Abstract
Δ9-Tetrahydrocannabinol (THC) and other cannabinoids are responsible for the psychoactive and medicinal properties of Cannabis sativa L. (marijuana). The first intermediate in the cannabinoid biosynthetic pathway is proposed to be olivetolic acid (OA), an alkylresorcinolic acid that forms the polyketide nucleus of the cannabinoids. OA has been postulated to be synthesized by a type III polyketide synthase (PKS) enzyme, but so far type III PKSs from cannabis have been shown to produce catalytic byproducts instead of OA. We analyzed the transcriptome of glandular trichomes from female cannabis flowers, which are the primary site of cannabinoid biosynthesis, and searched for polyketide cyclase-like enzymes that could assist in OA cyclization. Here, we show that a type III PKS (tetraketide synthase) from cannabis trichomes requires the presence of a polyketide cyclase enzyme, olivetolic acid cyclase (OAC), which catalyzes a C2–C7 intramolecular aldol condensation with carboxylate retention to form OA. OAC is a dimeric α+β barrel (DABB) protein that is structurally similar to polyketide cyclases from Streptomyces species. OAC transcript is present at high levels in glandular trichomes, an expression profile that parallels other cannabinoid pathway enzymes. Our identification of OAC both clarifies the cannabinoid pathway and demonstrates unexpected evolutionary parallels between polyketide biosynthesis in plants and bacteria. In addition, the widespread occurrence of DABB proteins in plants suggests that polyketide cyclases may play an overlooked role in generating plant chemical diversity.
Keywords: natural products, phytocannabinoid, terpenophenolic, aldolase, ferredoxin-like
Humans have used Cannabis sativa L. (marijuana, hemp; Cannabaceae) as a medicinal and psychoactive herbal drug for at least 2,500 y (1), and today it is the most widely consumed illicit drug worldwide (2). Its unique effects are due to the presence of cannabinoids, which include Δ9-tetrahydrocannabinol (THC) and more than 70 related metabolites (3). THC is responsible for the characteristic intoxication of marijuana and exhibits diverse pharmacological properties including analgesia, antiemesis, and appetite stimulation (4, 5). Medical marijuana and cannabinoid drugs are increasingly used to treat a range of diseases and conditions such as multiple sclerosis and chronic pain (6).
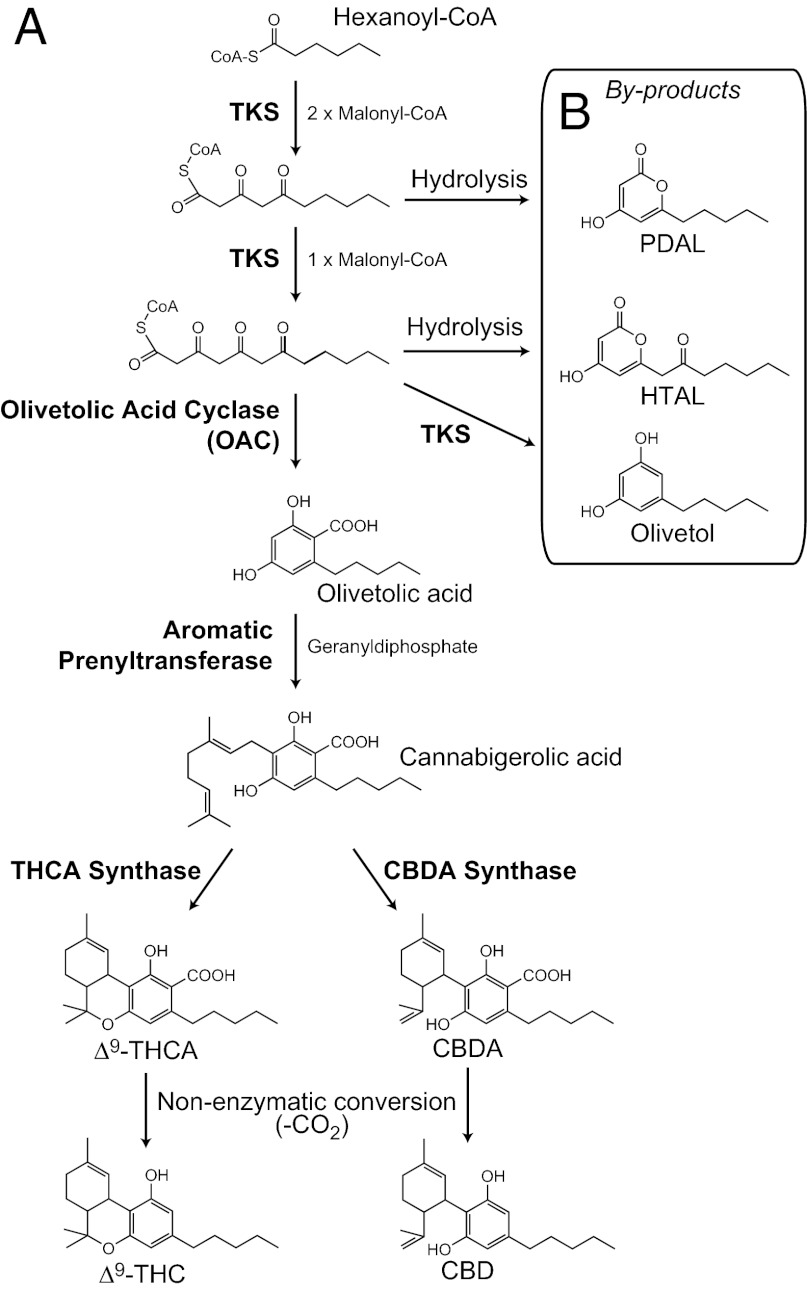
The biosynthesis of cannabinoids (Fig. 1), which are prenylated polyketides derived from fatty acid and isoprenoid precursors, is not completely understood at the molecular level. The first enzyme in the cannabinoid pathway is proposed to be a type III polyketide synthase (PKS) that catalyzes the condensation of hexanoyl-CoA with three molecules of malonyl-CoA to yield olivetolic acid (OA). This C2→C7 aldol cyclization reaction is noteworthy for its retention of the carboxylate moiety, which is rare in plant polyketides. OA is geranylated to form cannabigerolic acid (CBGA) (7), which is converted by oxidocyclase enzymes to the major cannabinoids, Δ9-tetrahydrocannabinolic acid (THCA) and cannabidiolic acid (CBDA) (8, 9). THCA and CBDA undergo nonenzymatic decarboxylation to their neutral forms, THC and cannabidiol (CBD), respectively.
Fig. 1.
The proposed cannabinoid biosynthetic pathway. (A) The pathway leading to the major cannabinoids Δ9-tetrahydrocannabinolic acid (THCA) and cannabidiolic acid (CBDA), which decarboxylate to yield Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD), respectively. (B) Recombinant TKS enzyme produces triketide (PDAL) and tetraketide (HTAL and olivetol) by-products in vitro.
Several groups have attempted to identify the PKS that synthesizes OA, but the enzymatic basis for this reaction remains unclear (10, 11). Taura et al. (10) assayed a type III PKS cloned from cannabis leaves and found that it produces olivetol and α-pyrones [pentyl diacetic acid lactone (PDAL) and hexanoyl triacetic acid lactone (HTAL)] but not OA (Fig. 1B). The formation of olivetol by this enzyme is puzzling because the requirement for acidic substrates by subsequent enzymes and the occurrence of cannabinoid acids in planta indicates that OA is the key pathway intermediate (7, 12). We renamed the “olivetol synthase” PKS as tetraketide synthase (TKS) to more accurately reflect its putative role in the cannabinoid pathway. HTAL and PDAL have not previously been identified in cannabis. α-Pyrones can form as aberrant products when the reactive poly-β-keto backbone produced by a PKS undergoes lactonization; e.g., bisnoryangonin (a triketide) and coumaroyl triacetic acid lactone (a tetraketide) are by-products of the type III PKS chalcone synthase (CHS) with p-coumaroyl-CoA (13). α-Pyrones are also produced by bacterial type II PKSs when polyketide cyclase enzymes essential for final cyclization reactions are absent. For example, the Streptomyces tetracenomycin PKS yields α-pyrones if the TcmN ARO/CYC cyclase is not present (14).
We hypothesized that the inability of TKS to synthesize OA was due to the absence of an accessory protein, such as a polyketide cyclase enzyme, which functions in polyketide assembly. Here, we use transcriptome analysis of cannabis trichome cells and biochemical assays of candidate proteins to identify olivetolic acid cyclase (OAC), which functions in concert with TKS to form OA. OAC is a dimeric α+β barrel (DABB) protein that is structurally similar to DABB-type polyketide cyclase enzymes from Streptomyces and to stress-responsive proteins in plants. The identification of OAC reveals a unique biosynthetic route to plant polyketides in which cyclases function cooperatively with type III PKSs to generate carbon scaffolds.
Results and Discussion
Identification of Polyketide Cyclase Candidates in the Trichome Transcriptome.
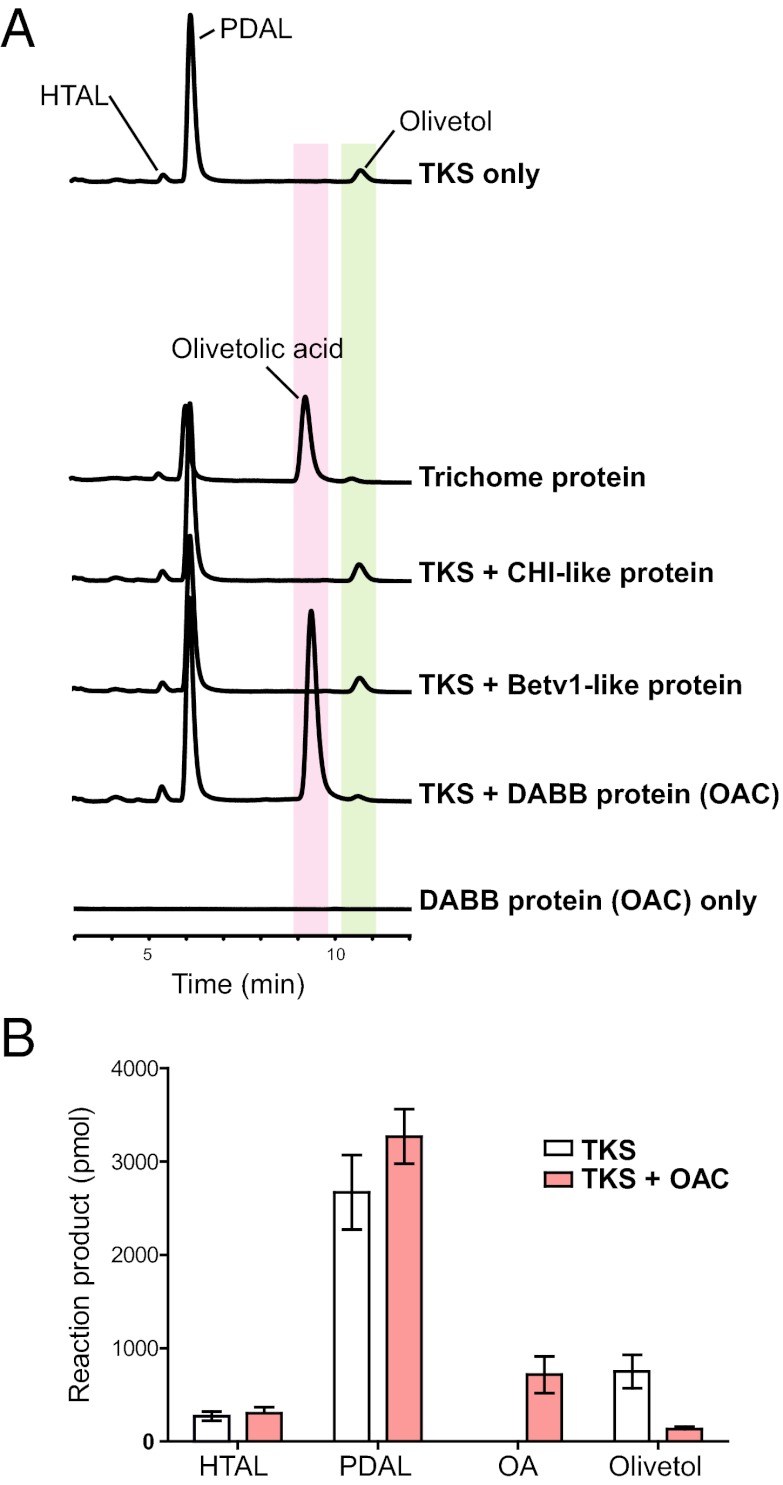
Cannabinoid biosynthesis occurs primarily in glandular trichomes that develop on female flowers and, to a lesser extent, leaves. We extracted proteins from trichome secretory cells isolated from a hemp cultivar of cannabis and tested their ability to catalyze the formation of OA from hexanoyl-CoA and malonyl-CoA. Crude trichome protein formed OA in addition to PDAL, HTAL, and olivetol (Fig. 2A). Comparing this result with the inability of the recombinant TKS to synthesize OA, we hypothesized that an OA-forming accessory protein is present in trichomes. To identify this protein, we analyzed a trichome-specific EST catalog from a potent marijuana strain of cannabis that contains a high number of ESTs corresponding to cannabinoid biosynthetic enzymes (e.g., TKS and THCA synthase) (15).
Fig. 2.
OA formation requires the presence of the DABB protein, olivetolic acid cyclase (OAC). (A) TKS produces the by-products PDAL, HTAL, and olivetol, but crude protein from hemp trichomes catalyzes OA formation. Assays of TKS together with polyketide cyclase candidate proteins shows OA is produced by the DABB protein OAC but not by Betv1-like and CHI-like proteins. (B) Comparison of polyketide product profiles in TKS assays performed with and without OAC (mean ± SD, n = 3). Reaction products in polyketide synthesis assays were analyzed by HPLC and identified by comparison with authentic standards (Fig. S6).
We reasoned that an OA-forming enzyme may be prominently represented in the trichome EST dataset and, therefore, searched for proteins with sequence or structural similarity to polyketide cyclases. This approach identified three candidates with high expression levels as determined by EST counts (Table 1). A chalcone isomerase (CHI)-like protein was selected based on the catalytic relationship of CHS with CHI (16, 17), which suggested that the cannabis CHI-like protein could partner with TKS to form OA, a possibility also discussed by Taura et al. (10). The second candidate was a member of the dimeric α+β barrel (DABB) protein superfamily with similarity to stress-responsive proteins from plants (18–20). The presence of this protein was intriguing because DABB proteins act as polyketide cyclases (e.g., TcmI cyclase) in Streptomyces species (21), although the bacterial cyclases show low sequence similarity to plant DABB proteins. The third candidate was a Betv1-like protein in the same protein family as the Streptomyces TcmN ARO/CYC polyketide cyclase (22). Several Betv1-protein family members function as enzymes in plant natural product biosynthesis (23). High numbers of ESTs corresponding to the DABB protein (9 ESTs) and the Betv1-like protein (6 ESTs) were found in the cannabis trichome EST dataset reported by Marks et al. (11).
Table 1.
Candidate polyketide cyclases identified in cannabis trichome EST dataset
| Protein name | No. of ESTs | Protein family, Pfam no. | Arabidopsis match*, accession no. | Identity, % |
| CHI-like protein | 51 | Chalcone-flavanone isomerase family, pfam02431 | Chalcone-flavanone isomerase family protein, At3g63170 | 31 |
| DABB protein | 42 | Stress responsive dimeric α+β barrel (DABB) domain family, pfam07876 | Heat stable protein 1 (AtHS1), At3g17210 | 48 |
| Betv1-like protein | 23 | Pathogenesis-related protein Betv1 family, pfam00407 | MLP-like protein (MLP423), At1g24020 | 66 |
*Blastx comparison with Arabidopsis proteins (TAIR10).
Trichome-Expressed DABB Protein Forms OA.
We expressed the three cyclase candidates in Escherichia coli and tested the purified proteins separately in polyketide synthesis assays containing recombinant TKS, hexanoyl-CoA, and components for malonyl-CoA synthesis [malonyl-CoA synthetase (MCS), malonate, CoA and ATP]. MCS was used to produce malonyl-CoA in situ as has been reported for in vitro assays of type II PKSs (24). OA was present in assays containing the DABB protein but not with the Betv1-like or CHI-like proteins (Fig. 2A). This small protein (12 kDa, 101 amino acids), which we named olivetolic acid cyclase (OAC), had no intrinsic PKS activity, and only produces OA in the presence of TKS. OAC did not convert HTAL to OA, indicating that ring opening of a HTAL is not occurring. Production of the α-pyrones was similar whether OAC was present, but olivetol formation decreased in assays containing OAC (Fig. 2B).
Quantitative RT-PCR analysis of cannabis tissues and cell types from a hemp cultivar found that the highest transcript levels of OAC were in trichomes and, to a lesser extent, female flowers, which parallels the expression of the transcripts for TKS and CBDA synthase (Fig. S1). Using transient expression of fluorescent protein fusions in Nicotiana benthamiana leaves, TKS and OAC, which lack predicted signal peptides, were both localized to the cytoplasm (Fig. S1).
Functional analysis of OAC via RNAi was not possible because cannabis transformation has not been achieved and our attempts at virus-induced gene silencing were unsuccessful. To demonstrate OAC activity in vivo, we reconstituted OA biosynthesis in yeast. Yeast cultures expressing TKS and OAC fed sodium hexanoate produced 0.48 mg/L OA (mean, n = 3) and olivetol in the medium but no α-pyrones (Fig. S2). Surprisingly, we detected a trace amount of OA in the medium of cultures expressing TKS only. This result may be because yeast has some endogenous cyclase activity or the intracellular environment alters the catalytic properties of TKS.
OAC Does Not Physically Interact with TKS.
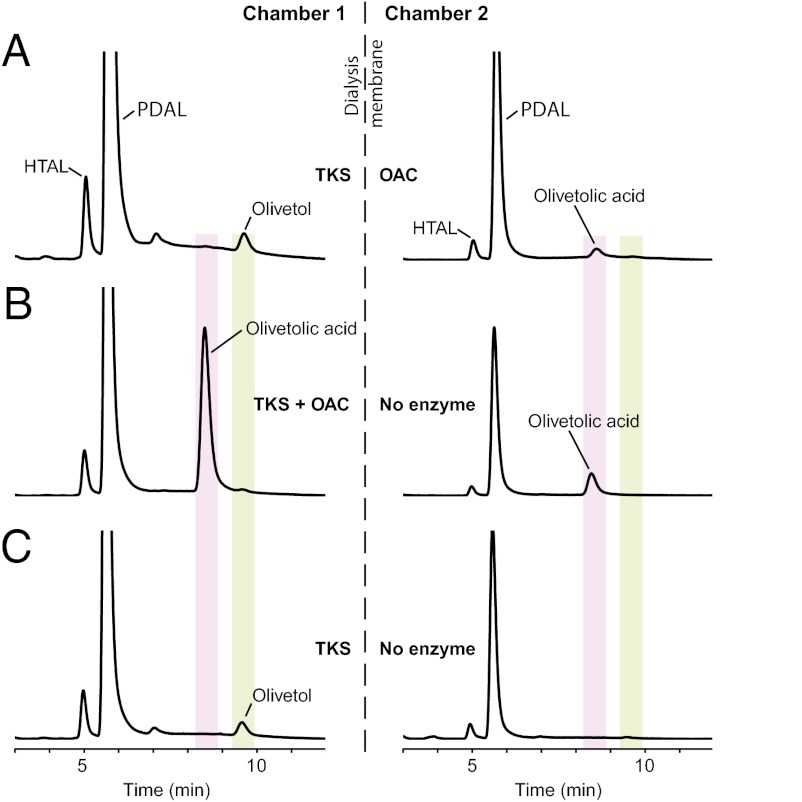
One explanation for the production of OA in reactions containing TKS and OAC is that OAC functions as an enzyme that acts on an intermediate produced by TKS. Alternatively, OAC may alter the catalytic properties of TKS through allosteric regulation, which allows TKS to itself form OA. To test the importance of physical interactions for OA formation, we separated TKS and OAC in 100-μl dialysis chambers by using a 5-kDa cutoff membrane that allowed substrates, intermediates, and reaction products to diffuse but not proteins. We also performed reactions in which one chamber contained both TKS and OAC (positive control) or TKS only (negative control) while the other chamber contained no enzyme. As shown in Fig. 3A, OA was formed in the OAC-containing chamber that was separated from TKS by the membrane. Large amounts of OA were formed in the positive control reaction containing TKS and OAC in the same dialysis chamber (Fig. 3B); OA was absent from the TKS only negative control (Fig. 3C). The reduced amount of OA formed when TKS and OAC were separated (Fig. 3A) compared with the positive control (Fig. 3B) may be due to the loss of the intermediate through conversion to olivetol before it reaches the OAC in chamber 2. In all three cases, there was diffusion of the reaction products from the TKS-containing chamber to the opposite side of the membrane during the 2-h assay.
Fig. 3.
Dialysis experiments show that physical interaction of TKS and OAC is not required for OA formation. Recombinant TKS and OAC were assayed in dialysis chambers separated by a 5-kDa cutoff membrane. (A) Assays with TKS and OAC in separate chambers resulted in the formation of HTAL, PDAL, and olivetol in the TKS-containing chamber 1 and HTAL, PDAL, and OA in the OAC-containing chamber 2. (B) Positive control assays with TKS and OAC in chamber 1 and no enzyme in chamber 2 produced large amounts of OA, in addition to HTAL and PDAL. (C) Negative control assays with TKS in chamber 1 and no enzyme in chamber 2 yielded only HTAL, PDAL, and olivetol. Chromatograms were extracted at 270 nm.
These results allow us to conclude that TKS synthesizes a diffusible intermediate that is converted to OA by OAC. We can exclude allosteric regulation of TKS because OA is produced when the two proteins are physically separated. We performed yeast two-hybrid analysis and found no evidence for the interaction of TKS and OAC (Fig. S3). It remains formally possible that OAC plays a chaperone-like role in guiding the folding of the tetraketide intermediate. We think it is more likely that OAC acts as an enzyme based on its structural similarities with bacterial DABB-type polyketide cyclases and the fact that aldol condensations in polyketide biosynthesis are enzyme catalyzed.
On the Nature of the OAC Substrate.
The novelty of the reaction catalyzed by OAC, and the reactivity of poly-β-keto intermediates produced by TKS, presents difficulties in determining its substrate. HTAL and PDAL production by TKS in the polyketide synthesis assays was similar whether OAC was present or not; however, OA formation was accompanied by a decrease in olivetol production (Fig. 2 A and B). We propose this pattern of products results from two co-occurring catalytic processes in the TKS-OAC coupled in vitro assay (Fig. 1): (i) hydrolytic release of poly-β-keto triketide and tetraketide intermediates from TKS, which are unacceptable substrates for OAC and undergo spontaneous lactonization to PDAL and HTAL, respectively; and (ii) TKS-catalyzed synthesis of a linear tetraketide-CoA intermediate, which is the substrate for OAC. In the absence of OAC, this intermediate undergoes aldol cyclization with decarboxylation to yield olivetol. Because HTAL, PDAL, and olivetol are in vitro products not known to be present in cannabis, it is likely that TKS only functions to produce the substrate for OAC in planta. The unstable nature of the putative tetraketide-CoA intermediate precludes its in vitro assay with OAC and its use in determining the kinetic parameters of OAC. We concluded that a comparison of the kinetics of TKS alone compared with the TKS-OAC coupled reaction, which may shed light on their respective catalytic roles, was complicated by the multiple products from TKS (PDAL, HTAL, olivetol, and the putative tetraketide-CoA intermediate). In addition, our uncertainty about the decarboxylation reaction that forms olivetol makes such experiments difficult to interpret. There are precedents for the release of CoA-linked intermediates in plant polyketide biosynthesis. A type III PKS from Curcuma longa forms a CoA-bound diketide that is transferred to a second type III PKS for further extension (25). Chalcone reductase has been postulated to act on a CoA-bound polyketide (26).
Structural and Mechanistic Analysis of OAC.
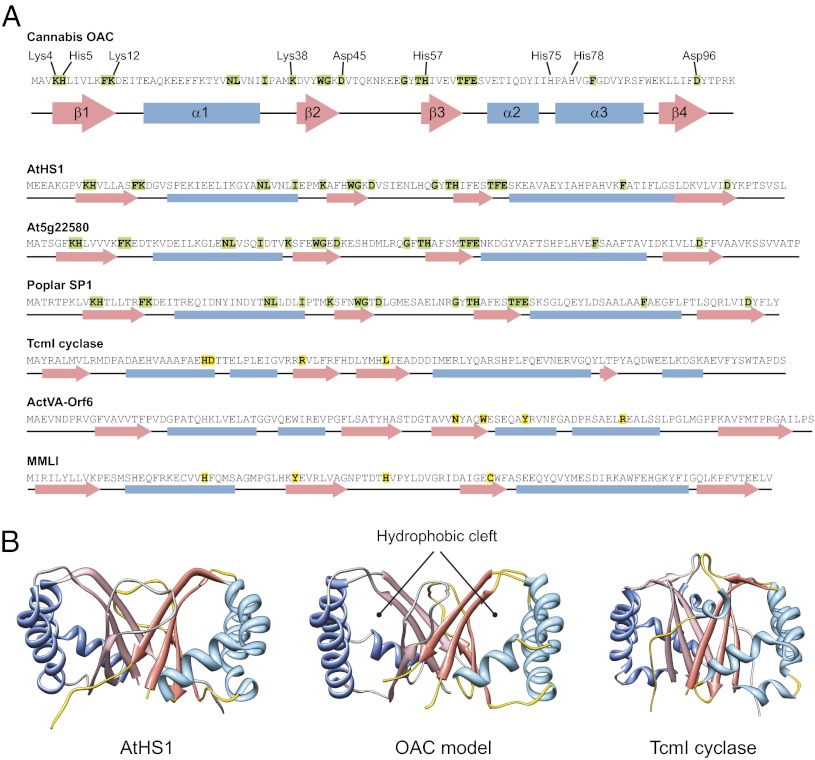
To gain insight into the mechanism by which OAC catalyzes OA synthesis through an intramolecular C2–C7 aldol condensation, we compared its structure with DABB proteins from bacteria and plants that have sequence or structural similarity to OAC. The structures of three plant stress-responsive DABB proteins are known [Arabidopsis heat stable 1 (AtHS1), the Arabidopsis At5g22580 gene product, and poplar stable protein 1 (SP1)] (27–29). The catalytic mechanisms of the bacterial DABB proteins TcmI cyclase, ActVA-Orf6 monooxygenase, and 4-methylmuconolactone methylisomerase (MLMI) have been investigated by using structural and biochemical approaches (21, 30, 31). These proteins possess a ferredoxin-like fold with an intermolecular β-barrel and a deep hydrophobic cleft in each monomer where the α2- and α3-helices arch over the β-sheets. This cleft forms the active site in TcmI, ActVA-Orf6, and MLMI and was suggested to be the putative active site in AtHS1 and the At5g22580 gene product.
OAC is predicted to have the characteristic β-α-β-β-α-α-β topology and possesses amino acid residues that are conserved in plant stress-responsive DABB proteins (Fig. 4A).We used homology modeling to generate the structure of OAC by comparison with AtHS1 (PDB ID code 1q53), which shares 48% identity with OAC. The OAC model exhibits the same overall structure as other DABBs (Fig. 4B), with a hydrophobic cleft that likely serves as the active site for the cyclization reaction.
Fig. 4.
Comparison of the OAC structure with other DABB proteins. (A) A schematic representation of the secondary structures of OAC and representative DABB proteins from plants and bacteria showing the characteristic β-α-β-β-α-α-β topology. Conserved residues in the plant proteins are indicated in bold with green background. Active-site residues in bacterial DABBs are indicated in bold with yellow background. The OAC residues targeted for site-directed mutagenesis are labeled. (B) Ribbon diagrams of AtHS1 (PDB ID code 1Q53), the homology model of OAC, and TcmI cyclase (PDB ID code 1TUW). The hydrophobic cleft formed between the α-helices and β-sheets in each monomer, which is the active site of TcmI cyclase, is present in all three proteins.
The catalytic mechanism and active-site residues involved in the OAC aldol condensation remain to be elucidated. OAC possesses three conserved lysines (Lys4, Lys12, Lys38; Fig. 4A) that could form Schiff bases in a type I aldolase reaction. However, Lys4Ala, Lys12Ala, and Lys38Ala mutants created by site-directed mutagenesis were active (Table S1). A type II aldolase mechanism involving a metal ion can be excluded because 10 mM EDTA was not inhibitory. A mechanism for OAC is suggested by the base-catalyzed aldol condensations of Streptomyces cyclases, which involve enolate intermediates (22, 32, 33). OAC has several conserved aspartate and histidine residues that could act as catalytic bases. Asp45Ala and Asp96Ala mutants were active, but single-residue mutants in which His5, His57, or His78 were replaced by Ala led to complete loss of activity; the His75Ala mutant had 1% of wild-type activity (Table S1). We are attempting to determine the structure of OAC to better understand its catalytic mechanism.
Evolution of OAC Function.
To investigate the evolution of OAC, we identified OAC homologs in diverse plant genomes by BLAST and keyword searching of the Phytozome database. We also identified DABB proteins in the cannabis genome and from hop (Humulus lupulus) ESTs (34). We included representatives from dicot and monocot lineages and the basal plants Selaginella moellendorffii and Physcomitrella patens. DABB proteins with sequence similarity to OAC are present in all of the plant genomes we analyzed including four DABB-encoding genes in Arabidopsis thaliana, 8 in cannabis, and 12 in Populus trichocarpa. In some cases, the OAC homologs code for proteins with a single DABB domain (e.g., OAC), whereas others have a duplicated DABB domain (e.g., AtDABB1, At1g51360). OAC homologs are also found in bacteria including Rhizobium leguminosarum and members of the enigmatic Planctomycetes-Verrucomicrobia-Chlamydieae superphylum. The function of these bacterial DABB proteins is unknown.
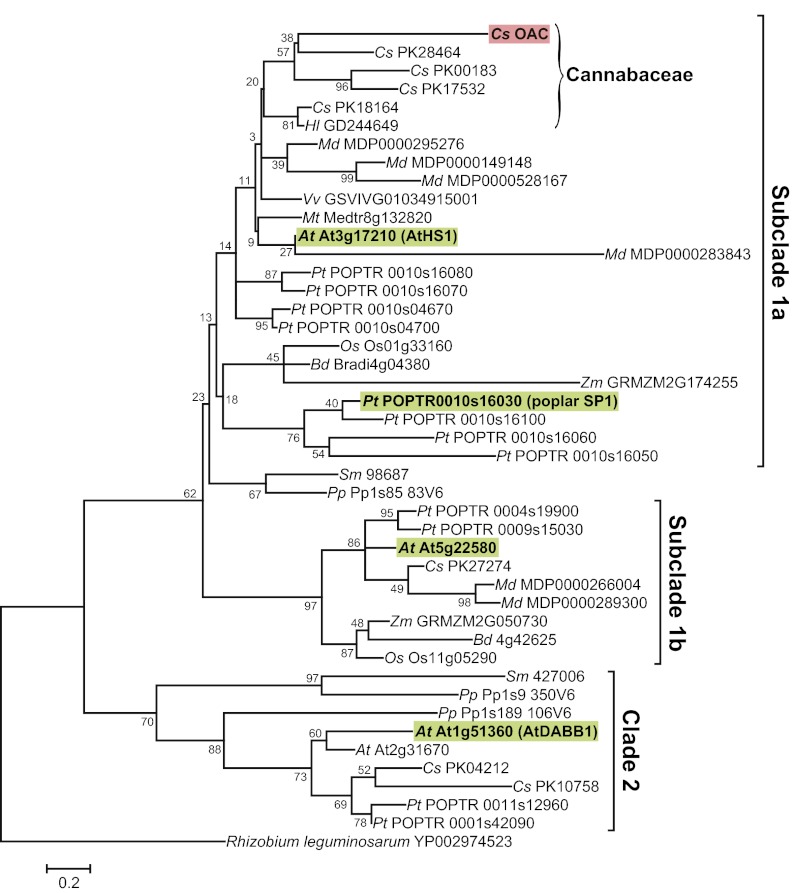
We constructed a phylogenetic tree of the DABB proteins by using the maximum-likelihood method (35), which we rooted on a single-domain DABB protein from R. leguminosarum (Fig. 5). A tree with similar topology was inferred by using the neighbor-joining method (Fig. S4). Phylogenetic analysis was made challenging by the small size of OAC and its homologs, most of which are ∼100 amino acids, and some of the nodes have low bootstrap values. The single-domain and double-domain DABB proteins formed separate clades, with the single-domain proteins further divided into subclades 1a and 1b. OAC was positioned in subclade 1a, where it clustered with a diverse group of the single-domain DABBs from dicots, monocots, and basal plants. In many cases, the bootstrap values in clade 1a were <50% and the relationships must therefore be considered tentative. However, it is clear that there has been an expansion of DABB proteins in Cannabaceae, with OAC grouped with four other cannabis proteins and the single hop protein. It is worth noting that the biosynthesis of the major polyketides in hop (e.g., humulone) by type III PKSs does not appear to require the involvement of a polyketide cyclase, and our analysis of hop trichome EST datasets did not find cDNAs corresponding to DABB proteins to be highly abundant. Plant stress-responsive DABB proteins such as AtHS1 and poplar SP1 are also present in subclade 1a. Most of the taxa that we used for our analysis were represented in clade 1b, with one gene from each of Arabidopsis, cannabis, corn, rice and Brachypodium, and two from apple and poplar. This result suggests its members may have a conserved function that is distinct from the proteins in subclade 1a. Clade 2 contained the double-domain DABB proteins such as AtDABB1 (At1g51360).
Fig. 5.
A phylogenetic tree of DABB proteins from plants inferred using the maximum-likelihood method. OAC and proteins that have been structurally or functionally characterized are highlighted. Branch lengths are proportional to the number of amino acid substitutions per site. The tree is rooted by a Rhizobium DABB protein. Species abbreviations: At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Cs, Cannabis sativa; Hl, Humulus lupulus; Md, Malus domestica; Mt, Medicago truncatula; Os, Oryza sativa; Pa, Physcomitrella patens; Pt, Populus trichocarpa; Rl, Rhizobium leguminosarum; Sm, Selaginella moellendorffii; Vv, Vitis vinifera; Zm, Zea mays. The details of the sequences are provided in Table S3.
The structural similarity of OAC with the bacterial DABB-type cyclases demonstrates that plants and bacteria exploit the same α+β barrel fold for polyketide cyclization. However, the low sequence similarity between OAC and the bacterial enzymes indicates that they are not homologous. Rather we suggest that OAC and the bacterial DABB-type cyclases are an example of convergent evolution with polyketide cyclizing activity arising independently in plants and bacteria. The ferredoxin-like fold is known to be suitable for ligand binding and as a structural framework for diverse catalytic functions (36), of which polyketide cyclization is but one. A likely evolutionary scenario is that OAC evolved from a plant DABB protein that was not involved in polyketide biosynthesis.
Role of Cyclase Enzymes in Plant Polyketide Pathways.
The current model of plant polyketide biosynthesis is that carbon scaffolds are assembled exclusively by the activity of type III PKS enzymes (37). The discovery of OAC indicates a variant catalytic route in plants in which cyclase enzymes function cooperatively with type III PKSs. It also demonstrates that polyketide cyclases, which until now were only known to partner with type II PKSs in bacterial polyketide pathways, are also found in biosynthetic pathways involving type III PKSs. Although cannabinoid biosynthesis may be unique in its requirement for a cyclase enzyme, we speculate that other plant DABB proteins may act as polyketide cyclases because some possess the hydrophobic cleft and conserved residues that we implicate in OAC function (Fig. 4 A and B). However, polyketide synthesis assays with TKS and recombinant AtHS1 show the latter possesses no OAC activity (Fig. S5), and gene expression databases show no evidence for interactions of the four Arabidopsis DABB proteins and their encoding genes with flavonoid/polyketide pathways (SI Methods and Materials). The role of such enzymes may be confined to the formation of polyketide products that retain a carboxylic acid moiety. Examples of plant metabolites formed by C2–C7 aldol condensation with carboxylate retention are the anacardic acids in cashew and gingko and stilbene carboxylates in Hydrangea species and liverworts (38, 39). It is worth noting that a rice type III PKS synthesizes long-chain alkylresorcinolic acids without the need for a cyclase enzyme (40). Another indication that some plant polyketide pathways may require cyclases is the formation of α-pyrones when recombinant type III PKSs enzymes are assayed in vitro e.g., Hypericum perforatum octaketide synthase (41).
Conclusion
The identification of OAC clarifies the polyketide phase of cannabinoid biosynthesis and provides an explanation for why the type III PKS (TKS) found in cannabis trichomes cannot produce OA on its own. Since THCA and CBDA synthases have been cloned (8, 9), the last step of the cannabinoid pathway to be cloned and characterized is the aromatic prenyltransferase enzyme that forms CBGA. The expanding interest in the cannabinoids as therapeutic agents suggests that metabolic engineering of the cannabinoid pathway in microorganisms may be worthwhile as a means to produce cannabinoids of high purity, and to make novel derivatives via combinatorial biosynthesis approaches. With the identification of OAC and our demonstration of the efficient OA synthesis in yeast (Fig. S2), the molecular tools for manipulating cannabinoid production are increasingly available.
Materials and Methods
Full details are provided in SI Materials and Methods.
Assay of Trichome Protein.
Trichome cells isolated from female flowers of the hemp cultivar ‘Finola’ using the Beadbeater method were homogenized in buffer, centrifuged to remove cell debris, desalted, and concentrated. Protein extracts were assayed for polyketide synthesis activity as described below.
Protein Expression and Assay.
TKS, OAC, Betv1-like, CHI-like, and MCS were amplified (primers in Table S2), cloned and expressed in E. coli, and proteins were purified with Talon resin (Clontech). Enzyme assays (50 μL) contained 20 mM Hepes at pH 7.0, 5 mM DTT, 0.2 mM hexanoyl-CoA, 12 μg of MCS, 0.2 mM CoA, 0.4 mM ATP, 2.5 mM MgCl2, 8 mM sodium malonate, TKS and either OAC, Betv1-like, or CHI-like. Reactions were incubated at 20 °C for 60 min, and products were analyzed by HPLC-photodiode array (PDA)/MS.
Dialysis Assay of TKS and OAC.
Assays were performed by using Fast Micro-Equilibrium Dialyzers with 100-μL chambers separated by a 5-kDa MWCO cellulose acetate membrane (Harvard Apparatus). Each chamber contained 20 mM Hepes at pH 7.0, 5 mM DTT, 200 μM hexanoyl-CoA, and 600 μM malonyl-CoA. Reactions consisted of TKS and OAC in separate chambers or together, or a TKS-only control. Reactions were incubated at 10 °C for 2 h, and products were analyzed by HPLC-PDA/MS.
Structural Analysis.
The homology structure of OAC was obtained from the 1q53 template by using comparative modeling with SWISS-MODEL (42). The OAC model had a QMEAN4 score of 0.45 (z-score of −3.25).
OAC Site-Directed Mutagenesis.
Single amino acid changes in OAC were introduced by gene synthesis (DNA 2.0) or, in some cases, using site-directed mutagenesis. The constructs were cloned directly into the plasmid vector pJExpress 411 (DNA 2.0). OAC mutants were expressed, purified, and assayed as above.
Phylogenetic Analysis.
A tree was inferred by the maximum likelihood method with a WAG matrix-based model (35) and 1,000 bootstrap replicates by using MEGA5 software (43).
Supplementary Material
Acknowledgments
We thank R. Taschuk, S. Polvi, and N. Theaker for technical assistance; S. Whitfield for assaying AtHS1; B. Haug for R. leguminosarum DNA; and P. Covello and M. Loewen for critical comments on the manuscript. This research was supported by funding from the Natural Sciences and Engineering Research Council of Canada and the National Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. OAC, JN679224; Betv1-like, JN679225; CHI-like, JN679226).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200330109/-/DCSupplemental.
References
- 1.Russo EB, et al. Phytochemical and genetic analyses of ancient cannabis from Central Asia. J Exp Bot. 2008;59:4171–4182. doi: 10.1093/jxb/ern260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.United Nations Office on Drugs and Crime 2010. World Drug Report 2010 (United Nations Office on Drugs and Crime, Vienna, Austria)
- 3.Elsohly MA, Slade D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005;78:539–548. doi: 10.1016/j.lfs.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Gaoni Y, Mechoulam R. Isolation, structure, and partial synthesis of an active constituent of hashish. J Am Chem Soc. 1964;86:1646–1647. [Google Scholar]
- 5.Joy JE, Watson SJ. In: Marijuana and Medicine: Assessing the Science Base. Benson JA, editor. National Academy, Washington, DC; 1999. [PubMed] [Google Scholar]
- 6.Ware MA, et al. Smoked cannabis for chronic neuropathic pain: A randomized controlled trial. CMAJ. 2010;182:E694–E701. doi: 10.1503/cmaj.091414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellermeier M, Zenk MH. Prenylation of olivetolate by a hemp transferase yields cannabigerolic acid, the precursor of tetrahydrocannabinol. FEBS Lett. 1998;427:283–285. doi: 10.1016/s0014-5793(98)00450-5. [DOI] [PubMed] [Google Scholar]
- 8.Sirikantaramas S, et al. The gene controlling marijuana psychoactivity: Molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J Biol Chem. 2004;279:39767–39774. doi: 10.1074/jbc.M403693200. [DOI] [PubMed] [Google Scholar]
- 9.Taura F, et al. Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett. 2007;581:2929–2934. doi: 10.1016/j.febslet.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 10.Taura F, et al. Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 2009;583:2061–2066. doi: 10.1016/j.febslet.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 11.Marks MD, et al. Identification of candidate genes affecting Δ9-tetrahydrocannabinol biosynthesis in Cannabis sativa. J Exp Bot. 2009;60:3715–3726. doi: 10.1093/jxb/erp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoyama Y, Yagi M, Nishioka I. Biosynthesis of cannabinoid acids. Phytochemistry. 1975;14:2189–2192. [Google Scholar]
- 13.Yamaguchi T, et al. Cross-reaction of chalcone synthase and stilbene synthase overexpressed in Escherichia coli. FEBS Lett. 1999;460:457–461. doi: 10.1016/s0014-5793(99)01403-9. [DOI] [PubMed] [Google Scholar]
- 14.Shen Y, et al. Ectopic expression of the minimal whiE polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes. Proc Natl Acad Sci USA. 1999;96:3622–3627. doi: 10.1073/pnas.96.7.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stout JM, Boubakir Z, Ambrose SJ, Purves RW, Page JE. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J. February 21, 2012 doi: 10.1111/j.1365-313X.2012.04949.x. [DOI] [PubMed] [Google Scholar]
- 16.Jez JM, Bowman ME, Dixon RA, Noel JP. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol. 2000;7:786–791. doi: 10.1038/79025. [DOI] [PubMed] [Google Scholar]
- 17.Burbulis IE, Winkel-Shirley B. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W-X, Pelah D, Alergand T, Shoseyov O, Altman A. Characterization of SP1, a stress-responsive, boiling-soluble, homo-oligomeric protein from aspen. Plant Physiol. 2002;130:865–875. doi: 10.1104/pp.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S-C, et al. Characterization of a heat-stable protein with antimicrobial activity from Arabidopsis thaliana. Biochem Biophys Res Commun. 2007;362:562–567. doi: 10.1016/j.bbrc.2007.07.188. [DOI] [PubMed] [Google Scholar]
- 20.Lee JR, et al. Functional characterization of pathogen-responsive protein AtDabb1 with an antifungal activity from Arabidopsis thaliana. Biochim Biophys Acta. 2008;1784:1918–1923. doi: 10.1016/j.bbapap.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Thompson TB, Katayama K, Watanabe K, Hutchinson CR, Rayment I. Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase. J Biol Chem. 2004;279:37956–37963. doi: 10.1074/jbc.M406144200. [DOI] [PubMed] [Google Scholar]
- 22.Ames BD, et al. Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclization specificity of aromatic polyketides. Proc Natl Acad Sci USA. 2008;105:5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Tang Y. In vitro analysis of type II polyketide synthase. Methods Enzymol. 2009;459:367–393. doi: 10.1016/S0076-6879(09)04616-3. [DOI] [PubMed] [Google Scholar]
- 25.Katsuyama Y, Kita T, Funa N, Horinouchi S. Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J Biol Chem. 2009;284:11160–11170. doi: 10.1074/jbc.M900070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bomati EK, Austin MB, Bowman ME, Dixon RA, Noel JP. Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J Biol Chem. 2005;280:30496–30503. doi: 10.1074/jbc.M502239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bingman CA, et al. Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana. Proteins. 2004;57:218–220. doi: 10.1002/prot.20215. [DOI] [PubMed] [Google Scholar]
- 28.Cornilescu G, et al. Solution structure of a homodimeric hypothetical protein, At5g22580, a structural genomics target from Arabidopsis thaliana. J Biomol NMR. 2004;29:387–390. doi: 10.1023/B:JNMR.0000032525.70677.16. [DOI] [PubMed] [Google Scholar]
- 29.Dgany O, et al. The structural basis of the thermostability of SP1, a novel plant (Populus tremula) boiling stable protein. J Biol Chem. 2004;279:51516–51523. doi: 10.1074/jbc.M409952200. [DOI] [PubMed] [Google Scholar]
- 30.Sciara G, et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 2003;22:205–215. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marín M, Heinz DW, Pieper DH, Klink BU. Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase. J Biol Chem. 2009;284:32709–32716. doi: 10.1074/jbc.M109.024604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen B, Hutchinson CR. Tetracenomycin F2 cyclase: Intramolecular aldol condensation in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry. 1993;32:11149–11154. doi: 10.1021/bi00092a026. [DOI] [PubMed] [Google Scholar]
- 33.Sultana A, et al. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 2004;23:1911–1921. doi: 10.1038/sj.emboj.7600201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Bakel H, et al. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011;12:R102. doi: 10.1186/gb-2011-12-10-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 36.Russell RB, Sasieni PD, Sternberg MJ. Supersites within superfolds. Binding site similarity in the absence of homology. J Mol Biol. 1998;282:903–918. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- 37.Yu O, Jez JM. Nature’s assembly line: Biosynthesis of simple phenylpropanoids and polyketides. Plant J. 2008;54:750–762. doi: 10.1111/j.1365-313X.2008.03436.x. [DOI] [PubMed] [Google Scholar]
- 38.Kozubek A, Tyman JHP. Resorcinolic lipids, the natural non-isoprenoid phenolic amphiphiles and their biological activity. Chem Rev. 1999;99:1–26. doi: 10.1021/cr970464o. [DOI] [PubMed] [Google Scholar]
- 39.Gorham J. The Biochemistry of the Stilbenoids. Chapman & Hall, London; 1995. [Google Scholar]
- 40.Matsuzawa M, Katsuyama Y, Funa N, Horinouchi S. Alkylresorcylic acid synthesis by type III polyketide synthases from rice Oryza sativa. Phytochemistry. 2010;71:1059–1067. doi: 10.1016/j.phytochem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Karppinen K, Hokkanen J, Mattila S, Neubauer P, Hohtola A. Octaketide-producing type III polyketide synthase from Hypericum perforatum is expressed in dark glands accumulating hypericins. FEBS J. 2008;275:4329–4342. doi: 10.1111/j.1742-4658.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 42.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.