Abstract
Studies of lipid rafts, ordered microdomains of sphingolipids and cholesterol within cell membranes, are essential in probing the relationships between membrane organization and cellular function. While in vitro studies of lipid phase separation are commonly performed using spherical vesicles as model membranes, the utility of these models is limited by a number of factors. Here we present a microfluidic device that supports simultaneous electrical measurements and confocal imaging of on-chip bilayer lipid membranes (BLMs), enabling real-time multi-domain imaging of membrane organization. The chips further support closed microfluidic access to both sides of the membrane, allowing the membrane boundary conditions to be rapidly changed and providing a mechanism for dynamically adjusting membrane curvature through application of a transmembrane pressure gradient. Here we demonstrate the platform through the study of dynamic generation and dissolution of ordered lipid domains as membrane components are transported to and from the supporting annulus containing solvated lipids and cholesterol.
Introduction
There is growing recognition that, rather than behaving merely as inert and homogeneous boundaries for cell organelles or platform for membrane proteins, cell membranes possess rich heterogeneities including asymmetric leaflets,1, 2 lipid caveolaes,3–6 and lateral dynamic nanoscale organizations7–10 that play central roles in cellular function. In particular, various membrane components, including sphingsolipids and cholesterol, segregate into microdomains, termed lipid rafts, by intermolecular interactions including van der Waals interactions between fully saturated acyl chains and hydrogen bonding between adjacent glycosyl moieties of glycosphingolipids.7 Membrane proteins can be either incorporated in or excluded from lipid rafts based on their physical properties. Rafts in the exoplasmic leaflet of the plasma membrane serve as a platform for controlling interactions with the cell’s fluidic surroundings, playing important roles in protein sorting,11–13 ion channel regulation,14 membrane traffic,15 and cell signaling.16–19 Therefore, understanding the biophysical properties and biological roles of lipid rafts may provide critical insights toward cell function and disease progression.
Because lipid rafts in biological cells are small domains on the order of 10–200 nm,27 they are below the diffraction limit for light microscopy, preventing direct visualization using standard optical methods. As a result, alternative approaches are needed to more fully elucidate the characteristics of lipid rafts. Detergent extraction of lipid raft components from cells has proven to be a valuable technique for evaluating raft-associated molecules following the exposure of cells to various inputs, but this approach does not provide direct observation of the lipid microdomains and may be subject to artifacts that limit its ability to accurately assess lipid raft composition.28 To avoid these limitations, in vitro model bilayer membranes produced using predetermined lipid mixtures are invaluable for elucidating the mechanisms of lateral heterogeneity in cell membranes. In particular, giant unilamellar vesicles (GUVs) on the order of 10 ~ 100 μm in diameter generated by electroformation29, 30 are commonly used for exploring the behaviors and interactions of ordered lipid domains.31 The use of GUVs enables lateral phase separation into liquid disordered (Ld) and liquid ordered (Lo) domains with micrometer length scales that can be directly observed by confocal microscopy. Like lipid rafts, the Lo domains are rich in cholesterol and lipids with high melting temperature lipids such as sphingolipids and 1,2-saturated phospholipids, while the Ld domains mimic the disordered phase of cell membranes which are primarily composed of phospholipids with at least one unsaturated acyle chain. Phase behavior can be directly observed by 3D reconstruction with z-slicing using confocal microscopy.31–39
Despite the benefits of GUVs, there are several important drawbacks that limit their utility for lipid domain studies. Because vesicles are closed elements, with no direct fluidic access to the inner compartment of the vesicle, the chemical or biochemical composition can only be readily changed at the outer membrane leaflet. Similarly, isolation of the inner vesicle compartment also prevents effective electrical characterization of the membrane, which would otherwise offer an orthogonal measurement dimension to reveal details of average membrane structure accompanying domain changes that cannot be monitored solely by optical methods. Furthermore, changing the conditions at the outer compartment is a slow diffusive process, making it impossible to evaluate dynamic processes with time constants faster than several minutes. Even when investigating the relationships between static boundary conditions and lipid raft behaviors and interactions, GUVs require laborious experimental efforts to adjust the concentrations of selected solvents and solutes, limiting the density of data that may be extracted from these studies. In addition, GUV surface tension is defined by the vesicle radius, which is difficult to reliably control during vesicle synthesis and cannot be adjusted dynamically. More generally, GUVs are by definition closed equilibrium systems, very different from cell membranes in which lipids are dynamically transported between the plasma membrane and intracellular compartments.40 Finally, since optical imaging of the spherical vesicles requires three-dimensional scanning microscopy, the frame rate limitations can complicate studies of dynamic phase behaviors such domain formation, coalescence, and dissolution. As an alternative to vesicles as model membranes, planar supported lipid bilayers enable full-field imaging of lipid microdomains including access by atomic force microscopy.31, 41, 42 However, because supported lipid bilayers provide direct fluidic access on only one side of the membrane, they suffer from many of the same issues as GUVs. In addition, the behaviors of lipid microdomains within supported bilayers do not accurately reflect the dynamics within biological cells since the supporting substrate impacts diffusion kinetics of both lipids and proteins by an order of magnitude or more.43 Solid supports also interfere sterically with conformational changes in proteins, and artificially impact the flip-flop dynamics of lipids and membrane-bound components across the membrane leaflets.
In contrast to both GUVs and solid-supported membranes, annularly-supported BLMs represent a promising membrane model for lipid domain imaging that is compatible with a variety of microfluidic technologies.44–50 BLMs are formed across an aperture fabricated in a thin hydrophobic film which separates two buffer chambers. Unlike both GUVs and supported membranes, BLMs provide direct fluidic and electrical access to both membrane leaflets. In addition, horizontal BLM systems compatible with direct microscope observation have been employed to optically and electrically study physics of lipids51, 52 and ion channels53–55 simultaneously. It is well known that BLMs also present certain limitations, particularly in terms of relatively low membrane stability and challenges in achieving true solvent-free bilayers. Despite these disadvantages, BLMs offer unique opportunities as a model membrane system for lipid domain studies. Previous efforts toward the characterization of lipid phase separation using macro-scale BLM chambers included real time observation of domain merger, dissolution of lipid domains under elevated temperature,56 and simultaneous electrical and optical measurements.51 Liquid ordered domains have also been formed using an asymmetric planar bilayer system,57 offering utility for observing phase behavior mimicking rafts in the exoplasmic leaflet of cell membranes.
A universal challenge of model membranes as an alternative to native cell membrane studies lies in the variable lipid composition inherent to both vesicles and BLMs. Although the composition of GUVs can be estimated, for example by matching the phase diagram for binary phospholipid mixtures58 or comparing the bending elasticity of membranes reconstituted from erythrocyte lipid extracts59 such characterization methods are indirect. Indeed, it remains a key challenge to form a multi-component GUV and directly determine its lipid content.30 While the lipid composition in BLMs is also uncertain, it can be directly quantified by a mercury droplet extraction method,60, 61 an approach which has been used to confirm that lipid content in the BLM can differ from the bulk annulus and may change over the lifetime of BLM. However, due to the experimental effort required to perform these various analysis, both indirect and direct methods for determining lipid content of model membranes are seldom employed. Despite this limitation of established model membrane systems including both GUVs31, 62 and BLMs51, 57, these models are routinely employed to study phase behavior of multi-component lipid mixtures without explicit knowledge of the lipid composition by labelling domains with phase-segregating dyes. With a range of fluorescence dyes known to segregate into either Lo or Ld phases,63 local lipid structure can be qualitatively determined, enabling the evaluation of key membrane characteristics including domain morphology,64 diffusion behaviour,65 phase diagrams,66 and inter-leaflet domain induction57.
Here we report the extension of a miniaturized and integrated BLM platform50 enabling precise control over membrane curvature and fluidic boundary conditions toward studies of lipid phase separation. The thermoplastic microfluidic chips support the in situ formation of annularly-supported planar BLMs that can be monitored by full-field confocal epifluorescent microscopy, allowing direct imaging of lipid microdomains together with the simultaneous characterization of transmembrane impedance for correlated electrical and optical membrane observations. The microfluidic system supports the rapid control of static and dynamic (bio)chemical conditions in either side of membrane via direct perfusion, and also supports controllable modulation of membrane curvature, enabling the impact of surface tension on lipid-lipid and lipid-protein interactions to be readily explored. Collectively, these capabilities serve to extend the available model membrane toolkit toward new studies of both transient and equilibrium-state lipid domain dynamics and interactions.
Materials and Methods
Reagents
Isobutanol, hexadecane, potassium chloride, piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), N-palmitoyl-D-erythro-sphingosylphosphorylcholine (PSM), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). Texas Red labeled 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-DHPE) was purchased from Invitrogen (Carlsbad, CA). Poly(methyl methacrylate) (PMMA) sheets and Polyvinylidene chloride (PVDC) film were procured from US Plastics (Lima, Ohio) and Sheffield Plastics (Sheffield, MA), respectively. Aqueous buffer solutions were prepared from 0.25M KCl and 20mM PIPES (pH 6.9).
Lipid Solutions
Membranes formed with POPC/PSM/chol were prepared from a solution of 5 mg/mL POPC, 2.5 mg/mL PSM, 2.5 mg/mL cholesterol (molar ratio 2:1:2) and 0.05 mg/mL TR-DHPE. The DPPC/DPhPC/chol membranes were prepared from a solution of 4.3 mg/mL DPhPC, 3.7 mg/mL DPPC and 2.0 mg/mL cholesterol (molar ratio 1:1:1) with the addition of 0.05 mg/mL TR-DHPE.
Microfluidic system fabrication
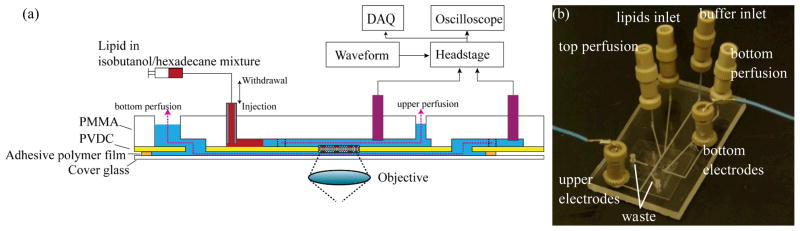
The electro-optical microfluidic BLM chips were fabricated from a hybrid glass/thermoplastic multilayer system. Microchannels (460 μm wide, 150 μm deep) were patterned in a 2.38 mm thick PMMA chip by direct computer numerical control (CNC) micromachining to create a microfluidic network supporting in situ lipid membrane formation and reagent perfusion. The PMMA chip was then thermally bonded to a 12 μm thick PVDC film containing a 50~100 μm diameter aperture fabricated using a thermal ablation method.50 To render the chip compatible with high-magnification confocal optical imaging, a 100 μm thick patterned adhesive polymer layer was patterned on a glass cover slip with average thickness of 110 μm, forming a lower microchannel beneath the membrane site, and bonded to the back side of the PVDC layer. The total distance between the lipid membrane to the bottom of the cover slip averages 210 μm, which enables high magnification confocal imaging of the membrane. Ag/AgCl electrodes were sealed into reservoirs connecting to both the upper and lower perfusion microchannels using a previously-reported process,50 enabling simultaneous monitoring of transmembrane current. A cross-sectional schematic and photograph of a chip fabricated by this process are shown in Figure 1. Estimates of transmembrane pressure within this system were established by setting known buffer flow rates through one of the perfusion channels, with the resulting pressure difference across the membrane defined by the product of the volumetric flow rate and estimated hydrodynamic resistance of the microchannel connecting the membrane site with the downstream waste reservoir. Large and balanced reservoirs at each channel outlet minimized the contributions of water column height and capillarity on the overall pressure gradient.
Fig. 1.
(a) Schematic of a microfluidic BLM chip supporting simultaneous electrical measurements and confocal optical imaging together with active perfusion to either side of the bilayer. (b) A fabricated BLM chip. The chip is 6.0 cm long and 2.5 cm wide.
Optical and electrical interfacing
Optical measurements were performed by placing the BLM chip on the imaging stage of a Leica SP5 X confocal microscope. Standard objectives (20X/0.7 NA Plan Apo multi-immersion, 63X/1.3 NA Plan Apo glycerin) were used to provide imaging with up to 60 nm pixel resolution. A laser line at 595 nm was used for excitation with a photomultiplier tube (9624s; Hamamatsu Photonics, Japan) used for fluorescence detection. Frame rates up to 25 frames per second via resonance scanning at 512 by 512 resolution were readily achieved. Video imaging was performed at frame rates ranging from 0.76–1.32 Hz to improve contrast. Leica AF Lite software was used to measure circular domain diameter while irregularly shaped domains were analyzed using ImageJ software. Transmembrane current was converted to a voltage signal by a custom amplifier and monitored using a data acquisition system including a 60 Hz noise eliminator (Hum Bug; AutoMate Scientific, CA), filter (LPF-202A, fc=10 kHz; Warner Instruments, CT) and digitizer (sampling at 250KHz, Digidata 1440A; Axon, CA). A syringe pump (11 Elite; Harvard Apparatus, MA) with both infusion and withdrawal capability was used to deliver lipid and buffer solutions to the microchip via stainless steel needle interfaces.67 All data were acquired at a temperature of 27 °C.
Results and Discussion
In-situ membrane formation
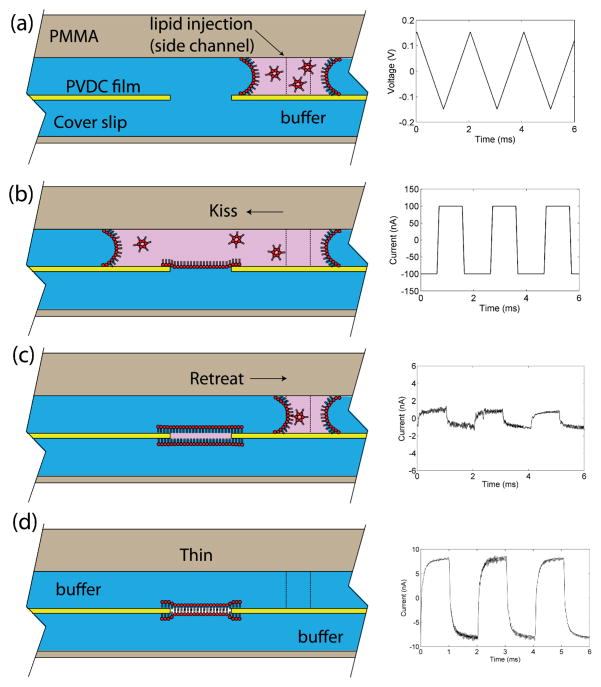
A novel technique termed “kiss and retreat” was developed for reliable and semi-automated in situ membrane formation (Figure 2). Briefly, lipid mixture is dissolved in solvent consisting of 1:4 v/v isobutanol and hexadecane. After the microchip is filled with buffer, the lipid mixture is delivered by syringe pump towards the BLM aperture at flow rates between 0.1–0.5 μl/min until the lipid front reaches the PVDC aperture (Figure 2a,b). Immediately after passing the aperture, as determined by optical observation or electrical monitoring of current through the aperture, the lipid solution is withdrawn back towards syringe, leaving a plug of lipid solution that remains attached to the rim of the aperture (Figure 2c). Isobutanol within the remaining lipid plug diffuses into the surrounding aqueous solution, thinning the lipids into a bilayer supported by a solvent-containing annulus (Figure 2d). If a bilayer fails to form, the inject/withdraw process is simply repeated until a stable bilayer is formed, as confirmed by electrical characterization of the membrane’s specific capacitance using the integrated Ag/AgCl electrodes. Compared to our previously-reported “diffusion painting” method48 this technique reliably produces on-chip bilayer membranes with a minimum of human operation. Moreover, like our previous demonstration of in situ BLM formation by two-sided lipid/solvent perfusion,68 the kiss and retreat technique is compatible with a closed chip format enabling controllable buffer exchange to both sides of the membrane, but with a significantly higher success rate above 90% for creating a robust BLM. Using this method, both dynamic and steady-state solution conditions at both sides of the membrane can be freely changed by rapid perfusion. Moreover, because the fluidic passages are only open at waste reservoirs distal from the membrane site, it is possible to control the transmembrane pressure by adjusting the relative perfusion flow rates across the upper and lower membrane compartments, allowing membrane curvature to be modified while maintaining the desired (bio)chemical boundary conditions.
Fig. 2.
Schematic of the kiss and retreat bilayer formation process and corresponding trans-membrane current traces measured with the integrated chip electrodes. (a) Lipid solution in isobutanol/hexadecane is injected from a side channel. (b) Lipid solution sweeps across the aperture, leaving a monolayer at the interface. (c) Lipid solution is withdrawn to the side channel, leaving a plug of lipid solution at the aperture. (d) As the lipid solution plug thins by solvent diffusion, a bilayer lipid membrane forms across the aperture. Electrical measurements were performed using a triangle wave with 150 mV amplitude and 500 Hz frequency. The stray capacitance of the chip was measured as 3.33 pF. The 60 μm diameter BLM was formed across an 80 μm diameter aperture. The final measured current corresponds to a membrane capacitance of 26.7 pF (0.94 μF/cm2) confirming the formation of a bilayer. The sealing resistance of BLM ranged from 1.5 – 2.5 GΩ.
Domain imaging and characterization
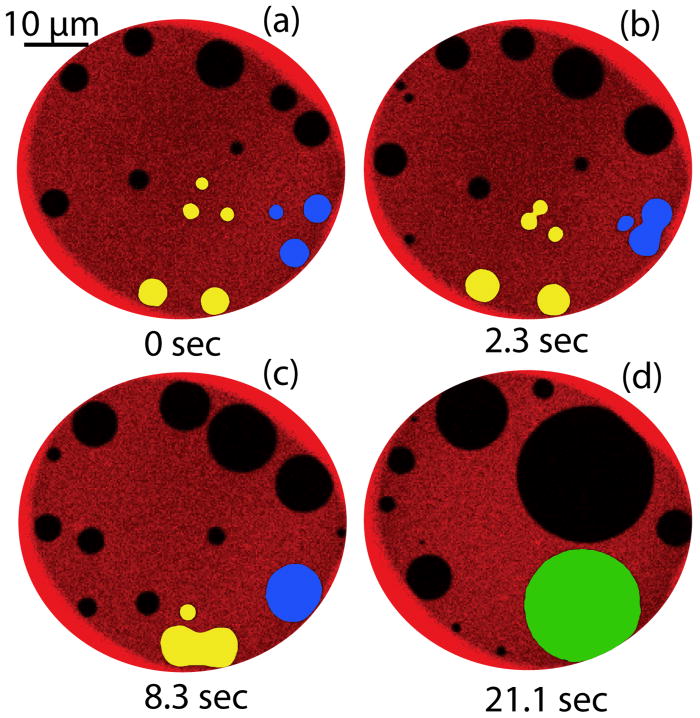
TR-DHPE has been widely employed to image the co-existence of lipid phases in solid-supported and GUV model membranes. In particular, membranes formed with DPPC/DPhPC/chol,69 POPC/PSM/chol, and DOPC/PSM/chol63, 70 exhibit highly efficient partition of dye to the liquid disordered phase. Similar dynamical evolution of coexisting Lo and Ld domains in both DPPC/DPhPC/chol and POPC/PSM/chol have been observed with TR-DHPE in this system. Figure 3 shows images at four sequential time points in a membrane formed by DPPC/DPhPC/chol. Selected domains (shaded in the figure) are tracked to reveal the dynamic movement and coalescence of discrete Lo domains using this ternary lipid system. While DPPC/DPhPC/chol mixtures can be readily used for on-chip membrane formation and phase separation experiments, other membrane compositions are of interest to provide greater biological relevance. Specifically, the remainder of the work described in this paper is focused on POPC/PSM/chol. POPC is a 1-saturated, 2-unsaturated PC, representing a large portion of naturally occurring PC.71 Sphingomeylin is the most abundant sphingolipid in plasma membranes, but has a rather complex thermotropic behavior. PSM was chosen for this study as it exhibits simpler thermotropic behavior but possesses a melting temperature very close to natural SM extract.66
Fig. 3.
The merger of ordered domains (dark regions) formed by DPPC/chol within a disordered phase composed primarily of DPhPC, with TR-labeled DHPE preferentially segregated to the disordered phase. Two sets of domains are presented in pseudo-color (yellow and blue) to highlight the evolution of multiple merger events leading to the formation of a single ~15 μm Lo domain (green).
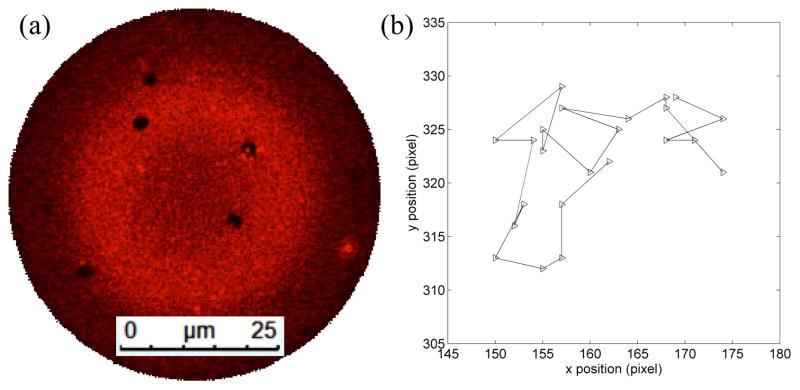
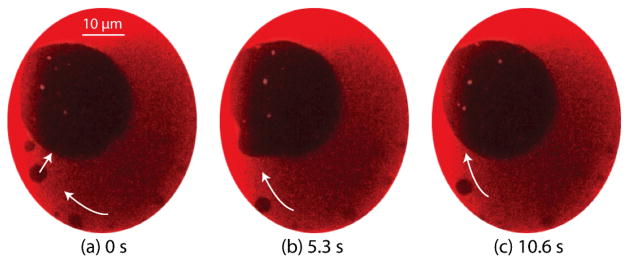
One characteristic of the Lo domains that is readily probed in the system is their behavior under free diffusion. Figure 4 shows the diffusion trajectory of a single Lo domain under the application of 0.96 Pa transmembrane pressure generated by buffer perfusion at 300 nL/min. Under these conditions, multiple domains with diameters of several microns were found within the center of the bilayer (Figure 4a). The domains were highly stable and remained detached from the supporting annulus, enabling the Brownian motion of the ordered structures to be observed over long time scales. An average diffusion coefficient of 0.125 μm2/s for ~1.5 μm diameter domains was determined in this experiment. In comparison, diffusion coefficients ranging from 0.09 ~ 0.20 μm2/s have been reported for Lo domains in GUVs using a different lipid composition (DOPC/DPPC/CHOL) with a lower cholesterol concentration in the initial lipid mixture.65
Fig. 4.
(a) Stable small ~1.5 μm diameter ordered domains in a membrane formed with molar ratio 2:1:2 POPC/DPPC/chol mixture. (b) Trajectory of a single Lo domain with 3.22 s between each time step.
Dynamic domain generation during membrane stabilization
In the case of GUV model membranes, lipid domain imaging is performed only after the vesicles are formed and fully stabilized. As a result, existing studies of lipid domains have generally focused on evaluating steady-state relationships such as lipid phase space rather than kinetic phenomena. In contrast, because our on-chip BLMs are formed in situ after positioning the chip on the microscope stage, the microfluidic system allows the full dynamic processes of ordered domain emergence, formation, and interaction during the early stages of membrane stabilization to be observed. An example revealing the typical behavior of these non-equilibrium processes in a POPC/PSM/chol membrane is shown in Figure 5 and SI-1.† Immediately after membrane formation across a 60 μm diameter aperture, Lo domains were spontaneously generated at the right side of the bilayer and observed to migrate along the interface of the bilayer and surrounding solvated lipid annulus to the left side, where a large Lo domain had formed immediately at the initial stage of bilayer formation. The large domain continued to expand as it absorbed incoming small domains until the composition of the bilayer reached equilibrium. In a typical experiment, Lo domain generation stops within 2–3 min after bilayer formation, with no further changes in membrane organization over an equivalent time period confirming equilibrium. Although different membranes exhibit different initial domain patterns at the moment of formation, the lipid exchange process involving dynamic domain generation and coalescence into a single large stable domain is consistently observed. While small domains can float freely within the bilayer, larger domains tend to attach to the surrounding annulus. This behavior can be interpreted as a measure to minimize the energy state by lowering the contacting circumference length between liquid ordered and disordered domains.
Fig. 5.
Growth of a large Lo domain in the absence of transmembrane pressure for a POPC/PSM/chol membrane. Ordered domains (dark regions) are generated from the right side of bilayer and driven to the pre-existing large domain, revealing that the composition of bilayer during the first 2–3 min following membrane formation is not in equilibrium with the surrounding annulus. Full video of the bilayer stabilization process can be found in SI-1.
Annulus-supported BLMs have been employed to study membrane biophysics for decades, with static steady-state partitioning of lipids between the bilayer and surrounding annulus routinely assumed. However, the composition the bilayer can differ from the initial lipid solution due to the dynamic partitioning process during the initial stages of membrane formation,60, 61 as well as dynamic changes that occur following membrane formation.61 Here we present direct observations of lipid exchange between the BLM and surrounding annulus in the early stages of membrane formation, manifested by lipid phase separation and domain stabilization, which we believe are the first reported visualizations of these dynamic processes. The microfluidic platform may further serve as a fluorescence-based alternative to the mercury droplet extraction method60, 61 for qualitatively studying membrane composition during these dynamic processes, and may be of particular utility toward examining membranes reconstituted from complex lipid mixture and natural lipid extracts.
Effect of transmembrane pressure on domain stability
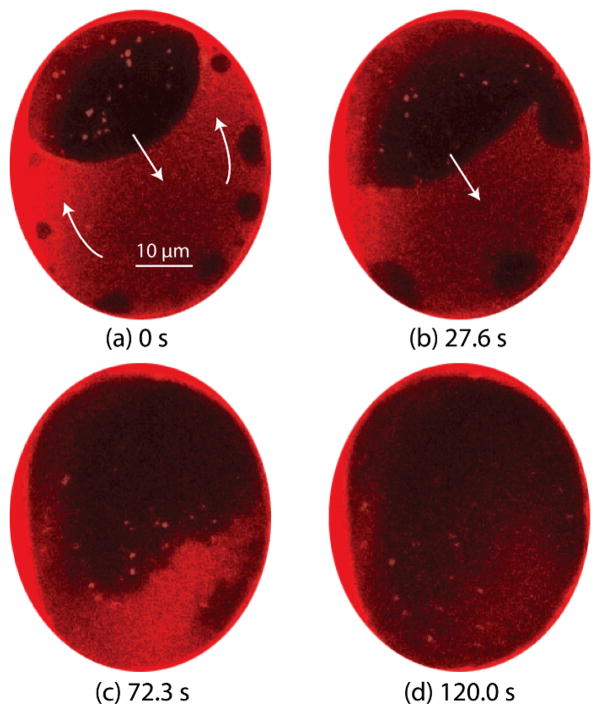
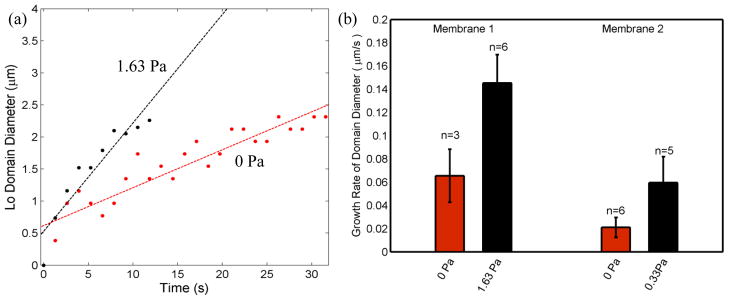
Surface tension directly affects the ordering of lipid membranes, and its impact on lipid domains is commonly studied in GUVs by micropipette aspiration or osmotic swelling72, 73 to control vesicle surface tension. Previous reports have revealed that small domains tend to grow and combine into larger domains as surface tension is increased. This observation can be interpreted as a thermodynamic effect, with increasing lateral tension caused by expansion of the vesicle surface resulting in increased line tension between adjacent lipid phases, encouraging a reduction in ordered domain size to minimize enthalpy. Unlike vesicle membranes, where surface tension of the closed system responds immediately to an osmotic pressure gradient, pressure-induced bulging of a BLM is a kinetic process.52, 74 As membrane curvature increases, lipids migrate dynamically from the annulus to the bilayer to maintain lower surface tension, thereby expanding the Plateau-Gibbs border that defines the edge of the annulus. This rate-dependent lipid transport process has previously been leveraged to enable high pulsatile perfusion rates while avoiding membrane rupture.50 In the present study, an estimated transmembrane pressure gradient of 1.63 Pa was defined by applying a constant 500 nL/min buffer flow rate to the microchannel positioned above a POPC/PSM/chol bilayer with an average diameter of 60 μm. Immediately upon perfusion, new Lo domains began to nucleate and grow adjacent to the annulus, followed by migration around the periphery of the membrane toward a large domain which had formed during initial membrane stabilization. Figure 6a shows a typical observation of this process, including the transport of lipids from the annulus to the bilayer over time. In this example, perfusion was initiated several minutes after complete stabilization of the membrane, so that all observed domain formation and migration was due solely to energy minimization under the influence of the applied pressure gradient. The generation of new Lo domains under pressure exhibits distinct characteristics compared to the membrane stabilization stage. Comparing the two cases, Lo domains are generated from 4–5x more nucleation sites with the application of transmembrane pressure. The maximum size of generated domains under pressure appears to be limited only by collisions with adjacent domains, while during zero-pressure stabilization the domains do not grow beyond a plateau of around 4 μm in diameter. In addition, the growth rate of individual domains under transmembrane pressure is significantly higher than the growth rate of domains during stabilization (Figure 7).
Fig. 6.
A POPC/PSM/chol membrane exhibiting clear phase separation experienced a topological change when 1.63 Pa pressure was applied across the membrane. (a) Domains generated at the bottom of the image migrate and merge with a large domain created during initial membrane formation. (b-c) As perfusion continues, the boundary between ordered and disordered domains becomes increasingly indistinct, until (d) the Lo and Ld regions become fully miscible. Full video of domain behaviors during this experiment can be found in SI-2.
Fig. 7.
(a) The growth of selected domains during initial membrane stabilization in the absence of external pressure (0 Pa) and applied transmembrane pressure (1.63 Pa). (b) Average domain growth rates extracted from multiple domains under different pressure states for two different membranes. The diameters of individual ordered domains were measured from nucleation at the bilayer boundary through departure from the boundary or merger with other domains. Membrane 1 has an area of ~2400 μm2, while membrane 2 has area of ~4400 μm2.
Following the initial growth and coalescence of Lo domains, continued application of the 1.63 Pa transmembrane pressure led to the boundaries between ordered and disordered phases becoming indistinct, until the discrete ordered domains ultimately dispersed, resulting in uniform fluorescence intensity across the bilayer (Figure 6c,d). The change from well-defined circular boundaries to irregular shapes, and finally phase miscibility, reflects a reduction in the contribution of line tension to the overall energy of the system. The reduction in line tension at the phase boundaries is presumably due to increasing surface tension in the bilayer that occurs when lipid migration from the annulus can no longer compensate for increasing membrane curvature. The higher surface tension provides larger space for the free motion of individual molecules within the bilayer, such that POPC, PSM and cholesterol can assume preferable interactions with water and lipid molecules without segregating to different phases. This observation is similar to the behavior of a Langmuir monolayer exposed to a change in surface pressure, where the increased space available to each lipid allows the monolayer to undergo a transition from Lo to Ld states.75 Since Lo domains maintain their circular shape in presence of high line tension to lower energy, reduced contribution of line tension to the overall enthalpy of the system would lead to reduced roundness of the domain shape. The process of Lo domain distortion, dispersion, and dissolution has been consistently observed over repeated experiments, together with the recovery of partially-dispersed Lo domains following removal of the applied pressure gradient, with recovery times on the order of several tens of seconds. A video showing the full process of pressure-induced domain generation, migration, and dispersion over the course of a typical experiment is provided in SI-2.†
Conclusions
Unlike GUVs and supported bilayers, planar BLMs better represent the native environment of biological membranes, with fluidic access to both membrane compartments, dynamic lipid exchange between the membrane and supporting annulus, and a free-standing configuration that does not impede the motion of lipid components within the membrane. The present work describes a microfluidic BLM system that supports in situ bilayer membrane formation with nearly perfect success rates, simultaneous electro-optical measurements of the bilayer, and rapid independent control over (bio)chemical boundary conditions at both sides of the membrane. To demonstrate the utility of the microfluidic system, phase-segregating dyes were introduced to the lipid solutions and used to successfully observe lipid phase separation in two different ternary lipid compositions, with high resolution confocal imaging of the resulting Lo domains. The chips have enabled, for the first time, direct optical observations of the initial lipid exchange between the bilayer and supporting annulus immediately after membrane formation, and full-field video of on-chip membranes have revealed details of the movements of Lo domains that provide a new view of energy minimization processes during membrane stabilization. Control of transmembrane pressure has been used to show that Lo domain generation and dissolution dynamics depend directly on membrane surface tension, and may allow future experiments to elucidate surface tension dependent phase diagrams for different lipid compositions or explore the response of mechanosensitive ion channels. By applying a moderate transmembrane pressure, the formation of small stable domains has also been demonstrated, allowing the diffusion coefficients of Lo domains undergoing free Brownian motion to be measured. Overall, the microfluidic-enabled platform represents a powerful and convenient model membrane system that can significantly extend the scope of biophysical and biochemical studies of lipid membranes, lipid rafts, and ultimately their interactions with membrane-bound proteins and exogenous agents delivered to the membrane by dynamic perfusion.
Supplementary Material
Acknowledgments
This research was supported by NIH grants R21EB009485 and R21EB011750, and by NSF grant CBET0966407. Helpful discussion with Dr. Marco Colombini in the Department of Biology is gratefully acknowledged. We also thank Amy Beaven with the University of Maryland Imaging Core Facility for help with confocal microscopy.
Footnotes
Electronic Supplementary Information (ESI) available:
SI-1: video of bilayer formation and stabilization process (http://mml.umd.edu/si/SI-1.mov).
SI-2: video revealing Lo domain growth and dissolution under application of 1.63 Pa transmembrane pressure (http://mml.umd.edu/si/SI-2.mov). See DOI: 10.1039/b000000x/
References
- 1.van Meer G. Annu Rev Cell Biol. 1989;5:247–275. doi: 10.1146/annurev.cb.05.110189.001335. [DOI] [PubMed] [Google Scholar]
- 2.Bretscher MS. Science. 1973;181:622–629. doi: 10.1126/science.181.4100.622. [DOI] [PubMed] [Google Scholar]
- 3.Parton RG. Curr Opin Cell Biol. 1996;8:542–548. doi: 10.1016/s0955-0674(96)80033-0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson RGW. Annual Review of Biochemistry. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 5.Parton RG, Simons K. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 6.Chidlow JH, Jr, Sessa WC. Cardiovasc Res. 2010;86:219–225. doi: 10.1093/cvr/cvq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 8.Brown DA, London E. Annual Review of Cell and Developmental Biology. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 9.Rietveld A, Simons K. Biochimica Et Biophysica Acta-Reviews on Biomembranes. 1998;1376:467–479. doi: 10.1016/s0304-4157(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 10.Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 11.Mellman I, Nelson WJ. Nature Reviews Molecular Cell Biology. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simons K, Lingwood D. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 13.Stockl M, Nikolaus J, Herrmann A. Methods Mol Biol. 2010;606:115–126. doi: 10.1007/978-1-60761-447-0_10. [DOI] [PubMed] [Google Scholar]
- 14.Dart C. J Physiol. 2010;588:3169–3178. doi: 10.1113/jphysiol.2010.191585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanzal-Bayer MF, Hancock JF. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Anderson RGW. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:10909–10913. doi: 10.1073/pnas.90.23.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simons K, Toomre D. Nature Reviews Molecular Cell Biology. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 18.Quest AFG, Gutierrez-Pajares JL, Torres VA. Journal of Cellular and Molecular Medicine. 2008;12:1130–1150. doi: 10.1111/j.1582-4934.2008.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel HH, Murray F, Insel PA. Annual Review of Pharmacology and Toxicology. 2008;48:359–391. doi: 10.1146/annurev.pharmtox.48.121506.124841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons K, Ehehalt R. Journal of Clinical Investigation. 2002;110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fittipaldi A, Ferrari A, Zoppe M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. Journal of Biological Chemistry. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 22.Toborek M, Zhong Y, Hennig B. Journal of Cerebral Blood Flow and Metabolism. 2010;30:522–533. doi: 10.1038/jcbfm.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakai Y, Kamiguchi H. Journal of Cell Biology. 2002;159:1097–1108. doi: 10.1083/jcb.200209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeaney NK, He M, Tang NF, Malouf AT, O’Riordan MA, Lemmon V, Bearer CF. Journal of Neurochemistry. 2009;110:779–790. doi: 10.1111/j.1471-4159.2009.06143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawarabayashi T, Shoji M, Younkin LH, Lin WL, Dickson DW, Murakami T, Matsubara E, Abe K, Ashe KH, Younkin SG. Journal of Neuroscience. 2004;24:3801–3809. doi: 10.1523/JNEUROSCI.5543-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rushworth JV, Hooper NM. International Journal of Alzheimer’s Disease. 2011;2011:14. doi: 10.4061/2011/603052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pike LJ. Journal of Lipid Research. 2008;50:S323. doi: 10.1194/jlr.R800040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown DA, Shogomori H. Biological Chemistry. 2003;384:1259–1263. doi: 10.1515/BC.2003.139. [DOI] [PubMed] [Google Scholar]
- 29.Angelova MI, Dimitrov DS. Faraday Discussions. 1986;81:303. [Google Scholar]
- 30.Meleard P, Bagatolli LA, Pott T. Methods in Enzymology Liposomes, Pt G. 2009;465:161–176. doi: 10.1016/S0076-6879(09)65009-6. [DOI] [PubMed] [Google Scholar]
- 31.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haverstick DM, Glaser M. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:4475–4479. doi: 10.1073/pnas.84.13.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veatch SL, Keller SL. Phys Rev Lett. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 34.Nag K, Pao JS, Harbottle RR, Possmayer F, Petersen NO, Bagatolli LA. Biophys J. 2002;82:2041–2051. doi: 10.1016/S0006-3495(02)75552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veatch SL, Keller SL. Biophys J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahya N, Scherfeld D, Bacia K, Poolman B, Schwille P. Journal of Biological Chemistry. 2003;278:28109–28115. doi: 10.1074/jbc.M302969200. [DOI] [PubMed] [Google Scholar]
- 37.Bernardino de la Serna J, Perez-Gil J, Simonsen AC, Bagatolli LA. Journal of Biological Chemistry. 2004;279:40715–40722. doi: 10.1074/jbc.M404648200. [DOI] [PubMed] [Google Scholar]
- 38.Veatch SL, Keller SL. Phys Rev Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 39.Klose C, Ejsing CS, Garcia-Saez AJ, Kaiser HJ, Sampaio JL, Surma MA, Shevchenko A, Schwille P, Simons K. Journal of Biological Chemistry. 2010;285:30224–30232. doi: 10.1074/jbc.M110.123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samsonoc AV, Mihakyav I, Cohen FS. Biophysics Journal. 2001;81:1486–1500. [Google Scholar]
- 41.Tokumasu F, Jin AJ, Feigenson GW, Dvorak JA. Biophys J. 2003;84:2609–2618. doi: 10.1016/S0006-3495(03)75066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin WC, Blanchette CD, Ratto TV, Longo ML. Biophys J. 2006;90:228–237. doi: 10.1529/biophysj.105.067066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonnleitner A, Schutz GJ, Schmidt T. Biophys J. 1999;77:2638–2642. doi: 10.1016/S0006-3495(99)77097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki H, Tabata K, Kato-Yamada Y, Noji H, Takeuchi S. Lab Chip. 2004;4:502–505. doi: 10.1039/b405967k. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki H, Tabata KV, Noji H, Takeuchi S. Biosens Bioelectron. 2007;22:1111–1115. doi: 10.1016/j.bios.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Zagnoni M, Sandison ME, Marius P, Lee AG, Morgan H. Lab Chip. 2007;7:1176–1183. doi: 10.1039/b703818f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandison ME, Zagnoni M, Morgan H. Langmuir. 2007;23:8277–8284. doi: 10.1021/la7007528. [DOI] [PubMed] [Google Scholar]
- 48.Hromada LP, Nablo BJ, Kasianowicz JJ, Gaitan MA, DeVoe DL. Lab Chip. 2008;8:602–608. doi: 10.1039/b716388f. [DOI] [PubMed] [Google Scholar]
- 49.Baaken G, Sondermann M, Schlemmer C, Ruhe J, Behrends JC. Lab Chip. 2008;8:938–944. doi: 10.1039/b800431e. [DOI] [PubMed] [Google Scholar]
- 50.Shao CR, Sun B, Colombini M, DeVoe DL. Annals of Biomedical Engineering. 2011;39:2242–2251. doi: 10.1007/s10439-011-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honigmann A, Walter C, Erdmann F, Eggeling C, Wagner R. Biophys J. 2010;98:2886–2894. doi: 10.1016/j.bpj.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picard G, Denicourt N, Fendler JH. J Phys Chem-Us. 1991;95:3705–3715. [Google Scholar]
- 53.Ichikawa T, Aoki T, Takeuchi Y, Yanagida T, Ide T. Langmuir. 2006;22:6302–6307. doi: 10.1021/la0535025. [DOI] [PubMed] [Google Scholar]
- 54.Borisenko V, Lougheed T, Hesse J, Fureder-Kitzmuller E, Fertig N, Behrends JC, Woolley GA, Schutz GJ. Biophys J. 2003;84:612–622. doi: 10.1016/S0006-3495(03)74881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harms G, Orr G, Lu HP. Appl Phys Lett. 2004;84:1792–1794. [Google Scholar]
- 56.Samsonov AV, Mihalyov I, Cohen FS. Biophys J. 2001;81:1486–1500. doi: 10.1016/S0006-3495(01)75803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins MD, Keller SL. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:124–128. doi: 10.1073/pnas.0702970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fidorra M, Hartel S, Ipsen JH, Bagatolli LA. Do GUVs composed of binary lipid mixtures obey the lever rule? A quantitative microscopy imaging approach. 2008 [Google Scholar]
- 59.Meleard P, Gerbeaud C, Pott T, Fernandez Puente L, Bivas I, Mitov MD, Dufourcq J, Bothorel P. Biophys J. 1997;72:2616–2629. doi: 10.1016/S0006-3495(97)78905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pagano RE, Ruysscha Jm, Miller IR. J Membrane Biol. 1972;10:11–30. doi: 10.1007/BF01867845. [DOI] [PubMed] [Google Scholar]
- 61.Bunce AS, Hider RC. Biochim Biophys Acta. 1974;363:423–427. doi: 10.1016/0005-2736(74)90081-9. [DOI] [PubMed] [Google Scholar]
- 62.Bagatolli LA. Biochimica Et Biophysica Acta-Biomembranes. 2006;1758:1541–1556. doi: 10.1016/j.bbamem.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 63.Baumgart T, Hunt G, Farkas ER, Webb WW, Feigenson GW. Biochimica Et Biophysica Acta-Biomembranes. 2007;1768:2182–2194. doi: 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bagatolli LA, Gratton E. Biophys J. 2000;79:434–447. doi: 10.1016/S0006-3495(00)76305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cicuta P, Keller SL, Veatch SL. J Phys Chem B. 2007;111:3328–3331. doi: 10.1021/jp0702088. [DOI] [PubMed] [Google Scholar]
- 66.de Almeida RF, Fedorov A, Prieto M. Biophys J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CF, Liu J, Hromada LP, Tsao CW, Chang CC, DeVoe DL. Lab Chip. 2009;9:50–55. doi: 10.1039/b812812j. [DOI] [PubMed] [Google Scholar]
- 68.Hromada L, Kasianowicz JJ, Gaitan MA, Millard R, DeVoe DL. presented in part at the Miniaturized Systems for Chemistry and Life Sciences; Tokyo, Japan. Nov. 5–9, 2006. [Google Scholar]
- 69.Veatch SL, Gawrisch K, Keller SL. Biophys J. 2006;90:4428–4436. doi: 10.1529/biophysj.105.080283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veatch SL, Keller SL. Biochim Biophys Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Zachowski A. Biochemical Journal. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ayuyan AG, Cohen FS. Biophys J. 2008;94:2654–2666. doi: 10.1529/biophysj.107.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baumgart T, Hess ST, Webb WW. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 74.Tien HT. J Phys Chem-Us. 1967;71:3395. doi: 10.1021/j100870a006. [DOI] [PubMed] [Google Scholar]
- 75.Kaganer VM, Mohwald H, Dutta P. Reviews of Modern Physics. 1999;71:779–819. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.