptide was tested in at least ten donors specifically allergic to each allergen source. We reasoned that this number of donors would be suitable to identify the more dominant, and more frequently recognized, epitopes, while at the same time allowing us to screen for a broad and diverse set of allergen sources. Nearly a quarter (322/1405= 22.9%) of the predicted peptides were positive in at least one of the donors tested, thus further validating the approach, and illustrating its broad applicability to human inhalant and contact allergens, including fungus, tree, grass, weed and animal allergens.
The significance of the observations is highlighted by the comparison of these newly identified epitopes with those previously identified and described in the scientific literature and curated in the IEDB (40, 44). The epitopes identified for 14 of the allergen sources were totally novel, in that no human class II/CD4 epitope could be found in the IEDB that overlapped with any of the epitopes described herein. For 11 other allergen sources, which have been more extensively studied, 84% of the epitopes identified were still novel. Conversely, when the data was analyzed to pinpoint the fraction of IEDB-contained epitopes that were re-identified in the course of the present study, we found that, on average, 38% of the known epitopes were re-identified. This is consistent with the Timothy Grass and German Cockroach studies noted above, which estimated that the predictive approach would identify the most dominant and prevalently recognized epitopes, corresponding to about 50% of the total T cell response.
The epitopes most frequently recognized were characterized in terms of their HLA binding capacity to a panel of 35 different HLA class II molecules. This panel was chosen to be representative of the most common allelic variants in the general population at the HLA class II DR, DP and DQ loci (41-43). This data is of relevance, as the quantitative binding data to the various common HLA class II variants can be used to project the potential coverage by the various epitopes in patient populations of different ethnicities (43, 45, 46). The issue of restriction was addressed in experiments where the HLA class II restricting locus was determined by antibody inhibition experiments. It was found that about a third of the epitopes were restricted by multiple loci. Furthermore, we estimate that at least 60% of the remaining epitopes are restricted by multiple allelic variants at a given locus. These results are not unexpected, given the fact that predicted promiscuous binding was utilized as a selection criterion for identifying candidate epitopes. Nonetheless, these observations underline the relevance of promiscuous epitope recognition in the context of HLA class II allergen-specific T cell responses.
A notable result of our side-by-side survey is that the identified epitopes account for a variable fraction of the response to a given allergen source, and that this fraction further correlates with how many different allergenic proteins have been described for that source. Thus, this data suggests that, at the T cell level, allergic responses target a relatively large number of antigens. Future experiments will have to address whether this observation may reflect the existence, for the allergen sources where only one or few specific sequences have been reported, of additional allergens recognized by IgE, and not yet identified. Alternatively, this may also reflect an incomplete overlap between the targets of T cell and IgE responses.
While some studies show a correlation between IL-5 levels and IgE (47), other studies do not (48). Thus, IgE or wheal responses may or may not be related to Th2 (i.e. IL-5) responses, and in our study these appear to depend upon the allergen studied. We also observed that different allergen sources appear to elicit patterns of responses that are differentially polarized in terms of their Th1/Th2 balance, at least as judged by IL-5 (Th2) or IFN-γ (Th1) production. Strikingly, even within an individual donor, responses to different allergens could be differentially polarized, with responses to one allergen dominated by Th1 responses, and to a different allergen dominated by Th2 responses. A similar phenomenon was observed with different allergenic proteins in the Timothy Grass system (11) following restimulation with pollen extract. A comparison of the LPS content of the various extracts used with the associated Th1/Th2 balances revealed no correlation, suggesting that the differential polarization observed is antigen specific, and not due to differential LPS content in the various extracts. The molecular basis for this effect is presently unknown, and might reflect differences in the relative concentrations and accessibility of the different allergens in the pollen and extract, their processing and presentation, and potentially the presence of distinct co-stimulatory signals associated with each allergen. The study of this mechanism might suggest avenues to influence or alter the lymphokine balance of Th responses, and thus potentially the outcome of responses in terms of IgE titers.
The epitopes identified herein might be of use in several respects. First, we expect that they might be used to characterize T cell responses associated with allergic reactions, including characterization of changes in responding T cell phenotype/T cell plasticity as a function of seasonality, as a result of SIT treatment, or as a function of varying disease severity (asthma versus rhinitis). Secondly, it is possible to speculate that T cell epitopes might be used to develop immunotherapeutic SIT treatments that could target T cell responses without the risks connected to administration of whole allergens capable of binding IgE, and that thus pose potential safety risks. Further studies could investigate the use of the epitopes identified in the present study in animal models sensitized to the various allergens. The use of HLA class II transgenic mice could overcome the different epitope repertoire of murine (H-2) versus human (HLA) class II molecules. Finally, the epitopes identified herein could be utilized for the generation of tetrameric reagents, useful for detailed characterization of the associated immune response. However, the generation of tetramers specific for the various epitopes poses several challenges. In general, HLA class II tetramers have been more difficult to produce than their human class l counterparts. This obstacle is further compounded by the fact that the dominant epitopes identified here appear to be promiscuous and would, in that respect, require generation of multiple tetrameric staining reagents. In this light, it seems that either experimental techniques based on pools of epitopes followed by intracellular cytokine staining, or staining with multiple tetramers (49, 50) would be desirable. Our laboratory is currently investigating these two alternative options.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carrie Moore, Kate Bradley, and Duy Le for performing the MHC binding assays. La Jolla Institute for Allergy and Immunology & Kyowa Hakko Kirin California publication number 1496.
Footnotes
This study was supported by Nation Institutes for Health (NIH) - National Institute Of Allergy And Infectious Diseases (NIAID) contract HSN272200700048C (AS, LIAI) and grant U19AI100275 (AS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health.
REFERENCES
- 1.Locksley RM. Asthma and allergic inflammation. Cell. 2010;140:777–783. doi: 10.1016/j.cell.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. 2011;378:2112–2122. doi: 10.1016/S0140-6736(11)60130-X. [DOI] [PubMed] [Google Scholar]
- 3.Barrett NA, Austen KF. Innate cells and T helper 2 cell immunity in airway inflammation. Immunity. 2009;31:425–437. doi: 10.1016/j.immuni.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt PG, Sly PD. Interaction between adaptive and innate immune pathways in the pathogenesis of atopic asthma: operation of a lung/bone marrow axis. Chest. 2011;139:1165–1171. doi: 10.1378/chest.10-2397. [DOI] [PubMed] [Google Scholar]
- 5.Minnicozzi M, Sawyer RT, Fenton MJ. Innate immunity in allergic disease. Immunological Reviews. 2011;242:106–127. doi: 10.1111/j.1600-065X.2011.01025.x. [DOI] [PubMed] [Google Scholar]
- 6.Burton OT, Oettgen HC. Beyond immediate hypersensitivity: evolving roles for IgE antibodies in immune homeostasis and allergic diseases. Immunological Reviews. 2011;242:128–143. doi: 10.1111/j.1600-065X.2011.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu LC. Immunoglobulin E receptor signaling and asthma. The Journal of biological chemistry. 2011;286:32891–32897. doi: 10.1074/jbc.R110.205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton RG. Clinical laboratory assessment of immediate-type hypersensitivity. The Journal of allergy and clinical immunology. 2010;125:S284–296. doi: 10.1016/j.jaci.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just TH2 cells. Nat Rev Immunol. 2010;10:838–848. doi: 10.1038/nri2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palomares O, Yaman G, Azkur AK, Akkoc T, Akdis M, Akdis CA. Role of Treg in immune regulation of allergic diseases. European Journal of Immunology. 2010;40:1232–1240. doi: 10.1002/eji.200940045. [DOI] [PubMed] [Google Scholar]
- 11.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci U S A. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui H-H, Frahm N, Brander C, Peters B, Grey H, Sette A. A Quantitative Analysis of the Variables Affecting the Repertoire of T Cell Specificities Recognized after Vaccinia Virus Infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 14.Botten J, Alexander J, Pasquetto V, Sidney J, Barrowman P, Ting J, Peters B, Southwood S, Stewart B, Rodriguez-Carreno MP, Mothe B, Whitton JL, Sette A, Buchmeier MJ. Identification of protective Lassa virus epitopes that are restricted by HLA-A2. J Virol. 2006;80:8351–8361. doi: 10.1128/JVI.00896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotturi MF, Botten J, Sidney J, Bui H-H, Giancola L, Maybeno M, Babin J, Oseroff C, Pasquetto V, Greenbaum JA, Peters B, Ting J, Do D, Vang L, Alexander J, Grey H, Buchmeier MJ, Sette A. A Multivalent and Cross-Protective Vaccine Strategy against Arenaviruses Associated with Human Disease. PLoS Pathog. 2009;5:e1000695. doi: 10.1371/journal.ppat.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mothe BR, Stewart BS, Oseroff C, Bui H-H, Stogiera S, Garcia Z, Dow C, Rodriguez-Carreno MP, Kotturi M, Pasquetto V, Botten J, Crotty S, Janssen E, Buchmeier MJ, Sette A. Chronic Lymphocytic Choriomeningitis Virus Infection Actively Down-Regulates CD4+ T Cell Responses Directed against a Broad Range of Epitopes. J Immunol. 2007;179:1058–1067. doi: 10.4049/jimmunol.179.2.1058. [DOI] [PubMed] [Google Scholar]
- 17.Moutaftsi M, Bui H-H, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, Grey H, Sette A. Vaccinia Virus-Specific CD4+ T Cell Responses Target a Set of Antigens Largely Distinct from Those Targeted by CD8+ T Cell Responses. J Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 18.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 19.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oseroff C, Peters B, Pasquetto V, Moutaftsi M, Sidney J, Panchanathan V, Tscharke DC, Maillere B, Grey H, Sette A. Dissociation between Epitope Hierarchy and Immunoprevalence in CD8 Responses to Vaccinia Virus Western Reserve. J Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assarsson E, Bui H-H, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. Immunomic Analysis of the Repertoire of T-Cell Specificities for Influenza A Virus in Humans. J. Virol. 2008;82:12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blythe M, Zhang Q, Vaughan K, de Castro R, Salimi N, Bui H-H, Lewinsohn D, Ernst J, Peters B, Sette A. An analysis of the epitope knowledge related to Mycobacteria. Immunome Research. 2007;3:10. doi: 10.1186/1745-7580-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan K, Blythe M, Greenbaum J, Zhang Q, Peters B, Doolan DL, Sette A. Meta-analysis of immune epitope data for all Plasmodia: overview and applications for malarial immunobiology and vaccine-related issues. Parasite Immunol. 2009;31:78–97. doi: 10.1111/j.1365-3024.2008.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughan K, Greenbaum J, Blythe M, Peters B, Sette A. Meta-analysis of all immune epitope data in the Flavivirus genus: inventory of current immune epitope data status in the context of virus immunity and immunopathology. Viral Immunol. 2010;23:259–284. doi: 10.1089/vim.2010.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarebski LM, Vaughan K, Sidney J, Peters B, Grey H, Janda KD, Casadevall A, Sette A. Analysis of epitope information related to Bacillus anthracis and Clostridium botulinum. Expert Rev Vaccines. 2008;7:55–74. doi: 10.1586/14760584.7.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidney J, Southwood S, Oseroff C, Del Guercio MF, Sette A, Grey H. Current Protocols in Immunology. John Wiley & Sons, Inc.; 1998. Measurement of MHC/Peptide Interactions by Gel Filtration. pp. 18.13.11–18.13.19. [DOI] [PubMed] [Google Scholar]
- 29.Gulukota K, Sidney J, Sette A, DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 31.Kotturi M, Botten J, Maybeno M, Sidney J, Glenn J, Bui H-H, Oseroff C, Crotty S, Peters B, Grey H, Altmann D, Buchmeier M, Sette A. Polyfunctional CD4+ T cell responses to a set of pathogenic arenaviruses provide broad population coverage. Immunome Research. 2010;6:4. doi: 10.1186/1745-7580-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, Kolla RV, De Silva AD, de Silva AM, Grey H, Peters B, Shresta S, Sette A. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. Journal of immunology. 2011;187:4268–4279. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arlehamn CS, Sidney J, Henderson R, Greenbaum JA, James EA, Moutaftsi M, Coler R, McKinney DM, Park D, Taplitz R, Kwok WW, Grey H, Peters B, Sette A. Dissecting Mechanisms of Immunodominance to the Common Tuberculosis Antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ). Journal of immunology. 2012 doi: 10.4049/jimmunol.1103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotturi M, Peters B, Buendia-Laysa F, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier M, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, Oseroff C, Tscharke DC, Irvine K, Bennink JR, Peters B, Southwood S, Cerundolo V, Grey H, Yewdell JW, Sette A. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J Immunol. 2005;175:5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- 37.Wilson CC, Palmer B, Southwood S, Sidney J, Higashimoto Y, Appella E, Chesnut R, Sette A, Livingston BD. Identification and Antigenicity of Broadly Cross-Reactive and Conserved Human Immunodeficiency Virus Type 1-Derived Helper T-Lymphocyte Epitopes. J. Virol. 2001;75:4195–4207. doi: 10.1128/JVI.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotturi MF, Swann JA, Peters B, Arlehamn CL, Sidney J, Kolla RV, James EA, Akondy RS, Ahmed R, Kwok WW, Buchmeier MJ, Sette A. Human CD8(+) and CD4(+) T cell memory to lymphocytic choriomeningitis virus infection. Journal of virology. 2011;85:11770–11780. doi: 10.1128/JVI.05477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. Journal of immunology. 2011;187:3111–3120. doi: 10.4049/jimmunol.1101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic Acids Res. 2009;38:D854–862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol. 2010;185:4189–4198. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. 2010;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaughan K, Greenbaum J, Kim Y, Vita R, Chung J, Peters B, Broide D, Goodman R, Grey H, Sette A. Towards defining molecular determinants recognized by adaptive immunity in allergic disease: an inventory of the available data. J Allergy (Cairo) 2010;2010:628026. doi: 10.1155/2010/628026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Middleton D, Menchaca L, Rood H, Komerofsky R. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. New allele frequency database: http://www.allelefrequencies.net. [DOI] [PubMed] [Google Scholar]
- 46.Meyer D, Singe R, Mack S, Lancaster A, Nelson M, Erlich H, Frenandez-Vina M, Thomson G. Single Locus Polymorphism of Classical HLA Genes.. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference; Seattle, WA. 2007. pp. 653–704. [Google Scholar]
- 47.Borger P, Ten Hacken NH, Vellenga E, Kauffman HF, Postma DS. Peripheral blood T lymphocytes from asthmatic patients are primed for enhanced expression of interleukin (IL)-4 and IL-5 mRNA: associations with lung function and serum IgE. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 1999;29:772–779. doi: 10.1046/j.1365-2222.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- 48.Wurtzen PA, van Neerven RJ, Arnved J, Ipsen H, Sparholt SH. Dissection of the grass allergen-specific immune response in patients with allergies and control subjects: T-cell proliferation in patients does not correlate with specific serum IgE and skin reactivity. The Journal of allergy and clinical immunology. 1998;101:241–249. doi: 10.1016/S0091-6749(98)70389-6. [DOI] [PubMed] [Google Scholar]
- 49.Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.