Abstract
Using chemical germ-line mutagenesis, we screened mice for defects in the humoral immune response to a type II T-independent immunogen and an experimental alphavirus vector. A total of 26 mutations that impair humoral immunity were recovered, and 19 of these mutations have been positionally cloned. Among the phenovariants were bumble, cellophane, and Worker ascribed to mutations in Nfkbid, Zeb1, and Ruvbl2, respectively. We show that IκBNS, the nuclear IκB-like protein encoded by Nfkbid, is required for the development of marginal zone and peritoneal B-1 B cells and additionally required for extrafollicular antibody responses to T-independent and -dependent immunogens. Zeb1 is also required for marginal zone and peritoneal B-1 B-cell development as well as T-cell development, germinal center formation, and memory B-cell responses. Finally, Ruvbl2 is required for T-cell development and maximal T-dependent antibody responses. Collectively, the mutations that we identified give us insight into the points at which disruption of an antibody response can occur. All of the mutations identified to date directly affect lymphocyte development or function; none have an exclusive effect on cells of the innate immune system.
Where it is known, the protective correlate of successful vaccines is a robust humoral immune response. Some vaccines, such as polysaccharide vaccines, stimulate humoral responses by directly activating B cells independently of T-cell help (1). However, the humoral response to most vaccines is dependent on helper T cells. During a T cell-dependent antibody response, antigen-presenting cells in the T-cell regions of secondary lymphoid tissues activate CD4+ T cells. A subset of these cells migrates to the outer T-cell region, where they interact with cognate B cells. Some of the responding B cells differentiate into short-lived, extrafollicular antibody-secreting cells (ASCs), whereas others form germinal centers (GCs), in which long-lived, high-affinity ASCs and memory B cells develop (2).
The development of B and T cells and their ability to mount protective immune responses are complex processes potentially dependent on many individual proteins with nonredundant functions. To identify such proteins, we carried out a forward genetic screen of N-ethyl-N-nitrosourea (ENU)-mutagenized mice carrying point mutations in both heterozygous and homozygous states. Adult animals were immunized with the type II T-independent stimulus (4-hydroxy-3-nitrophenylacetyl)-Ficoll (NP-Ficoll) and an experimental viral vector based on Semliki Forest virus (SFV), which induces robust class-switched antibody responses (3). Here, we describe the mutations that scored in the screen and characterize three mutants in detail: bumble, cellophane, and Worker.
Results
Forward Genetic Screen for Mutations That Impair Humoral Immune Responses.
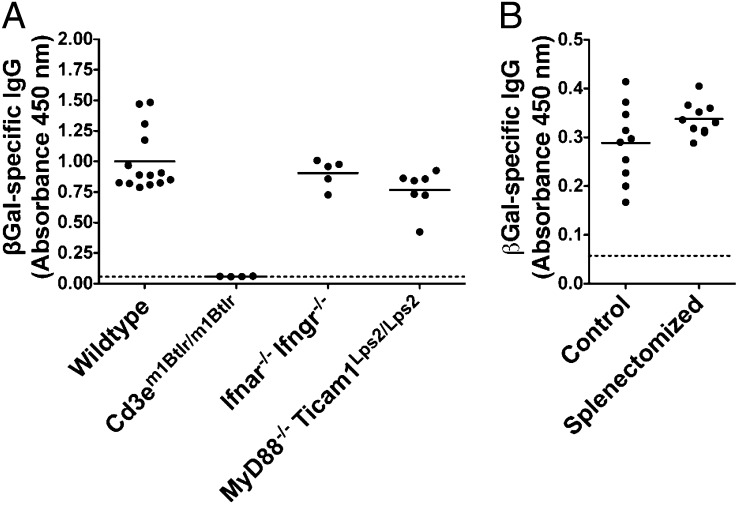
To screen for mutations that impair T-dependent antibody responses, we immunized ENU-mutagenized G3 mice with a recombinant SFV vector encoding the model antigen, β-gal (rSFV-βGal) (3). The class-switched antibody response to rSFV-encoded antigen was dependent on T-cell help (Cd3em1Btlr/m1Btlr) (Fig. 1A) but not an intact spleen (Fig. 1B), excluding a requirement for marginal zone (MZ) B cells in the response. It also occurred independently of IFN and Toll-like receptor (TLR) signaling, because mice deficient in both the IFN-α and -γ receptors (Ifnar−/− Ifngr−/−) (Fig. 1A) or MyD88 and Ticam1 (MyD88−/− Ticam1Lps2/Lps2) (Fig. 1A) made normal βGal-specific IgG responses after immunization with rSFV-βGal.
Fig. 1.
The antibody response to rSFV-encoded antigen is dependent on T-cell help but not IFN or TLR signaling or MZ B cells. Serum levels of βGal-specific IgG determined by ELISA 14 d after priming with rSFV-βGal in (A) WT mice and mice deficient in CD3ɛ, the IFN-α and -γ receptors, or MyD88 and Ticam1 and (B) control and splenectomized WT mice. Each point represents data from one mouse, and the bar indicates the mean of all values. Background (indicated by the dashed line) was determined by incubating pooled WT sera on uncoated wells. Results are representative of two to three independent experiments.
To simultaneously screen for impaired antibody responses to both T-dependent and -independent stimuli, mice were inoculated one time with rSFV-βGal, and 10 d later, most mice were also injected with the type II T-independent stimulus NP-Ficoll. On day 14 after rSFV-βGal (and day 5 after NP-Ficoll) inoculation, we measured serum βGal-specific IgG and NP-specific IgM by ELISA. In about one-half of the mice, we also stained peripheral blood lymphocytes (PBLs) with antibodies against CD4, CD8, CD44, and NK1.1 or against CD23, IgD, B220, and IgM to identify mutants with defects in lymphocyte development or maturation. Mice with low or undetectable levels of βGal-specific IgG or NP-specific IgM or with altered frequencies of CD44lo or CD44hi CD4+ and CD8+ T cells, NK1.1+ cells, or mature CD23hi IgDhi IgMlo B220hi B cells were considered hits in the screen and retained for breeding.
From among 7,343 G3 mice immunized with rSFV-βGal, 6,198 G3 mice immunized with NP-Ficoll, and 3,815 mice whose PBLs were analyzed by flow cytometry, we recovered 30 phenovariants, 26 of which were attributable to genetically transmissible mutations. Four mutants (15%) had significant B-cell deficiencies but were, nevertheless, able to make normal antibody responses after immunization. Eight mutants (31%) had selective defects in the T-independent IgM response to NP-Ficoll. Six mutants (23%) had selective defects in the T-dependent IgG response to rSFV-encoded antigen. Eight mutants (31%) made undetectable or suboptimal T-independent and -dependent responses after immunization with NP-Ficoll and rSFV-βGal. Transmissible phenotypes and their causative mutations, identified by candidate gene sequencing or a combination of bulk segregation analysis and whole-genome sequencing (4, 5), are summarized in Table S1. The identification and characterization of the bumble, cellophane, and Worker mutations, which affect genes with previously unknown roles in humoral immunity, are described in detail below.
Identification of the bumble, cellophane, and Worker Mutations.
bumble.
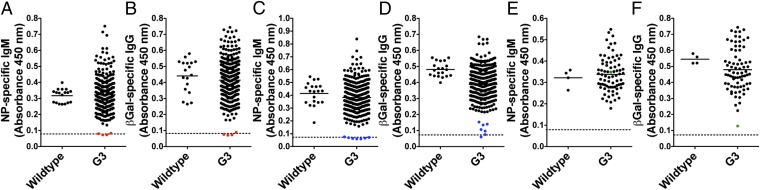
bumble was considered genetically transmissible because four siblings from the same founder shared an inability to make detectable T-independent IgM and T-dependent IgG responses to NP-Ficoll (Fig. 2A) and rSFV-βGal (Fig. 2B), despite normal frequencies of PBLs. To map the mutation, bumble males were outcrossed to C57BL/10J females, and the F1 progeny was intercrossed to generate F2 mice. Age-matched F2 animals were immunized with NP-Ficoll, and their serum NP-specific IgM responses were measured 5 d later. Of 47 F2 mice tested, 18 mice made no detectable NP-specific IgM, whereas 29 mice made responses within normal range (Fig. S1A). Equal amounts of genomic DNA isolated from all 18 mutant and 29 normal F2 mice were pooled separately, and a total of 127 SNPs that distinguish the C57BL/6J and C57BL/10J strains were sequenced in each pooled DNA sample. By analyzing linkage between a region of C57BL/6J alleles and the inability of mutant mice to respond to NP-Ficoll, we localized the bumble mutation to an ∼38-Mb region on chromosome 7 (Fig. S1B). Genotyping individual F2 mice for C57BL/6J and C57BL/10J alleles of the SNP markers in the critical region resulted in a peak logarithm of odds (LOD) score of 11.93 at 28,467,081 bp (Fig. S1B, Inset).
Fig. 2.
Recovery of the bumble, cellophane, and Worker phenovariants by forward genetic screening. Serum NP-specific IgM (A, C, and E) and βGal-specific IgG (B, D, and F) measured by ELISA in WT and G3 mice immunized 5 or 14 d before with NP-Ficoll and rSFV-βGal. (A and B) Results from 4 wk of screening are pooled with values for bumble index mice in red. (C and D) Results from 4 wk of screening are pooled with values for cellophane index mice in blue. (E and F) Results from 1 wk of screening are shown with values for the Worker index mouse in green. Each point represents data from one mouse. Background (indicated by the dashed line) was determined by incubating pooled WT sera on uncoated wells.
Within the critical region on chromosome 7, we identified a T→G transversion in the conserved donor splice site in the fourth intron of Nfkbid using the Applied Biosystems SOLiD 3 sequencing platform, which we confirmed by capillary sequencing (Fig. S1C). This mutation prevented the fourth intron from being removed from the Nfkbid transcript, which would result in the addition of glycine and valine residues and a premature stop codon after exon 4 (Fig. S1 D and E). Because this transcript would encode only 65 of 327 aa encoded by full-length Nfkbid, it would not be expected to retain any function, and indeed, animals with a targeted mutation in Nfkbid phenocopied bumble mice (Fig. S2).
cellophane.
cellophane was also considered transmissible because seven siblings from the same founder made undetectable NP-specific IgM and suboptimal βGal-specific IgG responses to NP-Ficoll (Fig. 2C) and rSFV-βGal (Fig. 2D). To map the mutation, cellophane males were outcrossed to C57BL/10J females, and the F1 progeny was intercrossed. Of 100 F2 mice tested, 18 mice made no detectable NP-specific IgM, whereas 82 mice made normal responses (Fig. S1F). Bulk segregation analysis localized the mutation to the proximal end of chromosome 18 with a peak LOD score of 7.9 at 15,408,257 bp (Fig. S1G). Using the Applied Biosystems SOLiD 3 sequencing platform, we identified a T→A transversion in the seventh exon of Zeb1. This mutation, confirmed by capillary sequencing (Fig. S1H), would replace the tyrosine at position 902 with a premature stop codon (Fig. S1I).
Worker.
The G3 index mouse from the Worker pedigree made a normal T-independent response to NP-Ficoll (Fig. 2E) but a suboptimal βGal-specific IgG response (Fig. 2F), and it had reduced frequencies of peripheral blood T cells, which expressed high surface levels of CD44. The T-cell deficiency in Worker mice was concordant with white belly spotting and white paws (Fig. S3). Both the immunological and pigmentation phenotypes were dominant. Therefore, to map the mutation, Worker males were outcrossed to C57BL/10J females, and the F1 progeny was directly phenotyped. Of 72 F1 mice tested, 9 mice made low βGal-specific IgG responses, had reduced frequencies of peripheral blood T cells (which expressed high levels of CD44), and had some degree of white belly spotting and white paws (Fig. S1J). Genomic DNA isolated from all 9 mutant and 63 normal F1 mice was pooled separately, and bulk segregation analysis localized the mutation to an ∼33-Mb region between 54,410,823 and 87,465,557 bp on chromosome 7, with a peak LOD score of 10.7 at 71,519,895 bp (Fig. S1K). Genotyping individual F1 mice for the C57BL/6J and C57BL/10J alleles of the SNP markers on chromosome 7 redefined the critical region as being between 38,216,957 and 71,519,895 bp, with strongest linkage (of both phenotypes) to 54,410,823 bp (LOD = 5.02) (Fig. S1K, Inset). Using the Applied Biosystems SOLiD 3 sequencing platform, we identified a heterozygous T→A transversion affecting a critical splice donor nucleotide in intron 2 of Ruvbl2 in the original Worker index mouse. This mutation, located at position 54,686,688 bp on chromosome 7, was confirmed by capillary sequencing (Fig. S1L).
When we genotyped Worker F1 mice for the Ruvbl2 mutation, we found that all of the mice with the Worker phenotype were heterozygous for the Ruvbl2 mutation, suggesting that homozygous mutants were not viable. Furthermore, of 61 normal F1 littermates, 18 littermates were heterozygous for the Ruvbl2 mutation, indicating that it was not fully penetrant. Sequencing thymic cDNA from heterozygous Worker mutants showed that these mice contained a mixture of WT and mutant Ruvbl2 transcripts. In mutant transcripts, exon 2 was skipped, resulting in splicing of exon 1 to exon 3, a frameshift error with the addition of 27 aberrant aa, and a premature stop codon downstream of the first exon (Fig. S1 M and N).
Impaired Lymphocyte Development in bumble, cellophane, and Worker Mice.
Nfkbid was recently implicated in ASC differentiation (6), but its role in regulating humoral responses is not well-understood. Although Zeb1 has been shown to be important for T-cell development in the thymus (7, 8), its role in B-cell development and function has not been described. Finally, Ruvbl2 has never been functionally inactivated in mice, and its role in immunity has not been studied. To understand how mutations in these genes affected antibody responses, we first analyzed lymphocyte development in these mice.
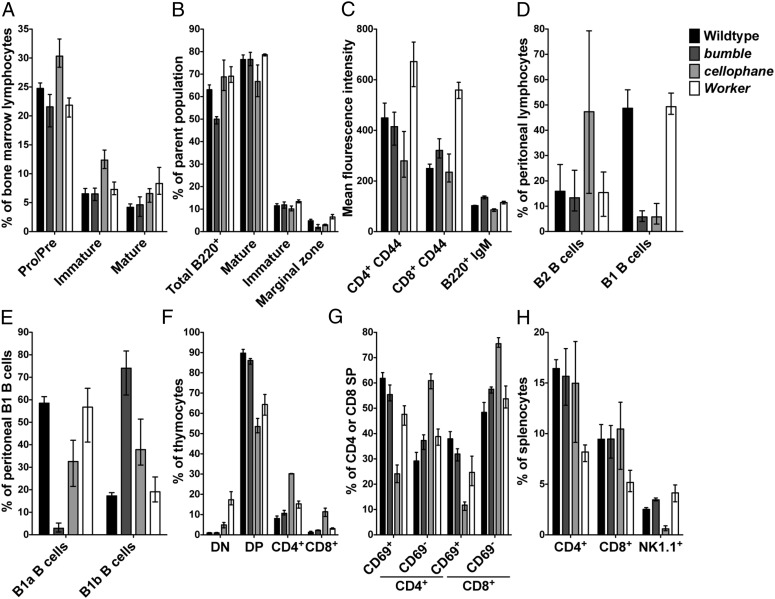
B-cell development in the bone marrow of bumble, cellophane, and Worker mice was normal in that the frequencies of pro-/pre-, immature, and mature B cells were equivalent in age- and sex-matched WT and mutant mice (Figs. 3A and 4A). Consistent with this finding, the frequencies of immature and mature follicular B cells were normal in the spleens of these mice (Fig. 3B), although follicular B cells in bumble mice expressed slightly higher surface levels of IgM (Figs. 3C and 4B). bumble and cellophane mice lacked discrete populations of MZ B cells (Figs. 3B and 4C), and both of these mutants had severely reduced frequencies of peritoneal B1 B cells (Figs. 3D and 4D). Of the peritoneal B1 B cells that developed in bumble mice, almost all were B1b B cells (Figs. 3E and 4E), whereas the frequencies of B1a and B1b B cells within the B1 B-cell population in cellophane mice were equivalent (Figs. 3E and 4E).
Fig. 3.
Lymphocyte development is impaired in bumble, cellophane, and Worker mice. Single-cell suspensions from WT and mutant mice were analyzed by flow cytometry. (A) IgM− B220int pro/pre, IgM+ B220int immature, and IgM+ B220high mature B cells as a percent of bone marrow lymphocytes. (B) B220+ B cells as a percent of splenocytes and IgDlow IgMhigh B220+ immature, IgDhigh IgMint B220+ mature follicular, and CD21/35high CD23low B220+ MZ B cells as a percent of splenic B220+ B cells. (C) Mean fluorescence intensity of CD44 on CD4+ and CD8+ T cells and IgM on B220+ B cells in spleen. (D) CD19int B220high B2 and CD19high B220low/int B1 B cells as a percent of peritoneal cavity lymphocytes. (E) CD5high CD43high CD19high B220low/int B1a and CD5low CD43low CD19high B220low/int B1b B cells as a percent of peritoneal cavity B1 B cells. (F) CD4− CD8− DN, CD4+ CD8+ DP, and CD4+ or CD8+ SP cells as a percent of thymocytes. (G) CD69− and CD69+ TCRβ+ CD4+ or CD8+ cells as a percent of CD4+ or CD8+ thymocytes, respectively. (H) CD4+ and CD8+ T cells and NK1.1+ cells as a percent of splenocytes. Bars indicate the mean (± range) of three to seven mice per genotype analyzed, except for A, in which the mean (± range) is shown for two WT mice.
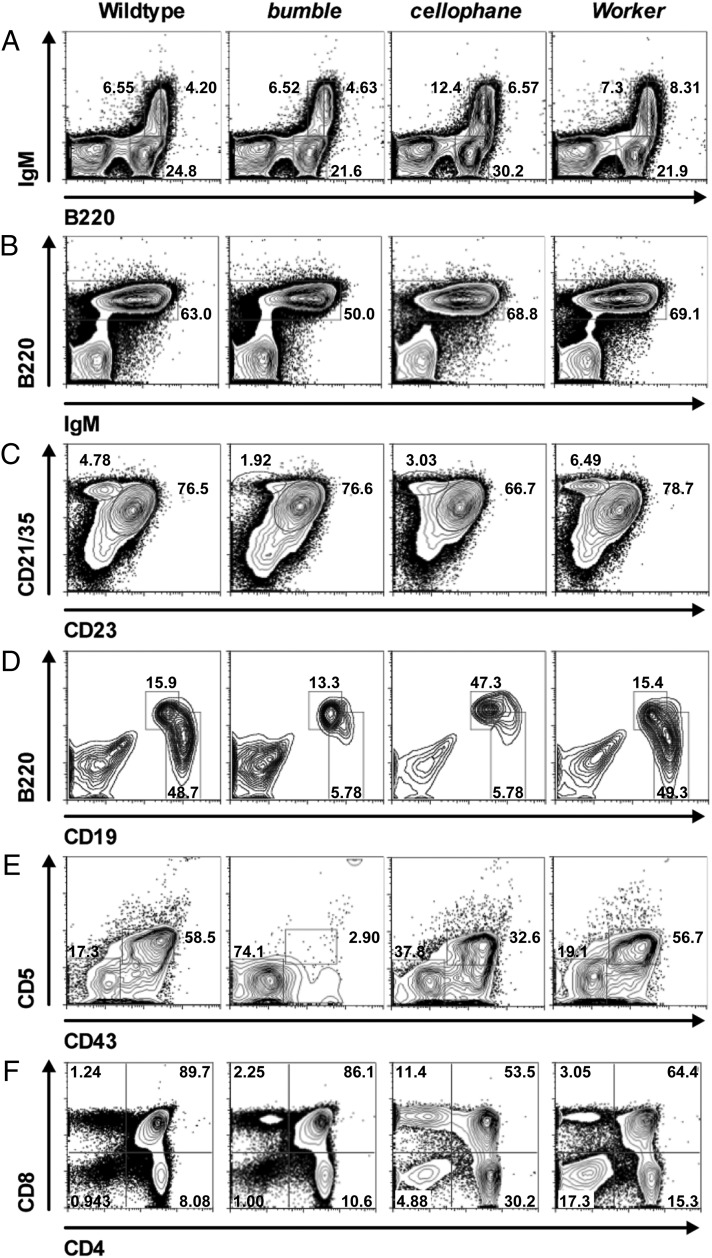
Fig. 4.
Representative staining for flow cytometric analysis of lymphocyte development in mutant mice. Single-cell suspensions from WT and mutant mice were analyzed by flow cytometry. (A) B220 vs. IgM in bone marrow. (B) IgM vs. B220 in spleen. (C) CD23 vs. CD21/35 on B220+ splenocytes. (D) CD19 vs. B220 on peritoneal lymphocytes. (E) CD43 vs. CD5 on CD19high B220low/int B1 B cells in the peritoneal cavity. (F) CD4 vs. CD8 in thymus. Flow cytometric plots and the indicated mean percentages of gated populations are representative of three to seven mice per genotype analyzed, except for A, which is representative of two WT mice.
The frequencies of double-negative (DN), double-positive (DP), and CD4 and CD8 single-positive (SP) thymocytes (Figs. 3F and 4F) as well as splenic CD44lo and CD44high CD4+ and CD8+ T cells (Fig. 3H) were equivalent in WT and bumble mice. By contrast, cellophane and Worker mice displayed significant alterations in thymic subsets. Compared with WT controls, the frequencies of DN thymocytes were increased, and the frequencies of DP thymocytes were decreased in the thymi of cellophane and Worker mice (Figs. 3F and 4F). cellophane mice also had expanded populations of SP thymocytes (Figs. 3F and 4F), and the frequencies of CD69+ TCRβ+ cells within the SP populations were reduced (Fig. 3G). Despite their thymic phenotype, the frequencies of splenic T cells were normal in cellophane mice, although these animals had a striking paucity of splenic NK cells (Fig. 3H). Finally, as expected, given their thymic phenotype and deficiency of peripheral blood T cells, Worker mice had fewer splenic CD4+ and CD8+ T cells (Fig. 3H), and these cells expressed high levels of surface CD44 (Fig. 3C).
Basal Levels of IgM and IgG3 Are Reduced in bumble Mice.
Consistent with their inability to make T-independent antibody responses to NP-Ficoll and reduced frequencies of MZ and B1a B cells, bumble mice had significantly lower basal levels of IgM and IgG3 (Fig. S4) compared with WT mice. Surprisingly, however, given that bumble mice were unable to make detectable βGal-specific antibody responses of any isotype for up to 42 d after priming with rSFV-βGal, they had normal serum levels of IgA, IgG1, IgG2a, and IgG2b (Fig. S4). By contrast, cellophane and Worker mice had normal basal levels of all of the Ig isotypes measured, with the possible exception of IgG2a, which was low in the former strain (Fig. S4). These data show that cellophane mice can make IgM, although not in response to NP-Ficoll, and that bumble, cellophane, and Worker mice all support class-switched antibody responses under some conditions but not in response to priming with rSFV-βGal.
Shared Defects in Extrafollicular Antibody Responses and a Unique Role for Zeb1 in Affinity Maturation.
In normal mice, immunization with NP-Ficoll fails to induce GCs, and the antibody response is mediated by extrafollicular ASCs (9). Likewise, we were unable to find evidence of βGal-specific GC formation in the lymphoid tissues of WT mice primed with rSFV-βGal, suggesting that the primary response to rSFV-encoded antigen is also carried out primarily by extrafollicular ASCs. We therefore hypothesized that the humoral defects in bumble and cellophane mice may be caused, at least in part, by impaired differentiation of extrafollicular ASCs during both T-independent and -dependent responses and that the humoral defect in Worker mice is caused by a defect in the differentiation of these cells during T-dependent responses. To test this hypothesis, we immunized WT and mutant mice with NP-chicken gamma globulin (NP-CGG), a hapten-protein carrier conjugate that induces both extrafollicular ASCs and GCs with well-defined kinetics (10).
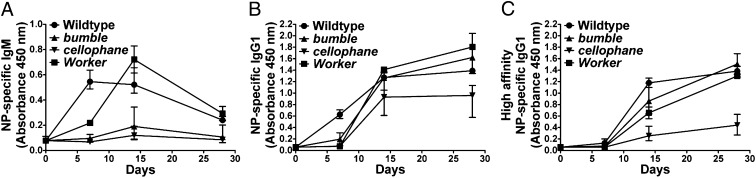
On day 7 after immunization with alum-precipitated NP-CGG, at the peak of the extrafollicular response (10), bumble, cellophane, and Worker mice had low or undetectable levels of NP-specific IgM (Fig. 5A) and IgG1 (Fig. 5B), consistent with impaired extrafollicular responses in all of these mice. By day 14 of the response, bumble mice still had no detectable NP-specific IgM (Fig. 5A), but NP-specific IgG1 reached WT levels (Fig. 5B). Because most of the NP-specific IgG1 detected at this time point was high affinity (Fig. 5C), it was most likely produced by ASCs exported from GCs (11). Collectively, these data suggest a selective defect in extrafollicular ASC responses in bumble mice. cellophane mice also had no detectable NP-specific IgM on day 14 of the response to alum-precipitated NP-CGG (Fig. 5A), and the levels of NP-specific IgG1 were reduced approximately twofold in cellophane mice compared with WT controls at this time point (Fig. 5B). Most of the NP-specific IgG1 in cellophane mice was low affinity (Fig. 5C), indicating that, in addition to impaired extrafollicular responses, cellophane mice did not support the formation of functional GCs. Finally, Worker mutants made normal (or slightly higher) levels of NP-specific IgM (Fig. 5A) and total NP-specific IgG1 (Fig. 5B) on day 14 after immunization, indicating that extrafollicular responses are delayed in Worker mice. By day 28 of the response, most of the NP-specific IgG1 in Worker mice was high affinity (Fig. 5C), showing that affinity maturation takes place in these animals.
Fig. 5.
The primary T-dependent antibody response is impaired in bumble, cellophane, and Worker mice. WT and mutant mice were immunized with alum-precipitated NP-CGG. Serum levels of NP-specific IgM (A), total NP-specific IgG1 (B), or high-affinity NP-specific IgG1 (C) were measured by ELISA. Each point represents the mean (± range) of greater than or equal to three mice per genotype, except for the day 14 and 28 time points for Worker, with two mice analyzed.
GC Formation and Secondary Antibody Responses Are Impaired in cellophane Mice.
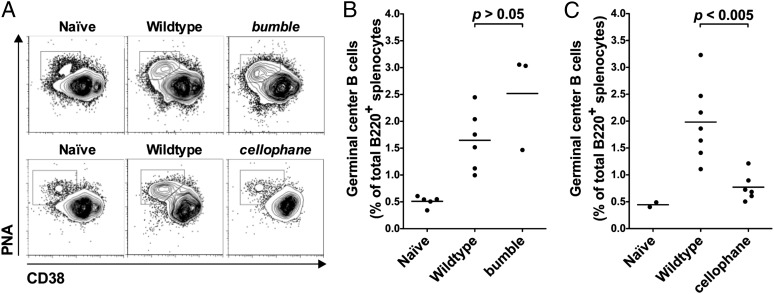
Consistent with their ability to make high-affinity NP-specific IgG1 responses (Fig. 5C), bumble mice had normal frequencies of GC B cells on day 14 after immunization with alum-precipitated NP-CGG (Fig. 6 A and B). In addition, splenic B cells isolated from immunized bumble mice transferred high-affinity NP-specific memory to naïve recipients (Fig. 7). Given their lack of high-affinity NP-specific IgG1 after immunization with alum-precipitated NP-CGG (Fig. 5C), it was not surprising that the frequency of GC B cells (Fig. 6 A and C) and the ability of splenic B cells to transfer NP-specific memory to naïve recipients (Fig. 7) were reduced in cellophane mice. Splenic B cells isolated from immunized Worker mice transferred high-affinity NP-specific memory to naïve recipients (Fig. 7).
Fig. 6.
cellophane mice have a defect in GC formation. (A) CD38 vs. Peanut Agglutinin (PNA) binding on IgDlow B220high splenocytes from naïve WT or WT and bumble mice immunized 14 d before (Upper) or WT and cellophane mice immunized 11 d before (Lower) with alum-precipitated NP-CGG. Plots are representative of two to five naïve WT and three to seven immunized mice per genotype. (B) GC B cells as a percent of total B220high splenocytes in naïve WT or immunized WT and bumble mice. WT vs. bumble, P > 0.05, Mann–Whitney U test. (C) GC B cells as a percent of total B220high splenocytes in naïve WT or immunized WT and cellophane mice. WT vs. cellophane, P < 0.005, Mann–Whitney U test. In B and C, each point represents data from one mouse, and the bar indicates the mean of all values.
Fig. 7.
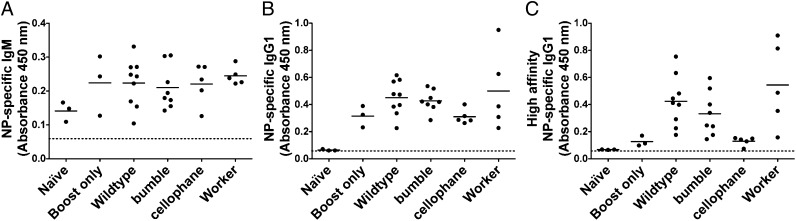
The memory B-cell response is reduced in cellophane mice. CD19+ B cells were enriched from the spleens of WT or mutant mice immunized 28 d before with alum-precipitated NP-CGG and transferred to CGG-primed WT mice. Immediately after cell transfer, the recipients were injected with NP-CGG in saline; 10 d later, serum levels of NP-specific IgM (A), total NP-specific IgG1 (B), and high-affinity NP-specific IgG1 (C) were measured by ELISA. As controls, NP-specific IgM and IgG1 levels were measured in naïve mice and mice that were immunized with NP-CGG in saline but did not receive memory B cells (boost only). In A and B, all of the recipients made IgM and IgG1 responses equivalent to boost-only controls, indicating that memory B cells were not required for these responses. In C, only mice that received functional memory B cells and were boosted with NP-CGG made high-affinity NP-specific IgG1 responses. Each point represents data from one mouse, and the bar indicates the mean of all values. Background (indicated by the dashed line) was determined by incubating pooled WT sera on uncoated ELISA wells.
Zeb1 Is Required for B-Cell Proliferation.
Because Nfkbid and Zeb1 were induced quickly after B-cell receptor cross-linking (Fig. S5 A and B), we next determined whether either of these genes was required for B-cell proliferation. Compared with WT cells, B cells isolated from the spleens of bumble mice proliferated as well (or slightly better) than WT B cells after stimulation with F(ab′)2αIgM for 72 h, even when the concentration of F(ab′)2αIgM was limiting (Fig. S5 C and D). The only notable difference between WT and bumble B cells cultured in vitro was that, in the absence of stimulation, more bumble B cells died within 72 h. A requirement for Nfkbid for B-cell survival during extrafollicular ASC formation seems unlikely, however, because Fas deficiency failed to rescue the humoral defects observed in bumble mice (Fig. S6). B cells isolated from the spleens of cellophane mice were impaired in their ability to proliferate in response to B-cell receptor cross-linking when the amount of F(ab′)αIgM was limiting (Fig. S5 C and D). B cells from cellophane mice did not, however, have an intrinsic defect in proliferation, because they divided as well as WT B cells after stimulation with CpG oligonucleotides (Fig. S5 C and D). The proliferation of splenic B cells from both bumble and cellophane mice in response to LPS was impaired (Fig. S5 C and D), consistent with the reduced frequencies of MZ B cells in both of these mutants.
Discussion
Using a forward genetic screen, we have probed the molecular requirements for humoral immune responses to T-independent and -dependent challenges. We have recovered a total of 26 transmissible mutations that impair responses to either or both challenges or that affect the development or maturation of lymphocytes. One mutation was lost because of infertility, whereas 19 mutations were identified in a total of 17 genes. Five of the 19 mutations (emptyhive, spelling, bumble, cellophane, and Worker) affected four genes that were not known to participate in immune cell development or humoral responses at the time that our work began. These four genes are Atp11c, Nfkbid, Zeb1, and Ruvbl2. The first gene has been described elsewhere (12), whereas the other three genes are described in this paper. Because our screen has detected approximately one unknown gene for every three known genes, we suggest that a large number of proteins with nonredundant function in the humoral immune response have yet to be identified.
None of nineteen mutations that we have identified to date prevent humoral immune responses by means of a deleterious effect on the innate immune response; on the contrary, all, at minimum, directly disrupt the development or function of lymphocytes. Of the seven mutations not yet found, six mutations present clear defects in lymphocyte development. Moreover, compound homozygous mutations that disrupt both IFN-α and -γ signaling or TLR signaling failed to score in our screens. This finding may indicate functional redundancy of innate immune mechanisms that support adaptive immune activation or alternatively, autonomy of the adaptive immune system in addressing the requirements of the screen.
IκBNS (revealed by bumble) belongs to the nuclear IκB-like family of proteins, which also includes Bcl-3 and IκBζ (13). These proteins contain ankyrin repeats and are classified as inhibitors of NF-κB (13). However, unlike the classical IκB proteins, which reside in the cytoplasm and regulate the translocation of NF-κB subunits to the nucleus, they reside in the nucleus and are thought to act as activators or inhibitors of NF-κB–dependent gene expression depending on the cell type in which they are expressed and the stimuli encountered (13). For example, whereas IκBNS positively regulates IL-2 transcription in stimulated T cells (14, 15), it negatively regulates the expression of IL-6 and IL-12p40 in LPS-stimulated macrophages (16).
Recently, Touma et al. (6) noted impaired B-cell development and function in IκBNS KO mice, with diminished B-cell proliferative responses to LPS and anti-CD40, reduced frequencies of MZ and B1 B cells, and lowered serum levels of IgM and IgG3. The mice failed to make T-independent responses to (2,4,6-trinitrophenyl)-Ficoll (TNP-Ficoll), and after immunization with alum-precipitated TNP-keyhole limpet hemocyanin (TNP-KLH), IκBNS KO mice failed to produce hapten-specific IgM and exhibited a delayed hapten-specific IgG1 response. In summary, their results are strikingly similar to our observations, with one exception: unlike bumble homozygotes immunized with NP-CGG, IκBNS KO mice failed to form GCs after immunization with sheep red blood cells (6). The reason for this discrepancy is unclear, but because we observed normal GC formation in immunized bumble mice bred and housed in two separate animal facilities, we conclude that IκBNS is not required for either GC formation or memory B-cell responses but rather, is required for the differentiation of extrafollicular ASCs that mediate T-independent antibody responses and the early antibody response to T-dependent challenge. The primary genes regulated by IκBNS and the mechanism by which it controls the differentiation of B cells into extrafollicular plasma cells remain to be determined.
Zeb1 (revealed by cellophane) was originally identified as an enhancer binding factor of the chicken δ1-crystallin gene (17). It contains two highly conserved Krüppel type-C2H2 zinc finger clusters near the N- and C-termini separated by a homeobox domain (18). The C-terminal zinc finger clusters are required for DNA binding activity and transcriptional repressor activity (19). Some studies suggest that Zeb1 might also activate transcription in a cell type- and context-dependent manner (20, 21). Mice homozygous for a Zeb1 KO allele exhibit multiple skeletal defects beginning at embryonic day 15 (E15) and small, hypocellular thymi at E18.5, and they die shortly after birth (8). Truncation of Zeb1 at residue 727 (and removal of the C-terminal zinc finger clusters) in Zeb1ΔC727/ ΔC727 mice does not lead to skeletal malformation, but it is still associated with a high level of mortality (7). These mice also have small, hypocellular thymi caused by a reduction in c-kit+ early T-cell precursors (7).
The cellophane allele of Zeb1 encodes a 901-aa protein lacking the C-terminal zinc finger domains. Like the Zeb1ΔC727/ ΔC727 mice, cellophane homozygotes have small, hypocellular thymi with fewer DP thymocytes and expanded populations of SP thymocytes. In addition, cellophane mice have fewer splenic NK and MZ B cells and an almost complete lack of peritoneal B-1 B cells. In contrast to the normal proliferation of Zeb1ΔC727/ ΔC727 T cells stimulated with ConA (7), splenic B cells from cellophane mice proliferate poorly in response to B-cell receptor-dependent stimuli. This most likely underlies the inability of cellophane mice to mount antibody responses and form GCs. Recently, Zeb1 was shown to synergize with FOXO transcription factors to activate growth suppressive genes in B cells (21) and form a repressive complex with C-terminal binding protein (CtBP) at a distal promoter element of Bcl6 (22), which is required for B-cell commitment to the GC pathway. Based on these studies, one would expect B-cell proliferation and GC formation to be enhanced in the absence of functional Zeb1, but we observed the opposite in cellophane mice. Thus, cellophane mice provide an in vivo model to dissect the mechanisms by which Zeb1 controls B-cell proliferation during humoral immune responses.
Ruvbl2 (revealed by Worker) is an ATP-binding protein that belongs to the AAA+ family of ATPases. It contains two AAA domains separated by Walker A, insertion, and Walker B domains, and it is endowed with ssDNA-stimulated ATPase activity and ATP-dependent DNA helicase activity (23). Ruvbl2 (and its homolog Ruvbl1) are components of the Ino80 and SRCAP complexes in mammals (24). The former functions in the repositioning of nucleosomes in regions of active transcription, whereas the latter facilitates replacement of the canonical nucleosome protein H2A with the variant H2AZ, which is also associated with active transcription. These two complexes may also be involved in DNA damage responses and DNA replication. A separate set of requirements for Ruvbl2 has been identified in the assembly and stabilization of small nucleolar ribonucleoprotein complexes and as a component of the R2TP complex, which acts as a cytoplasmic chaperone to stabilize members of the phosphoinositide kinase kinase family (24).
Homozygosity for the Worker allele was never observed despite efforts to produce a stock from heterozygous mice, which is suggestive of a homozygous lethal effect of the mutation. Moreover, heterozygotes were born at less than the expected Mendelian ratio, indicating that the locus is haploinsufficient for viability. No KO allele of Ruvbl2 has been described, and it is not known whether it would be viable either in the homozygous or heterozygous state. Worker heterozygotes have normal frequencies of B cells and respond as well as WT mice to T-independent challenge, indicating that their reduced ability to make class-switched antibody responses to rSFV-βGal and delayed response to alum-precipitated NP-CGG do not reflect a B cell-intrinsic defect. Instead, the Worker phenotype is consistent with limiting T-cell help during T-dependent humoral immune responses secondary to impaired thymocyte development. We hypothesize that Ruvbl2 functions in the repair of DNA double-strand breaks during V(D)J recombination in developing thymocytes and that repair is partially disrupted by heterozygosity for the Worker mutation. This hypothesis is consistent with the observation that DN thymocytes are expanded in the thymi of Worker mice, reminiscent of the partial DN→DP block in mice with a hypomorphic allele of Rag1 (25). Worker mice provide the first opportunity to study the in vivo functions of Ruvbl2 in mammalian development and immunity.
Nfkbid, Zeb1, and Ruvbl2 are each represented by orthologous human genes, and we predict that alleles similar to the alleles described here may cause immunodeficiency. In understanding the function of the encoded proteins, we may assemble a more complete picture of the molecular events that must occur to permit robust antibody responses to infection and immunization.
Materials and Methods
Animals.
Mice were housed and bred in The Scripps Research Institute vivarium. C57BL/6J, C57BL/10J, and C57BL/6.MRL-Faslpr/J were purchased from The Jackson Laboratory. B6NDen;B6N-Nfkbidtm1a(EUCOMM)Wtsi/Cnrm embryos were purchased from European Mouse Mutant Archive. MyD88−/− Ticam1Lps2/Lps2 mice have been described elsewhere (26). Ifnar−/− and Ifngr−/− mice, provided by J. Sprent and C. Surh (The Scripps Research Institute, La Jolla, CA), respectively, were intercrossed to generate Ifnar−/− Ifngr−/− mice. ENU mutagenesis was performed on C57BL/6J mice as described (26). All studies were performed in accordance with the guidelines established by the Institutional Animal Care and Use Committee of The Scripps Research Institute.
Splenectomy.
Splenectomy was performed on 8-wk-old C57BL/6J mice, which were maintained on water supplemented with trimethroprim-sulfamethoxazole antibiotics.
Recombinant SFV.
Recombinant SFV encoding βGal was prepared and titered as described (3).
Immunizations.
G3 mice were injected i.p. with 2 × 106 IU rSFV-βGal in 200 μL 0.9% saline; 10 d later, the mice were injected i.p. with 50 μg NP28-AECM-Ficoll (Biosearch Technologies) in 200 μL 0.9% saline. On day 14 after immunization with rSFV-βGal, blood was collected in serum separator tubes (Becton Dickinson). Up to 75 μL blood were subjected to two rounds of red blood cell lysis (Sigma-Aldrich) before staining and flow cytometric analysis. The remaining blood was centrifuged to separate serum. To characterize the T-dependent antibody response in greater detail, mice were injected i.p. with 50 μg NP39-CGG (Biosearch Technologies) or CGG (Jackson ImmunoResearch) and 5 μg LPS [from Salmonella minnesota R595 (Re); Alexis Biochemicals] mixed 1:1 with Imject alum (Pierce).
Adoptive Transfer.
Six- to eight-week-old mice were immunized with alum-precipitated NP-CGG. On day 28 after immunization, splenic B cells were positively enriched using the Mouse CD19 B-cell Isolation Kit (Miltenyi Biotec); 5 × 106 cells in 200 μL 0.9% saline were transferred i.v. to C57BL/6J mice, which had been primed with alum-precipitated CGG 28 d before. Immediately after cell transfer, the recipients were injected i.p. with 50 μg NP-CGG in a total volume of 200 μL 0.9% saline.
Flow Cytometry.
Cells were incubated with mAb to CD16 and CD32 (BD Pharmingen) before labeling with biotinylated PNA (Vector Laboratories) and/or biotin- or fluorochrome-conjugated mAb recognizing the following murine cell surface markers: CD4, CD5, CD8, CD19, CD21/35, CD23, CD38, CD43, CD69, B220, BP1, and TCRβ (BD Pharmingen); CD24, CD44, IgM, and NK1.1 (eBioscience); and IgD (Southern Biotechnology). The following secondary reagents were used: FITC- or allophycocyanin-streptavidin (BD Pharmingen). Data were acquired on a FACSCalibur and analyzed with FlowJo software (Treestar).
ELISA.
Polyvinyl chloride microtiter 96-well round-bottom plates (Fisher Scientific) were coated with 2 μg/mL βGal (Roche), 5 μg/mL NP23-BSA, or 5 μg/mL NP4-BSA (Biosearch Technologies). Plates were blocked with 5% milk, and serum samples were serially diluted in 1% milk. Plates were incubated with HRP-conjugated goat anti-mouse IgM, IgG, or IgG1 (Southern Biotechnology), developed with SureBlue TMB Microwell Peroxidase Substrate and TMB Stop Solution (KPL), and read at 450 nm on a MAXline Emax Microplate Reader (Molecular Devices). Basal serum Ig levels were measured using the Mouse Isotyping Kit (BD Pharmingen).
DNA Sequencing and Bulk Segregation Analysis.
Whole-genome and validation sequencing and bulk segregation analysis were performed as described (4, 5).
RT-PCR.
Spleens or thymi from C57BL/6J and bumble or Worker mice were homogenized in TRIzol (Invitrogen). RNA was reverse-transcribed using oligo(dT) primers from the RETROscript kit (Ambion), and cDNA was amplified using JumpStart REDTaq ReadyMix (Sigma-Aldrich). Purified PCR products were sequenced on an ABI 3730 XL DNA analyzer.
In Vitro Stimulation and Proliferation.
Splenic B cells were negatively enriched using the Mouse B-Cell Isolation Kit (Miltenyi Biotec) and labeled with carboxyfluorescein succinimidyl ester (Invitrogen). Labeled cells were resuspended at a concentration of 2 × 106 cells/mL in RPMI-1640 supplemented with 10% heat-inactivated FBS, penicillin and streptomycin, nonessential amino acids, sodium pyruvate, and β-mercaptoethanol (Sigma-Aldrich). A total of 2 × 105 cells were cultured in triplicate in 200 μL complete media supplemented with saline, 1–25 μg/mL F(ab′)2 fragment goat anti-mouse IgM (Jackson Immunoresearch), 10 μg/mL LPS, or 0.2 μM CpG oligodeoxynucleotides (IDT) for 72 h at 37 °C in 5% CO2.
Quantitative PCR.
Splenocytes were cultured at 5 × 106 cells/mL in complete RPMI-1640 supplemented with saline, 25 μg/mL F(ab′)2 fragment goat anti-mouse IgM, 10 μg/mL LPS, or 0.2 μM CpG oligodeoxynucleotides for up to 24 h at 37 °C in 5% CO2. Cells were pelleted and lysed in TRIzol. RNA and cDNA were prepared as described above. Levels of 18S rRNA, Nfkbid, and Zeb1 transcripts were analyzed by real-time PCR using 2× quantitative PCR mix (BioPioneer). Real-time PCR was carried out on an Applied Biosystems 7300 machine.
Supplementary Material
Acknowledgments
We thank Christine Domingo, Elizabeth Hanley, and Sara Kalina for production of G3 mice; Mercedes Gutierrez for animal husbandry; Chip Ross and Pei Lin for technical support; Nora Smart and Eva Moresco for writing the pages annotating the mutants identified in this screen, which can be found at http://mutagenetix.utsouthwestern.edu; and Diantha LaVine for artwork in Fig. S1. C.N.A. was supported by an Irvington Institute Fellowship of The Cancer Research Institute. This work was funded by a grant from The Bill and Melinda Gates Foundation (to G.B.K.H. and B.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209134109/-/DCSupplemental.
References
- 1.Defrance T, Taillardet M, Genestier L. T cell-independent B cell memory. Curr Opin Immunol. 2011;23:330–336. doi: 10.1016/j.coi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 3.Hidmark AS, et al. Humoral responses against coimmunized protein antigen but not against alphavirus-encoded antigens require alpha/beta interferon signaling. J Virol. 2006;80:7100–7110. doi: 10.1128/JVI.02579-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold CN, et al. Rapid identification of a disease allele in mouse through whole genome sequencing and bulk segregation analysis. Genetics. 2011;187:633–641. doi: 10.1534/genetics.110.124586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia Y, et al. Bulk segregation mapping of mutations in closely related strains of mice. Genetics. 2010;186:1139–1146. doi: 10.1534/genetics.110.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Touma M, et al. Impaired B cell development and function in the absence of IkappaBNS. J Immunol. 2011;187:3942–3952. doi: 10.4049/jimmunol.1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashi Y, et al. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 9.de Vinuesa CG, et al. Germinal centers without T cells. J Exp Med. 2000;191:485–494. doi: 10.1084/jem.191.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob J, Kassir R, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J Exp Med. 1991;173:1165–1175. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi Y, Dutta PR, Cerasoli DM, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J Exp Med. 1998;187:885–895. doi: 10.1084/jem.187.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siggs OM, et al. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat Immunol. 2011;12:434–440. doi: 10.1038/ni.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto M, Takeda K. Role of nuclear IkappaB proteins in the regulation of host immune responses. J Infect Chemother. 2008;14:265–269. doi: 10.1007/s10156-008-0619-y. [DOI] [PubMed] [Google Scholar]
- 14.Fiorini E, et al. Peptide-induced negative selection of thymocytes activates transcription of an NF-kappa B inhibitor. Mol Cell. 2002;9:637–648. doi: 10.1016/s1097-2765(02)00469-0. [DOI] [PubMed] [Google Scholar]
- 15.Touma M, et al. Functional role for I kappa BNS in T cell cytokine regulation as revealed by targeted gene disruption. J Immunol. 2007;179:1681–1692. doi: 10.4049/jimmunol.179.3.1681. [DOI] [PubMed] [Google Scholar]
- 16.Kuwata H, et al. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity. 2006;24:41–51. doi: 10.1016/j.immuni.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Funahashi J, Kamachi Y, Goto K, Kondoh H. Identification of nuclear factor delta EF1 and its binding site essential for lens-specific activity of the delta 1-crystallin enhancer. Nucleic Acids Res. 1991;19:3543–3547. doi: 10.1093/nar/19.13.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 19.Sekido R, et al. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda K, Kawakami K. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem. 1995;233:73–82. doi: 10.1111/j.1432-1033.1995.073_1.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Yusuf I, Andersen HM, Fruman DA. FOXO transcription factors cooperate with delta EF1 to activate growth suppressive genes in B lymphocytes. J Immunol. 2006;176:2711–2721. doi: 10.4049/jimmunol.176.5.2711. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulou V, Postigo A, Sánchez-Tilló E, Porter AC, Wagner SD. ZEB1 and CtBP form a repressive complex at a distal promoter element of the BCL6 locus. Biochem J. 2010;427:541–550. doi: 10.1042/BJ20091578. [DOI] [PubMed] [Google Scholar]
- 23.Kanemaki M, et al. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- 24.Jha S, Dutta A. RVB1/RVB2: Running rings around molecular biology. Mol Cell. 2009;34:521–533. doi: 10.1016/j.molcel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khiong K, et al. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J Clin Invest. 2007;117:1270–1281. doi: 10.1172/JCI30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.