Abstract
Gene expression differences are shaped by selective pressures and contribute to phenotypic differences between species. We identified 964 copy number differences (CNDs) of conserved sequences across three primate species and examined their potential effects on gene expression profiles. Samples with copy number different genes had significantly different expression than samples with neutral copy number. Genes encoding regulatory molecules differed in copy number and were associated with significant expression differences. Additionally, we identified 127 CNDs that were processed pseudogenes and some of which were expressed. Furthermore, there were copy number-different regulatory regions such as ultraconserved elements and long intergenic noncoding RNAs with the potential to affect expression. We postulate that CNDs of these conserved sequences fine-tune developmental pathways by altering the levels of RNA.
Keywords: copy number variation, evolution, genomics
Gene expression differences contribute substantially to the evolution of phenotypes within and between species. For example, differential expression of Bmp4 during development in finches alters beak morphology, Darwin’s classic example of phenotypic adaptation (1). Moreover, genetic variation in the transcription factor (TF) gene, FOXP2, appears responsible for neuronal gene expression differences that drive the development of complex spoken language, a uniquely human trait (2–4). These two genes, among others, indicate that differences in gene expression can lead to species-specific characteristics.
Genomic variation, including structural variation, can alter expression levels between species (5). Copy number differences (CNDs) represent gains and losses of orthologous genomic regions between species. CNDs differ from copy number variants (CNVs) (6–10), in that the former refers to interspecies variation, whereas the latter refers to intraspecies variation. Certain CNDs may have evolved under positive selection in primates (11–16). Furthermore, CNDs that overlap genes and nongenic regulatory elements can lead to divergent expression profiles and affect phenotypes. For example, genic CNVs between two inbred mouse strains are associated with differences in the expression levels of the Itlna gene that are correlated with abdominal weight and insulin levels (17). In another instance, a human-specific deletion of an enhancer alters the expression context of an androgen receptor during development, likely leading to the human specific loss of penile spines (18). Hence, CNDs, that overlap genic and nongenic regions can regulate gene expression and underlie phenotypic differences within and between species.
To understand better the evolution of primates, we investigated the impact of CNDs on gene expression profiles. We found that certain highly conserved, functionally relevant sequences are copy number different among primate species. We further demonstrated that these CNDs may affect the regulation of gene expression. We were able to identify multiple mechanisms by which CNDs might drive the divergence of primate gene expression.
Results
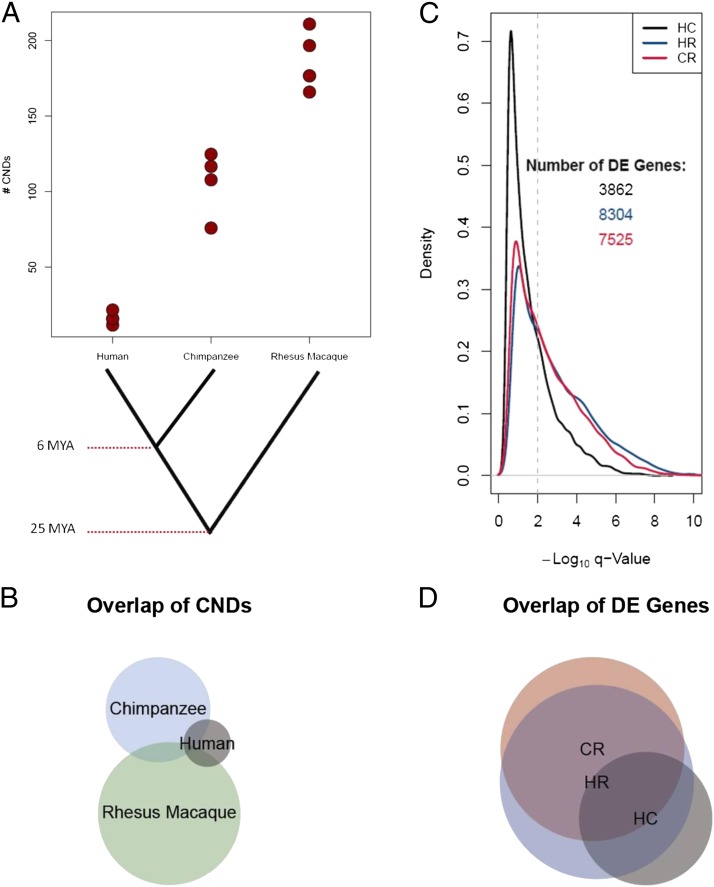
To evaluate CNDs in functionally relevant regions, we analyzed sequences that are 100% identical across human, chimpanzee, and rhesus macaque reference genomes by using a custom-designed array comparative genomic hybridization (aCGH) platform (Dataset S1). The use of such sequence-conserved probes produces an enrichment of functional elements, including exons, regulatory regions, and other conserved loci, while biasing away from segmental duplications, known human CNVs, and other loci that are genetically divergent at the nucleotide level (Fig. S1). We hybridized human (n = 4), chimpanzee (n = 4), and rhesus macaque (n = 5) lymphoblastoid cell line (LCL) derived DNA onto the described platform along with a single human reference DNA sample (HapMap sample NA10851). We also hybridized gorilla (n = 2) and orangutan (n = 2) samples to the same human reference to help rule out human-specific reference artifacts (Fig. S2). Altogether, these experiments revealed 964 CND events in the human, chimpanzee, and rhesus macaque samples that merge into 407 CND regions (Fig. 1A, Fig. S3, and Datasets S2–S4). As expected, the number of CNDs was found to be higher in samples with greater genetic distance from the human array reference (Fig. 1).
Fig. 1.
Copy number and gene expression levels differ between primate species. (A) The number of CND calls per sample. Detailed calling parameters are provided in SI Materials and Methods. Note the increase in the number of CND calls with the increased genetic distance from the human reference DNA (NA10851). Eighteen of the 33 CNDs found in human samples were previously known CNVs among humans. (B) Shared CNDs among species using a 50% reciprocal overlap criterion. (C) Density plot of the –log10 q-values for pairwise comparisons of species for DE genes by using RNAseq data. RNAseq read depth was used to determine the probability of each of ∼14,000 genes being DE between pairs of species: HC, HR, and CR. The vertical dashed line is a cutoff of q-value of 0.01 (FDR of 1%). All genes to the right of that line are considered to be DE for that pairwise comparison in subsequent analyses. Note how the red and blue lines (HR and CR) remain above the black line to the right of the significance level cutoff, indicating more DE genes in these pairwise comparisons. (D) Overlap of DE genes for each pairwise comparison (using an FDR of 1%).
We generated RNA sequencing (RNAseq) data for the nonhuman primate LCL samples and used previously published human RNAseq data from LCL samples for comparison of gene expression levels (19). Altogether, we analyzed 14,730 genes with human exon orthologues defined in chimpanzee and rhesus macaque (SI Materials and Methods). For this set of genes, we performed a pairwise analysis between the species as described previously (20) to determine which genes are differentially expressed (DE). By using a stringent false discovery rate (FDR) of 1%, there were 3,862, 8,304, and 7,525 genes that were DE between human and chimpanzee (HC), human and rhesus macaque (HR), and chimpanzee and rhesus macaque (CR), respectively (Dataset S5). Similar to the CND results, the number of gene expression differences observed between closely related species (i.e., HC) is much smaller than the number of gene expression differences between more distantly related species (i.e., HR and CR; Fig. 1).
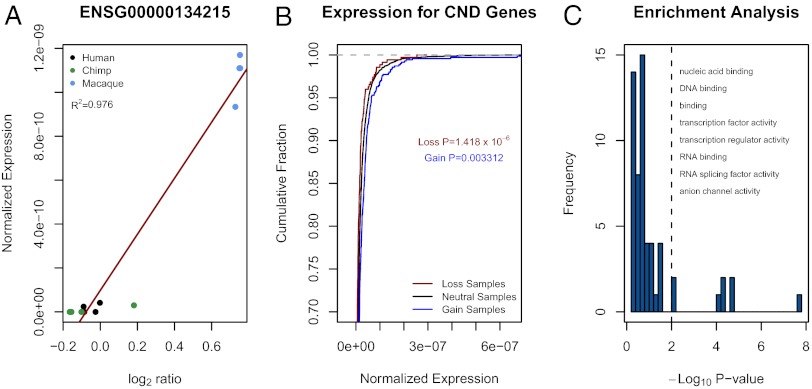
Some species-specific CNDs overlapping genes appear to affect the expression levels of those genes (Fig. 2A). As expected, for genes overlapped by CNDs, those samples with gains tend to have higher gene expression, whereas samples with losses tend to have lower gene expression and these differences are significant (one-sided Kolmogorov–Smirnov compared with copy number neutral gene samples, P < 0.01 for losses and gains; Fig. 2B). Surprisingly, 18 genes had expression levels that appear to be inversely correlated with copy number (R2 > 0.45). By examining genes with significant differential expression between pairs of species in LCLs (Fig. 1C) or in liver tissue (20), 366 of 9,576 (3.8%) DE genes can be explained by CNDs overlapping the gene in at least one of the two species being compared (Dataset S6).
Fig. 2.
Genes overlapped by CNDs can have differential gene expression. (A) An example of a gene overlapped by a CND in rhesus macaques as demonstrated by elevated aCGH log2 ratios in these samples. Samples with higher copy number also have higher expression of this gene. (B) Samples with genes overlapped by gains tend to have higher expression of those genes whereas samples with genes overlapped by losses tend to have lower expression of those genes. Shown is the cumulative fraction plots of normalized read counts. The difference from samples with neutral copy number (no change relative to NA10851) is significant for gains and losses by a one-sided Kolmogorov–Smirnov test. (C) PANTHER enrichment analysis was performed to determine whether CND genes were enriched for specific molecular functions. The P values are the corrected probabilities that a given category of genes would be enriched by chance. The vertical dashed line indicates a P value of 0.01. Categories to the right of the line were considered significantly enriched among CND genes. Categories are listed starting with the most enriched. Details on the background set and P values are provided in SI Materials and Methods.
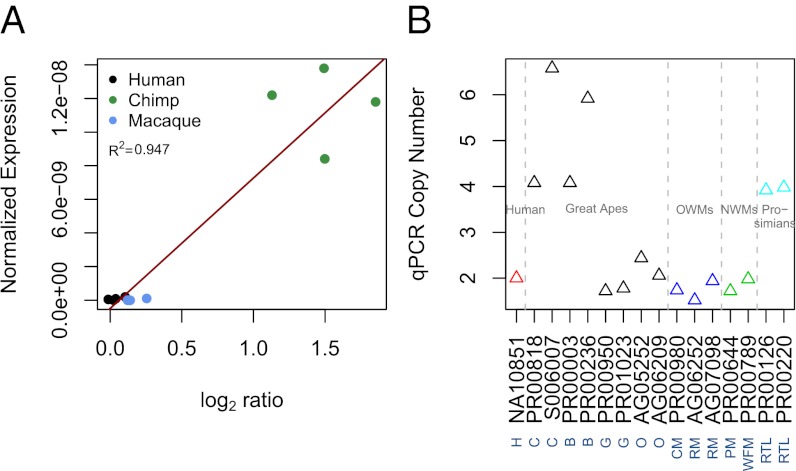
One gene in particular, KANK1, is gained in chimpanzee and also expressed higher in chimpanzee relative to human and rhesus macaque (Fig. 3). Based on PAML likelihood ratio estimates (21), the nucleotide level variation for the coding portion of KANK1 seems to be evolving under purifying selection (P = 0.004). It is possible that the protein function of KANK1 must be maintained between species, but the gene expression can be temporally, spatially, and quantitatively altered. By using quantitative PCR (qPCR), we examined the copy number of KANK1 across a panel of primate species. Bonobos also appear to have an increased copy number of KANK1 relative to human, but this increase may be variable among bonobos (SI Materials and Methods). The similarity between these two species may be expected, as chimpanzees and bonobos are closely related.
Fig. 3.
The KANK1 gene is gained in chimpanzee and DE between chimpanzee and other species. (A) The number of RNAseq reads mapping to KANK1 in each sample was normalized by the total number of reads per sample and the mappable length of the gene and plotted as a function of the mean log2 ratios for this gene from aCGH. (B) SYBR Green qPCR for KANK1 was performed across additional samples to determine whether other primate species also have copy number gains of the gene. The corresponding species are as follows: B, bonobo; C, chimpanzee; CM, colobus monkey; G, gorilla; H, human; O, orangutan; PM, pygmy marmoset; RM, rhesus macaque; RTL, ring-tailed lemur; WFR, white-fronted marmoset.
We performed an enrichment analysis for genes overlapping CNDs using PANTHER (22) taking into consideration the bias of the array platform (SI Materials and Methods). We discovered that CNDs tend to overlap TF genes and other regulatory genes more often than expected by chance (P = 0.000053; Fig. 2C). In our data, we identified 70 CND TF genes (Dataset S7 and Fig. S4). Likewise, TF genes were recently found to be enriched for differential expression in liver tissue among primates (23). We argue that some of the expression differences of TF genes may be a result of CNDs. In fact, the fraction of TF genes DE in LCLs or in liver that appear to be affected by CNDs is 5.3%, compared with 3.8% for all genes (Fig. S5).
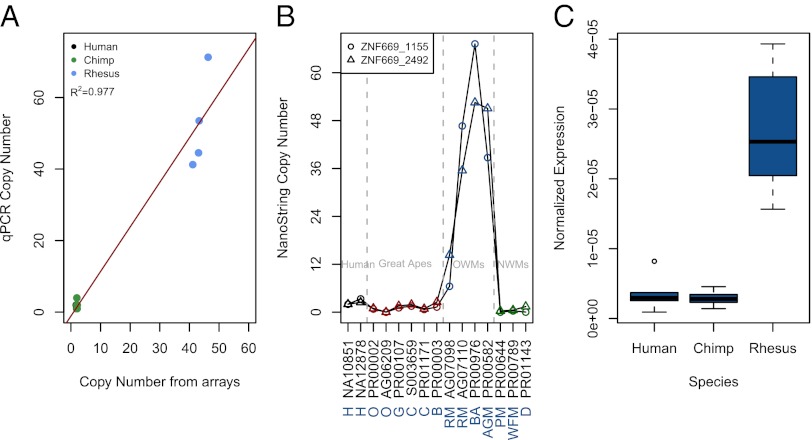
An extreme example of a CND TF is ZNF669. This gene has a massive increase in copy number associated with an increase in expression in rhesus macaques (Fig. 4). We further showed that ZNF669 is also gained in Savannah baboon and African green monkeys, but not in great apes or New World monkeys (Fig. 4B). Thus, expansion of the copy number of this TF initially occurred in the ancestor of Old World monkeys. It seems that this increase in copy number is accompanied by nucleotide variation shaped by positive selection between the additional copies in rhesus macaque based on likelihood ratio analysis from PAML (Bayes empirical P > 99%; Fig. S6) (21, 24).
Fig. 4.
TF genes are enriched for overlap with CNDs. (A) qPCR using an ABI TaqMan assay was performed for the ZNF669 gene and used to confirm the high copy number in rhesus macaques. The red line is the least-squares fit of the data. (B) NanoString copy number assay was used to confirm the additional copy number of ZNF669 across different primate species. Black samples are human. Red samples are nonhuman great apes. Blue samples are Old World monkeys. Green samples are New World monkeys. Coriell IDs are listed. The shapes of the data points are to indicate two different NanoString probes that were tested. Species are as follows: AGM, African green monkey; B, bonobo; BA, baboon; C, chimpanzee; D, dourocouli; G, gorilla; H, human; O, orangutan; PM, pygmy marmoset; RM, rhesus macaque; WFM, white-fronted marmoset. Note that all Old World monkeys assessed have some relative gain of ZNF669 compared with human. (C) Normalized read count for the human ZNF669 gene and its orthologues in chimpanzee and rhesus macaque. RNAseq read counts were normalized by the total number of reads per sample and the mappable length of the exons within the gene.
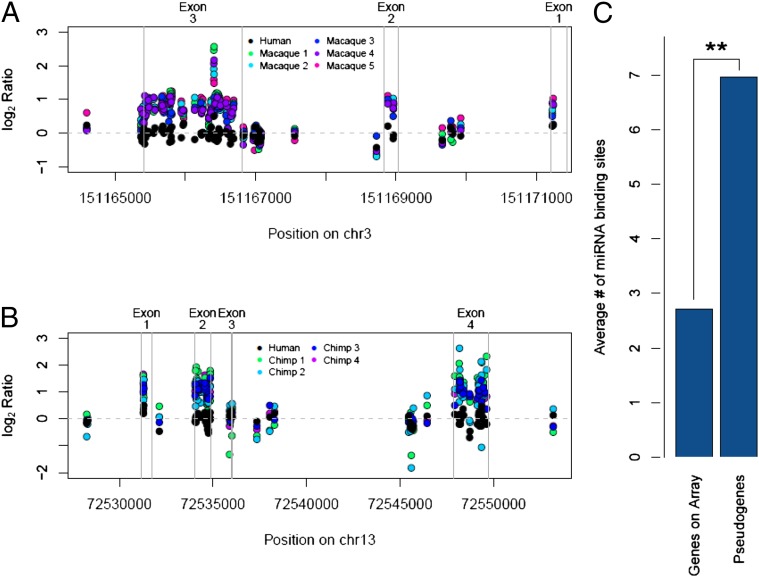
We observed that some of the genes overlapped by CNDs appeared to have gains in exonic sequence, but not in intronic sequence. Processed pseudogenes are DNA sequences that resemble known genes but are lacking introns, as they are copied from mRNA sequences and incorporated into the genome. With aCGH, the presence of processed pseudogenes can be determined by copy number gains in the exons of a given gene but with no similar gains for the corresponding intronic sequences (Fig. 5 and Fig. S7). By using a custom pipeline to look for this pattern, we found 127 genes with processed pseudogenes in human, chimpanzee, and rhesus macaque (SI Materials and Methods). Sixty of these processed pseudogenes were confirmed by their presence in their respective reference genomes. The remaining 67 have not been previously observed (Dataset S8).
Fig. 5.
Species-specific processed pseudogenes are common among primates. (A) The log2 ratios across the PFN2 gene for one human and five macaque samples. Exons are demarcated with vertical gray lines. Exons are in reverse order because the gene is on the minus strand relative to hg18. Note how probes are elevated in the exons of macaque samples, but not in the introns. The distribution of elevated probes indicates the presence of a processed pseudogene in macaques. (B) The log2 ratios across the KLF5 gene for one human and four chimpanzee samples. Note how probes are elevated in the exons of chimpanzee samples, but not in the introns. The distribution of probes indicates the presence of a processed pseudogene in chimpanzees. (C) The average number of miRNA binding sites per gene is enriched among processed pseudogenes. Genes on the array were defined by having at least one exon overlapped by three or more probes. The processed pseudogenes are a nonredundant list of the genes in Dataset S8. miRNA binding sites per gene were estimated by using overlap with the miRNA binding site track from the University of California, Santa Cruz, Genome Browser. The difference between the two gene sets was significant (P < 0.01, χ2 test with Yates continuity correction).
As a result of the mechanism of processed pseudogene formation, we observed an excess of 3′ UTR duplications relative to other exons within genes (P < 0.01, χ2 test taking into consideration the exons that could be assessed by our array platform; Fig. S7). As the majority of miRNA binding sites are found in 3′ UTRs and an enrichment of copy number variable miRNA binding sites was recently reported among humans (25), we examined whether copy number different processed pseudogenes, like human CNVs, are enriched for these sites. Indeed, we found an enrichment of miRNA binding sites within species-specific processed pseudogenes (relative to all genes that could be assessed by our array, P < 0.001, χ2 test; Fig. 5C and Dataset S9). By using RNAseq reads that align to informative sites (i.e., sequence differences between the processed pseudogene and the “parental” gene for those processed pseudogenes present in their reference genomes; Fig. S7), we identified eight expressed processed pseudogenes in rhesus macaque and six in chimpanzee (Fig. S7).
We also interrogated other noncoding regulatory regions, including ultraconserved elements (UCEs; regions of ≥200 bp that are identical in sequence across multiple species) (26). We identified 59 copy number-different UCEs (Dataset S10), six of which were present in their respective reference genomes. Some UCEs have been shown to have regulatory function (27) and, as such, we suspect that CNDs overlapping UCEs may be a prominent cause of gene expression differences for nearby and distant genes (Fig. S8). We also identified 200 long intergenic noncoding RNA genes (lincRNAs; as defined in ref. 28) that have different copy numbers across primate species (Dataset S11 and Fig. S8). lincRNAs are genes that do not appear to code for protein products, but, instead, their RNAs have multiple regulatory roles (28, 29). The vast majority (91%) of lincRNAs we could assess by our arrays were not expressed at a detectable level in LCLs based on the RNAseq data (the lincRNA had fewer than an average of five reads in each of the three species); thus, if CNDs overlapping lincRNA genes affect the expression levels of those genes (as appears to be the case for protein coding genes; Fig. 2), we would need to explore lincRNA gene expression levels in other tissues and cell types to observe this effect.
Discussion
Previously, species-specific aCGH platforms have been used to determine copy number variation between individuals within a given primate species (e.g., refs. 30–32). These arrays have also been used to perform cross-species comparisons (33, 34). However, interpretation of data from such studies can be complicated because of the substantial amount of noise that results from sequence mismatches between the probes and genomic DNA (reviewed in ref. 35). In this study, we used probe sequences that are 100% identical across the reference genomes of multiple primate species (Dataset S1). Thus, we were able to identify CNDs of sequences that are highly conserved among primate species at the DNA sequence level, but vary in copy number between species. The array should be considered as a targeted platform for conserved regions and not as an unbiased genome-wide array.
The duplication of a gene can have multiple possible consequences (reviewed in ref. 36), two of which are (i) an increase in the gene’s expression level as a result of dosage or (ii) the paralogues possibly diverging from each other, with one or both taking on new functions. In this study, we find examples of both scenarios among primates. The chimpanzee-specific duplication of KANK1 can best be described by the first scenario. Interestingly, deletions of this gene have been implicated in underdeveloped gonads (37). If deletions cause hypogonadism, it is possible that duplications would lead to enlarged gonads. Such “mirrored” phenotypes have been observed for CNVs in humans (38, 39). In fact, chimpanzees have substantially larger testes-to-body weight ratios than humans, related to the increased competition of chimpanzee males during mating (40). By using qPCR, we found that bonobos and ring-tailed lemurs may also have additional copies of KANK1, and that these additional copies are likely variable and not fixed (Fig. 3B and SI Materials and Methods). Both these species also have multimale mating habits and large testes-to-body weight ratios (40, 41). We did not observe a gain of KANK1 in any species with small testes-to-body weight ratios; however, additional primate species and samples within each species should be examined to determine whether KANK1 copy number and testes-to-body weight ratios are significantly associated. Taken together, these observations suggest that CNDs in KANK1 may play a role in the phenotypic variation of gonad size among primates.
The second scenario for gene duplications can best be described by Ohno’s theory of “neofunctionalization” in which gene duplications can take on new or specialized function (42). We observed such a scenario with the multiple gene duplications of the putative TF, ZNF669, in rhesus macaque. This TF gene not only experienced a massive increase in copy number among Old World monkeys, but the individual gene copies appear to be diverging from each other faster than expected under neutral conditions (Fig. S6). As such, not only has the expression level of ZNF669 (or highly similar genes) increased substantially in rhesus macaques, but members of this gene family are potentially taking on different, but related, functions. Of note, ZNF669 is substantially overexpressed in rhesus macaque brains compared with human and chimpanzee brains (43). Based on these observations, ZNF669 is an expanding gene family in Old World monkeys and may be an example of neofunctionalization.
In addition to ZNF669, we found that copy number different genes were enriched for regulatory functions including regulation of transcription, DNA binding, and RNA processing (Fig. 2C). TF genes are also enriched for differential expression levels across species in certain tissues (23). As the protein products of TF genes serve to regulate downstream genes, it is possible that the effects of single copy number different genes will be amplified in specific cellular and developmental contexts when the TF is most active. Interestingly, many of the regions we showed to be copy number different and DE between species are expressed highly in gonads (Fig. S9). It is possible that gene expression differences, driven by CNDs, alter the developing reproductive organs and may be related to sexual-selective pressures in primates, as is the case for the androgen receptor in humans (18).
Besides traditional protein coding genes, we found more than 100 species-specific processed pseudogenes, many of which contain miRNA binding sites and are expressed. Similarly, a processed pseudogene of the PTEN tumor suppressor gene was recently shown to contain miRNA binding sites and be expressed. As such, the pseudogene was able to titrate miRNAs away from binding to PTEN mRNA (44). This miRNA titration hypothesis has been implicated as a major regulator of changes in gene expression (28, 45–47). The varying copy number and expression level of processed pseudogenes among primates could affect the cross-talk between noncoding regulatory RNAs, ultimately leading to differing global gene expression profiles and phenotypic divergence.
In addition, we identified copy number-different UCEs among primates. One UCE that is gained in copy number in rhesus macaques is located immediately upstream of the SNX14 gene and in an alternatively spliced 3′ exon of the SYNCRIP gene (Fig. S8). Both these genes have differential expression levels when comparing chimpanzee and rhesus macaque, or human and rhesus macaque, but not when comparing human and chimpanzee (Fig. S8). It is possible that the copy number gain of this UCE in rhesus macaques may play a role in the expression differences of these two genes. Interestingly, the duplication of this UCE coincides with lower expression levels of both genes in rhesus macaque.
Conclusions
Altogether, sequences that are conserved at the nucleotide level between primate species are capable of differing in copy number and gene expression levels. Genomic CNDs may help drive species-specific gene expression profiles. Such expression differences can result from the direct overlap with CNDs or through the downstream effects of TF genes, miRNA titrators, and UCEs. As such, when considering the evolution of closely related species, we should examine genomes beyond just the nucleotide sequence level and include gene expression and copy number data, as these characteristics may also affect eventual phenotypes.
Materials and Methods
aCGHs.
Agilent catalog probes for human (hg18) were downloaded from eArray and aligned against the chimpanzee (panTro2) and rhesus macaque (rheMac2) reference genomes by using BLAT (48). Those probes that had at least one 100% match to both reference genomes were similarity filtered based on the human reference genome (i.e., probes with only one perfect hit to hg18 were considered for the array; Dataset S1). Agilent-recommended protocols were followed for DNA labeling and array hybridizations. Array data (Agilent feature extraction files) were imported into Nexus 5.0t and analyzed using the Rank Segmentation algorithm (SI Materials and Methods). All Feature Extraction files can be found in the Gene Expression Omnibus under accession no. GSE33960.
RNAseq.
RNA was extracted from LCLs by using standard protocols. cDNA libraries were created from polyadenylated RNA as described previously (49). Sequencing library preparation was executed using Illumina recommended protocols. cDNA was sequenced on an Illumina GA-II and aligned to the human (hg18), chimpanzee (panTro2), and rhesus macaque (rheMac2) reference genomes by using MAQ version 0.6.8 (50) with default parameters. Genes were analyzed for pairwise differential expression as described previously (20). RNAseq data can be found in the Gene Expression Omnibus under accession no. GSE38572.
qPCR.
Validations were performed by using Applied Biosystems TaqMan assays, NanoString Copy Count, and SYBR Green qPCR. SI Materials and Methods provides more details.
Supplementary Material
Acknowledgments
The authors thank Amy Langdon, Sunita Setlur, and Kim Brown for their insightful comments on earlier versions of this manuscript. This work was supported in part by National Institutes of Health Grants R01GM081533 and P41HG004221 (to C.L.) and training Grants F32AG039979 and 2T32HL007627-26.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE33960 (array comparative genomic hybridization) and GSE38572 (RNA sequencing data)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205199109/-/DCSupplemental.
References
- 1.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 2.Konopka G, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 5.Blekhman R, Oshlack A, Gilad Y. Segmental duplications contribute to gene expression differences between humans and chimpanzees. Genetics. 2009;182:627–630. doi: 10.1534/genetics.108.099960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad DF, et al. Wellcome Trust Case Control Consortium Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 8.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park H, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 11.Sudmant PH, et al. 1000 Genomes Project Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.She X, et al. NISC Comparative Sequencing Program A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson ME, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 15.Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 16.Niu AL, et al. Rapid evolution and copy number variation of primate RHOXF2, an X-linked homeobox gene involved in male reproduction and possibly brain function. BMC Evol Biol. 2011;11:298. doi: 10.1186/1471-2148-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orozco LD, et al. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet. 2009;18:4118–4129. doi: 10.1093/hmg/ddp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 22.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felekkis K, et al. Increased number of microRNA target sites in genes encoded in CNV regions. Evidence for an evolutionary genomic interaction. Mol Biol Evol. 2011;28:2421–2424. doi: 10.1093/molbev/msr078. [DOI] [PubMed] [Google Scholar]
- 26.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 27.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 28.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry GH, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee AS, et al. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 2008;17:1127–1136. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 32.Gazave E, et al. Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res. 2011;21:1626–1639. doi: 10.1101/gr.117242.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumas L, et al. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locke DP, et al. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gökçümen O, Lee C. Copy number variants (CNVs) in primate species using array-based comparative genomic hybridization. Methods. 2009;49:18–25. doi: 10.1016/j.ymeth.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Innan H, Kondrashov F. The evolution of gene duplications: Classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 37.Tannour-Louet M, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PloS ONE. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacquemont S, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harcourt AH, Harvey PH, Larson SG, Short RV. Testis weight, body weight and breeding system in primates. Nature. 1981;293:55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- 41.Schultz AH. The relative weight of the testes in primates. Anat Rec. 1938;8:309–394. [Google Scholar]
- 42.Ohno S. Evolution by Gene Duplication. London: Allen and Unwin; 1970. [Google Scholar]
- 43.Khaitovich P, et al. Positive selection on gene expression in the human brain. Curr Biol. 2006;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- 44.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.