Abstract
The phytohormone abscisic acid (ABA) plays an essential role in plant development and during the response of the plant to abiotic stress. In this study, we report that the R2R3-type transcription factor MYB30 is involved in the regulation of ABA signaling. Arabidopsis mutants lacking MYB30 are hypersensitive to ABA during germination and seedling growth. A K283R substitution in MYB30 blocks its SUMO E3 ligase SIZ1-mediated sumoylation in Arabidopsis protoplasts, indicating that MYB30 is sumoylated by SIZ1 and that K283 is the principal site for small ubiquitin-like modifier conjugation. Expression of MYB30K283R in myb30 partially rescues the mutant ABA-hypersensitive phenotype, but expression of wild-type MYB30 complements the mutant phenotype. Overexpression of MYB30 in wild-type results in an ABA-insensitive phenotype, whereas overexpression of MYB30 in the siz1 mutant does not alter siz1 hypersensitivity to ABA. The siz1-2 myb30-2 double-mutant exhibits greater ABA sensitivity than either single mutant, but a mutation in the SIZ1-sumoylated ABI5 transcription factor suppresses the ABA hypersensitivity of myb30-2 to wild-type levels. Our results suggest that coordination of ABI5 and MYB30 sumoylation by SIZ1 may balance gene expression, which is required for regulation of ABA signaling during seed germination.
Abscisic acid (ABA) plays important roles throughout the life cycle of the plant, including during seed development and dormancy, seed germination, early seedling development, flowering, and in response to abiotic and biotic stress. Genetic studies have revealed that a number of loci are essential for ABA biosynthesis (1–5), catabolism (6, 7), and signal transduction (8, 9) in plants.
A number of studies have focused on understanding ABA signaling from perception to response. Recently, three groups of ABA receptors (ABAR/GUN5, GRGC/GTG, and PYR/PYL/RCAR) have been identified in Arabidopsis and shown to bind ABA with high affinity (10–14). The various functions and cellular locations of these ABA receptors point to the complexity of ABA signaling. For example, PYR/PYL/RCAR-modulated ABA signaling has been studied extensively and found to involve a double-negative regulatory system that includes the receptor, protein phosphatases (PP2C), and protein kinases (SnRK2) (15, 16).
A group of basic leucine zipper transcription factors, including ABI5, serve as the downstream targets of SnRK2 and phosphorylation is correlated with ABI5 transcriptional activity and protein stability (17–19). ABI5 undergoes 26S proteasome-mediated degradation and ABA stabilizes the ABI5 protein (17). Two ABI5 interacting proteins, AFP (ABI5-interacting protein) and KEG (KEEP ON GOING, a RING-finger ubiquitin E3 ligase) are required for ABI5 degradation (20, 21). Arabidopsis lacking either gene is hypersensitive to ABA and an abi5 mutation suppresses the ABA sensitivity of both the afp-1 and keg mutants (20, 21). Recently it has been shown that ABI5 is sumoylated by the small ubiquitin-like modifier (SUMO) E3 ligase SIZ1 at amino acid K391, and this sumoylation is also required for the stability of ABI5. Consistent with these data, the siz1 mutant is hypersensitive to ABA and this phenotype is suppressed by the abi5 mutation (22).
Sumoylation affects protein–protein interaction, protein subcellular localization, enzymatic activity, and stability, which in turn regulate multiple biological processes, including the cell cycle, DNA repair, and transcriptional activity (23, 24). In addition, sumoylation is important for hormonal and environmental responses in plants (25, 26). The identified SIZ1 targets are mostly transcription factors, such as PHR1, ICE1, FLD, and ABI5, which function in numerous biological pathways (22, 27–30).
In this study, the myb30-2 transcription factor was identified in a mutant screen for components of ABA signaling in Arabidopsis. We show that myb30-2 is hypersensitive to ABA during germination compared with wild type or abi5-8, and that sumoylation of MYB30 at amino acid K283 by SIZ1 is crucial for its function in ABA signaling. In addition, we found that MYB30 and ABI5 regulate largely different sets of genes and the abi5-8 myb30-2 double-mutant exhibits wild-type–like ABA sensitivity. Our results indicate that MYB30 and ABI5 function in parallel pathways to regulate germination and that their SIZ1-dependent sumoylation results in coordinate regulation of germination in response to ABA.
Results
Genetic Screening for Arabidopsis Mutants Defective in Response to ABA.
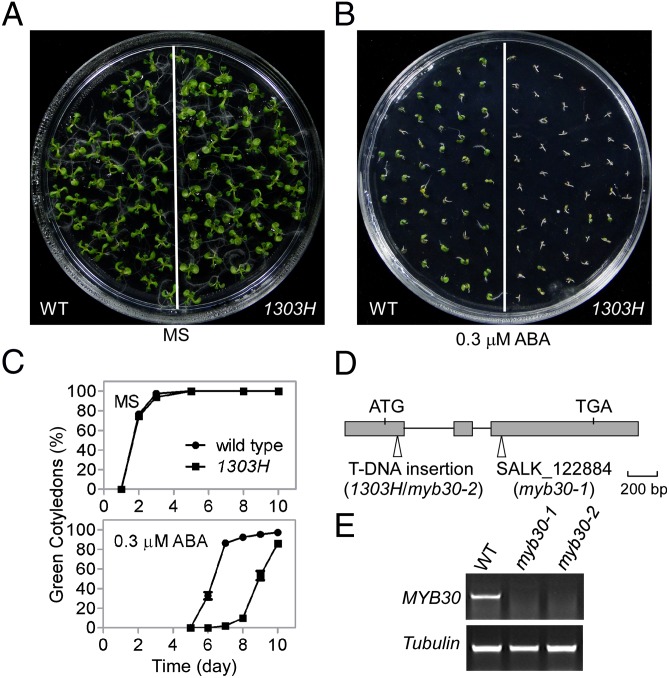
To identify components of ABA signaling in Arabidopsis, we screened a T-DNA (Agrobacterium-transferred DNA) insertion pool (Col-0 background) for changes in seed germination in the presence of exogenous ABA and identified the 1303H mutant, which was hypersensitive to ABA (Fig. 1 A–C). Insertion of the T-DNA fragment in the mutant was detected (TAIL-PCR) in exon one of At3g28910, which encodes the MYB30 transcription factor that has previously been shown to be involved in plant defense responses (31–36) (Fig. 1D). As a result, 1303H was renamed as myb30-2. The phenotype of myb30-1 (SALK_122884), an allelic mutant obtained from the Arabidopsis Biological Resource Center (ABRC), was similar to that of myb30-2 (Fig. 1D and Fig. S1). The expression of MYB30 was abolished in both myb30-1 and myb30-2 (Fig. 1E). Germination and primary root growth of the myb30 mutants was hypersensitive to ABA (0.1–0.5 μM), compared with these processes in wild-type (Fig. 1 A–C and Fig. S1). Cotyledon greenness of the myb30 mutants was also sensitive to mannitol. Because there was no significant difference in seedling growth and leaf water loss under osmotic/drought stress between the mutant and wild-type (Fig. S1), mutant sensitivity to mannitol postgermination provides additional evidence that MYB30 mainly functions in regulation of seed germination and very early seedling development.
Fig. 1.
myb30 is hypersensitive to ABA. (A and B) ABA sensitivity of wild-type and the 1303H/myb30-2 mutant. Photographs were taken 8 d after sowing seeds on MS medium without (A) or with 0.3 μM ABA (B). (C) Germination (green cotyledons) frequencies were scored at the indicated times and represent an average of 100 seeds from three independent experiments ± SE. (D) The gene structure of At3g28910. Filled boxes indicate exons and the lines indicate introns. Sites of insertions in the myb30-2 and myb30-1 mutants are marked. (E) Expression of MYB30 in wild-type, myb30-1, and myb30-2 mutant seedlings was analyzed by RT-PCR with Tubulin included as a loading control.
MYB30 and ABI5 Regulate the Expression of Largely Different Sets of Genes.
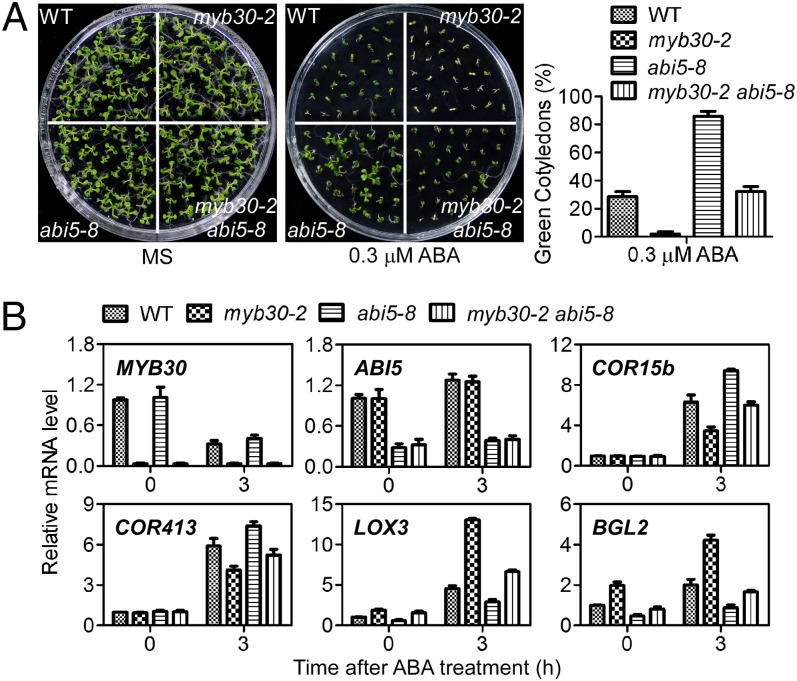
To determine if MYB30 functions in previously identified ABA signaling pathways, we crossed myb30-2 to abi1-3, abi2-2, abi3, abi4, and abi5-8 (all in Col-0) (37–41). Double-mutants of abi1-3 myb30-2 and abi2-2 myb30-2 exhibited enhanced ABA sensitivity relative to their single mutants, suggesting that MYB30 functions in parallel pathways with ABI1 or ABI2 during ABA signaling (Fig. S2). Double-mutants of abi3 myb30-2 and abi4 myb30-2 exhibited an ABA-insensitive phenotype similar to abi3 and abi4 single mutants (Fig. S2), suggesting that abi3 and abi4 are epistatic to myb30. However, double-mutants of abi5-8 myb30-2 exhibited a wild-type–like phenotype during seed germination (Fig. 2A), suggesting that ABI5 and MYB30 may function in parallel pathways to coordinately regulate ABA signaling.
Fig. 2.
MYB30 and ABI5 coordinately regulate ABA signaling. (A) The ABA sensitivity of myb30-2 was suppressed by abi5-8. Photographs were taken 9 d after seeds were sown on MS medium without (Left) or with 0.3 μM ABA (Right). The mean germination values for wild-type and the myb30-2, abi5 single, and abi5 double-mutants in response to 0.3 μM ABA is shown in the graph to the right of the images. Green cotyledons were scored after 6 d of growth and represent an average of 100 seeds from at least three independent experiments ± SE. (B) Seven-day-old seedlings of wild-type, myb30-2, abi5-8, and myb30-2 abi5-8 were treated without or with ABA (100 μM) for 3 h. Relative mRNA levels were determined by quantitative RT-PCR analysis. Transcript levels of MYB30, ABI5, COR15b, COR413, LOX3, and BGL2 are illustrated. Three independent experiments were performed; data represent means ± SD.
MYB30 has been shown to regulate defense and brassinosteroid (BR) responses by mediating the expression of genes involved in very long-chain fatty acid biosynthesis and the BES1 (bri1-Ethylmethane Sulphonate suppressor1) transcriptional activator in BR signaling, respectively (33, 34). To determine if MYB30 also regulates ABI5 target genes in response to ABA, expression of EM1, EM6, RAB18, ADH, and RD29 was examined in myb-30 mutants; no significant difference was observed between wild-type and myb30. Consistent with these results, the expression of ABI5 did not change in myb30, and expression of MYB30 was not affected by the abi5 mutation (Fig. 2B). These results suggest that ABI5 and MYB30 regulate different sets of genes and that ABI5 and MYB30 do not regulate each other.
The expression of MYB30 was reduced by ABA treatment (Fig. 2B). ABA induces the expression of many plant genes associated with germination and dehydration responses (42). To identify ABA-responsive genes regulated by MYB30, wild-type and myb30-2 mutant seedlings were treated with 100 μM ABA for 3 h, and ABA-responsive gene expression was examined by Solexa mRNA sequencing analysis. The expression of 120 vs. 41 genes was induced and reduced 1.5-fold in myb30-2 compared with expression in wild-type (Table S1). Expression differences of four stress-responsive genes, COR413, COR15B, LOX3, and BGL2, were confirmed by real-time PCR (Fig. 2B). In the myb30-2 mutant, the induction of COR413 and COR15B was substantially lower and the induction of LOX3 and BGL2 was significantly higher than their expression in wild-type. The expression of COR15B, LOX3, and BGL2 was regulated in the opposite manner by ABA in the abi5-8 and myb30-2 mutants (Fig. 2B). Consistent with the seed-germination phenotype in single and double myb30 and abi5 plants, the expression of COR413, COR15B, LOX3, and BGL2 in response to ABA in the abi5-8 myb30-2 double-mutant was similar to that in wild-type, suggesting that MYB30 and ABI5 may work in parallel to regulate the balance of gene expression during ABA signaling (Fig. 2B).
MYB30 Sumoylation at Residue K283 Is Crucial for the Response to ABA.
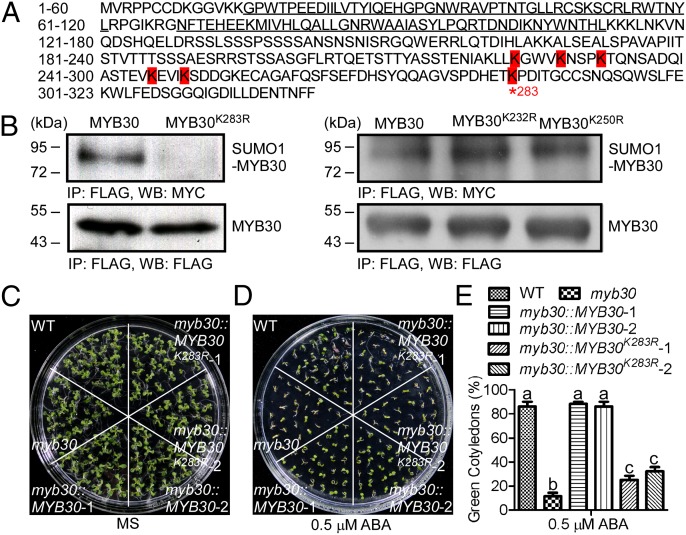
Okada et al. (43) used SUMO E1 and E2 enzymes to reconstitute the Arabidopsis sumoylation cascade in Escherichia coli and showed that the K283 of MYB30 was the major sumoylation site (Fig. 3A). To determine if MYB30 is sumoylated in Arabidopsis, and if the target is K283, we generated 35S:Myc-SUMO1, 35S:Flag-MYB30, 35S:Flag-MYB30K283R, 35S:Flag-MYB30K232R, and 35S:Flag-MYB30K250R constructs. Myc-SUMO1 plasmid was cotransformed with these Flag-MYB30 plasmids individually into wild-type protoplasts. SUMO1 conjugation of MYB30 (Fig. 3B) was detected in protein extracts isolated from MYB30, MYB30K232R, and MYB30K250R, but not in MYB30K283R. These results indicate that the sumoylation of MYB30 occurs in Arabidopsis at residue K283.
Fig. 3.
MYB30 is sumoylated at residue K283. (A) Amino acids marked with red indicate predicted sumoylation sites in MYB30. K283, the major sumoylation site in vitro (43), is marked by an asterisk. The MYB DNA binding domain is underlined. (B) Amino acid K283 is the sumoylation site in vivo. Combinations of Myc-SUMO1 and Flag-MYB30, Flag-MYB30K283R, Flag-MYB30K232R, or Flag-MYB30K250R were coexpressed in Arabidopsis protoplasts. Soluble extracts from protoplasts were immunoprecipitated with anti-Flag antibody (IP: FLAG) and the immunoprecipitated products were detected with anti-Myc antibody to monitor SUMO-MYB30 conjugates (Output: MYC). Protein loading was detected with anti-Flag antibody (Input: FLAG). (C and D) MYB30K283 sumoylation is required for the response to ABA. Photographs were taken 10 d after seeds were sown on MS medium without (C) or with 0.5 μM ABA (D). (E) Germination frequencies were scored after 10 d of growth and represent an average of 100 seeds from three independent experiments ± SE. Based on a Student t test, genotypes with different letters are statistically different, P < 0.05.
To examine if sumoylation of MYB30 is required for its function in ABA signaling, we transferred 35S:Flag-MYB30 or 35S:Flag-MYB30K283R plasmids into myb30-2 plants. Two transgenic lines with equivalent expression of MYB30K283R or MYB30 (Fig. S3) were tested for ABA sensitivity. Expression of MYB30 rescued the myb30-2 ABA-sensitive phenotype; however, expression of an equivalent amount of MYB30K283R only partially reduced myb30-2 ABA sensitivity (Fig. 3 C–E), suggesting that sumoylation of K283 is critical for MYB30 function during ABA signaling.
SIZ1 Mediates the Sumoylation of MYB30.
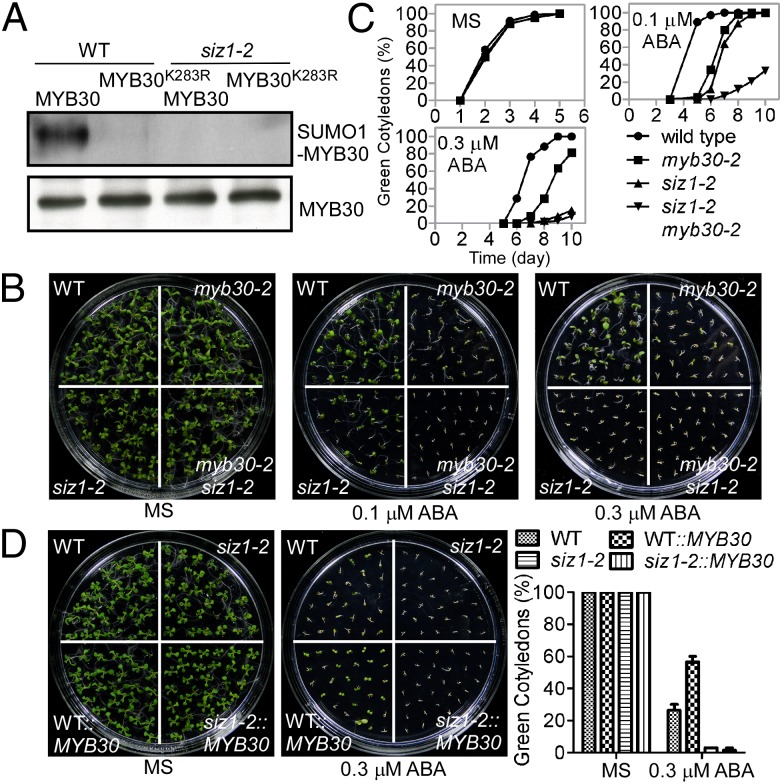
SIZ1 is a SUMO E3 ligase identified in Arabidopsis and shown to play an important role in responses to ABA through sumoylation of ABI5 (22). Our results showing that MYB30 is sumoylated in Arabidopsis and works in parallel with ABI5 suggested that MYB30 may also be a target for SIZ1. To test this hypothesis, 35S:Myc-SUMO1 and 35S:Flag-MYB30 or 35S:Flag-MYB30K283R were transferred into either wild-type or siz1-2 protoplasts, and the sumoylation of MYB30 was monitored. Sumoylation was detected in wild-type but not in the siz1-2 mutant, demonstrating that SIZ1 modulates the sumoylation of MYB30 (Fig. 4A).
Fig. 4.
SIZ1 mediates the sumoylation of MYB30. (A) SIZ1 mediates the sumoylation of MYB30 at K283. MYB30-SUMO1 conjugation was detected in wild-type but not in siz1-2 protoplasts and mutation of MYB30 at K283 (MYB30K283R) abolished this activity. (B) ABA phenotype of the myb30-2 siz1-2 double-mutant. Seeds were sown on MS medium without (Left), or with 0.1 μM (Center) or 0.3 μM (Right) ABA and photographs were taken after 9, 7, or 9 d, respectively. (C) Green cotyledons were scored at the indicated times and represent an average of 100 seeds from at least three independent experiments ± SE. (D) SIZ1 regulates MYB30 function during ABA signaling. Flag-MYB30 was expressed in wild-type and the siz1-2 mutant and T4 transgenic lines were used for phenotypic analyses. ABA sensitivity was monitored as described in (B and C). Seeds were sown on MS medium without (Left) or with 0.3 μM ABA (Right) and photographs were taken after 9 or 6 d, respectively. The mean germination values for wild-type, the siz1 mutant, and wild-type and the siz1 mutant overexpressing MYB30 in response to 0.3 μM ABA is shown in the graph to the right of the images. Green cotyledons were scored after 9 (MS) or 6 (0.3 μM ABA) d of growth and represent an average of 100 seeds from at least three independent experiments ± SE.
To determine if SIZ1-dependent sumoylation of MYB30 is critical for the response of the plant to ABA, we generated the siz1-2 myb30-2 double-mutant and expressed Flag-MYB30 in wild-type and siz1-2 plants. The double mutant exhibited enhanced ABA sensitivity compared with either single mutant (Fig. 4 B and C and Fig. S3). Overexpression of MYB30 in wild-type resulted in transgenic plants that were insensitive to ABA during germination (Fig. 4D); however, overexpression of MYB30 at a similar level in siz1-2 (Fig. S3) did not affect the ABA-sensitive phenotype of the mutant (Fig. 4D). The ABA-sensitive phenotype of the siz1 mutant may be because of the activation of ABI5 and inactivation of MYB30. In the siz1-2 myb30-2 double-mutant, MBY30 is absent and ABI5 is activated; therefore, the double-mutant would be expected to be more sensitive to ABA than either single mutant.
SIZ1 Stabilizes MYB30 During ABA Treatment.
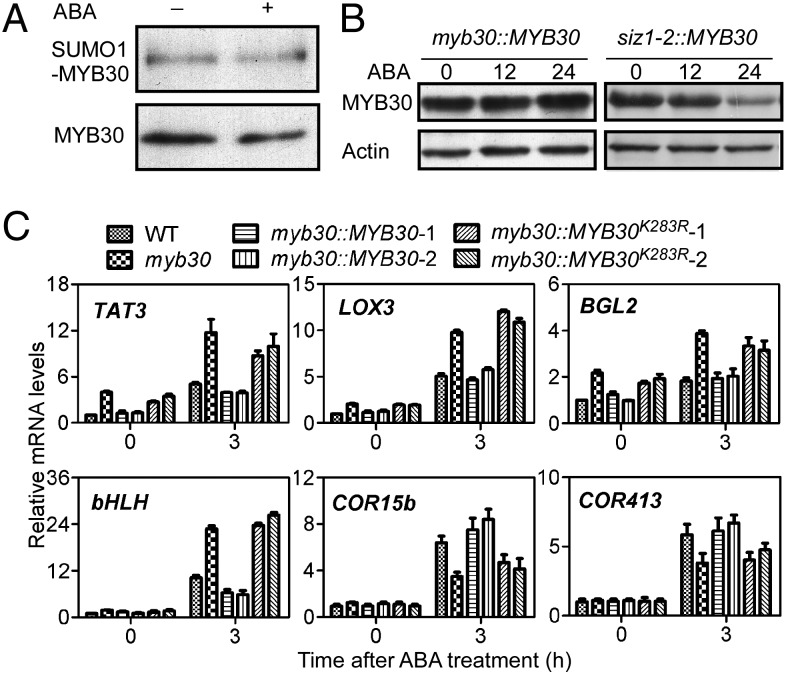
Sumoylation plays a role in protein–protein interaction, protein subcellular localization, enzymatic activity, and stability. To determine the biological function of MYB30 sumoylation, we expressed 35S:MYB30-GFP or 35S:MYB30K283R-GFP in wild-type protoplasts. Both MYB30-GFP and MYB30K283R-GFP localized to the nucleus (Fig. S4A). To examine if sumoylation changes MYB30 transcriptional activity, the Arabidopsis proteins were expressed in yeast, as previously reported (29). The MYB30K283R mutation did not significantly affect MYB30 transcriptional activity in yeast (Fig. S4B). The 35S:Flag-MYB30 transgenic wild-type and siz1-2 lines were then used to determine if sumoylation affects the stability of MYB30. The sumoylation of MYB30 was not significantly affected by ABA treatment (Fig. 5A); however, in the presence of ABA, MYB30 was less abundant in the siz1-2 seedlings than in the wild-type seedlings (Fig. 5B). These data suggest that sumoylation by SIZ1 stabilizes MYB30. The expression of the TAT3, LOX3, BGL2, bHLH, COR15b, and COR413 ABA responsive genes was rescued by expression MYB30 but not by MYB30K283R (Fig. 5C). These data, together with the results showing that MYB30K283R partially reduced myb30-2 ABA sensitivity, suggest that sumoylation of MYB30 by SIZ1 may also affect its transcriptional activity.
Fig. 5.
MYB30K283 sumoylation is required for the response to ABA. (A) Sumoylation of MYB30 was not significantly affected by ABA. Flag-MYB30 and Myc-SUMO1 were coexpressed in wild-type protoplasts. For the ABA treatment, the protoplasts were incubated with 40 μM ABA for 1 h. Sumoylation was detected as described in Fig 3B. (B) ABA-induced MYB30 degradation in the siz1-2 mutant. Flag-MYB30 was expressed in wild-type and the siz1-2 mutant. T4 transgenic lines were used to test protein stability. Eight-day-old seedlings were treated without or with ABA (100 μM) for 12 or 24 h. MYB30 levels in wild-type or siz1-2 seedlings were detected using immunoblots with anti-Flag antibody. Actin was used as a loading control. (C) Transcript abundance of TAT3, LOX3, BGL2, bHLH, COR15b, and COR413 in different transgenic lines was determined by quantitative PCR using 1-wk-old seedlings without or with 100 μM ABA treatment for 3 h. mRNA levels are expressed relative to the value of wild-type seedlings before ABA treatment. Data represent means ± SD for three independent experiments.
Discussion
Results from our studies demonstrate that MYB30 is a component of ABA signaling in Arabidopsis. MYB30 is a R2R3-MYB transcription factor that has been shown to be a positive regulator of the hypersensitive response (HR), programmed cell death associated with disease resistance in plants, and of BR signaling. Overexpression of MYB30 in Arabidopsis and tobacco increases resistance to different pathogens, whereas loss of MYB30 function in Arabidopsis reduces HR-mediated cell death and pathogen resistance (31). MYB30 modulates HR and pathogen resistance via regulation of salicylic acid (SA) levels and SA-associated gene expression (32). In addition to its role in the HR, MYB30 functions to amplify BR signaling by directly interacting with BES1 and optimizing BES1 activity (34). Our results demonstrate that MYB30 is also involved in ABA signaling. Loss of MYB30 function leads to hypersensitivity to ABA during germination. SIZ1 sumoylation of both MYB30 (at residue K283) and the ABI5 transcription factor leads to regulation of parallel pathways that coordinate germination in response to ABA.
Gene-expression analyses also suggest that MYB30 plays a role in ABA signaling in addition to its functions in HR and BR signaling. Eighteen genes were identified as possible targets of MYB30 during the HR response (33), 16 of which are also regulated by ABA treatment. More than 100 genes were reduced at least twofold in the myb30 mutant, with the expression of 13 changing significantly in response to BR (34) and 6 in response to ABA treatment. However, none of these genes changed expression in the myb30 mutant compared with their expression in wild-type in response to ABA, suggesting that MYB30 regulates different target genes during HR, BR, and ABA signaling. The expression of MYB30 is induced by bacterial pathogens (44) and Brassinolide (34) but reduced by ABA (Fig. 2B), providing additional evidence for MYB30 in these processes.
Functional and gene-expression studies link SIZ1 to MYB30 during ABA signaling. SIZ1 stabilizes MYB30 upon ABA treatment and is required for the sumoylation of MYB30 at K283, which is critical for MYB30 function in response to ABA. Overexpression of MYB30 in wild-type results in the transgenic plants that are insensitive to ABA during seed germination; however, this phenomenon is abolished in the siz1 mutant, suggesting that the sumoylation of MYB30 regulates its activity in Arabidopsis, although it is likely not the case in yeast (Fig. S4B).
SIZ1 has been shown to modulate gene expression in response to HR, BR, and ABA signaling, as well as during the transition to flowering (22, 26, 45–48). For example, SIZ1 regulates systemic-acquired resistance through both NONEXPRESSOR OF PR1 (NPR1) gene-dependent and SA accumulation-dependent pathways (45). Results from the present studies provide additional support for a link between SIZ1 and ABA signaling. Expression of ABA-responsive genes displays similar patterns in the siz1 and myb30 mutants (Fig. S5); however, MYB30 mediates SA levels and SA-associated gene expression, most likely independently of NPR1 (32), suggesting that MYB30 also regulates different sets of genes.
Several lines of evidence suggest that SIZ1 sumoylation of MYB30 and ABI5 regulates ABA signaling during germination. (i) Loss-of-function of the two transcription factors alters germination in response to ABA (Fig. 2A). (ii) The germination response of the abi5 myb30 double-mutant restores wild-type responses to ABA during germination (Fig. 2A). (iii) The stability of the two transcription factors is likely regulated by SIZ1 sumoylation in response to ABA (22) (Fig. 5B).
In addition, our results suggest that MYB30 and ABI5 regulate parallel pathways during germination in response to ABA. The two transcription factors regulate different sets of genes in response to SIZ1 sumoylation (22) (Fig. S5) and different subgroups of ABA-responsive genes (Table S1). Taken together, our results suggest that regulation of ABI5 and MYB30 by SIZ1 sumoylation could serve to balance gene expression in response to ABA (Fig. S6).
Materials and Methods
Plant Growth and ABA Treatment.
Arabidopsis seeds were sown on Murashige and Skoog (MS) medium with 0.3% agar, kept in the dark for 3 d at 4 °C to break dormancy, and then grown in a growth chamber at 23 °C in constant light. To monitor ABA sensitivity, seeds were sown on MS medium or MS medium containing the indicated concentrations of ABA (Sigma). Germination frequencies were obtained by scoring green cotyledons for an average of 100 seeds from three independent experiments ± SE. Stable WT::MYB30 and siz1-2::MYB30 T4 lines were used to monitor ABA-induced MYB30 degradation. Eight-day-old seedlings were treated with water (0 h) or ABA for 12 or 24 h. Seedlings were homogenized on ices in extraction buffer [10 mM Tris (pH 7.5), 0.5% Nonidet P-40, 2 mM EDTA, 150 mM NaCl, 1 mM PMSF, and 1% (vol/vol) protease inhibitor mixture (Sigma)]. Extracts were centrifuged at 12,000 × g for 15 min at 4 °C and the resulting supernatant was used to detect MYB30 levels in wild-type or siz1-2 seedlings by immunoblot analysis.
The Arabidopsis siz1-2 mutant was kindly provided by Paul M. Hasegawa (Purdue University, West Lafayette, IN). The myb30-1 (SALK_122884), abi1-3 (SALK_076309), abi2-2 (SALK_015166), abi3 (SALK_138922), abi4 (SALK_080095), and abi5-8 (SALK_013163) mutants were obtained from the ABRC.
Transgenic Plants.
The MYB30 or MYB30K283R coding region was cloned into the pCAMBIA1307-Flag binary vector driven by a cauliflower mosaic virus 35S promoter. MYB30 or MYB30 (K283R) overexpressing transgenic plants (in the Col-0, myb30-2 or siz1-2 backgrounds) were generated using Agrobacterium-mediated floral transformation. RT-PCR and immunoblot analysis were used to detect the abundance of the transgenes and proteins. Stable WT::MYB30, siz1-2::MYB30, myb30-2::MYB30 and myb30-2::MYB30K283R T4 lines were used for phenotypic analyses.
Quantitative RT-PCR Analysis.
Seven-day-old seedlings grown on MS plates were used for ABA treatments. The seedlings were harvested for RNA extraction after treatment with 100 μM ABA for 3 h. Total RNA isolation, reverse transcription and quantitative real-time PCR amplification were as described (27, 29). The primer pairs used are shown in Table S2. The value of wild-type without ABA treatment (0 h) was normalized to 1 with differences calculated as relative expression.
In Vivo SUMO Conjugation Assays.
For transient expression of MYB30 and SUMO1 in Arabidopsis protoplasts, the SUMO1 coding region was amplified and inserted into the pCAMBIA1205-Myc plasmid to produce the Myc-SUMO1 fusion protein.
The 35S:Flag-MYB30 and 35S:Myc-SUMO1 were coexpressed in wild-type or siz1-2 protoplasts by polyethylene glycol-mediated transformation (22). After a 20-h incubation at 23 °C, the protoplasts were harvested and lysed in 1 mL extraction buffer. For ABA treatment, protoplasts were incubated with ABA (at a final concentration of 40 μM) at 23 °C for 1 h (22).
Fifteen microliters of anti-Flag agarose conjugate (Sigma) were incubated with the extract supernatant for 2 h at 4 °C. After washing five times in 1 mL of extraction buffer, proteins were detected by immunoblot analysis (49). Anti-Myc (Sigma) antibody was used to detect the Flag-MYB30-Myc-SUMO1 conjugation. Anti-Flag (Sigma) antibody was used to detect the abundance of Flag-MYB30 as a control.
Supplementary Material
Acknowledgments
We thank Dr. Jingbo Jin for critical reading of the manuscript and stimulating discussions and the Arabidopsis Biological Resource Center for the T-DNA insertion lines. This work was supported by China National Funds for Distinguished Young Scientists Grant 31025003 (to Y.G.); National Basic Research Program of China Grant 2012CB114200 (to Y.G.); Foundation for Innovative Research Group of the National Natural Science Foundation of China Grant 31121002 (to Y.G.); National Transgenic Research Project Grant 2008ZX08009-002 (to Y.G.); and US Department of Energy/Energy Biosciences Grant DE-FG02-04ER15616 (to K.S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202630109/-/DCSupplemental.
References
- 1.Xiong LM, Ishitani M, Lee HJ, Zhu J-K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem. 2001;276:40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- 3.Xiong LM, Lee HJ, Ishitani M, Zhu J-K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- 4.González-Guzmán M, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng WH, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushiro T, et al. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KH, et al. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006;126:1109–1120. doi: 10.1016/j.cell.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 8.Mizoguchi M, et al. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010;51:842–847. doi: 10.1093/pcp/pcq041. [DOI] [PubMed] [Google Scholar]
- 9.Klingler JP, Batelli G, Zhu J-K. ABA receptors the START of a new paradigm in phytohormone signalling. J Biochem. 2010;276:40381–40384. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen YY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, et al. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 12.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 14.Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 15.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii H, Verslues PE, Zhu J-K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Molina L, Mongrand S, Chua N-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii H, Verslues PE, Zhu J-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima K, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura K, et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Miura K, Lee J, Miura T, Hasegawa PM. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 2010;51:103–113. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- 27.Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo CY, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura K, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin JB, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vailleau F, et al. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raffaele S, Rivas S, Roby D. An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 2006;580:3498–3504. doi: 10.1016/j.febslet.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Raffaele S, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20:752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froidure S, et al. AtsPLA2-α nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc Natl Acad Sci USA. 2010;107:15281–15286. doi: 10.1073/pnas.1009056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canonne J, et al. The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell. 2011;23:3498–3511. doi: 10.1105/tpc.111.088815. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Saez A, et al. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 2006;141:1389–1399. doi: 10.1104/pp.106.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 2011;156:550–563. doi: 10.1104/pp.111.176214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu PF, Chang WC, Wang YK, Chang HY, Pan RL. Signaling pathways mediating the suppression of Arabidopsis thaliana Ku gene expression by abscisic acid. Biochim Biophys Acta. 2008;1779:164–174. doi: 10.1016/j.bbagrm.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Kakizaki T, et al. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009;151:1339–1353. doi: 10.1104/pp.109.145987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.W-K Ng D, Hall TC. PvALF and FUS3 activate expression from the phaseolin promoter by different mechanisms. Plant Mol Biol. 2008;66:233–244. doi: 10.1007/s11103-007-9265-5. [DOI] [PubMed] [Google Scholar]
- 42.Weiner JJ, Peterson FC, Volkman BF, Cutler SR. Structural and functional insights into core ABA signaling. Curr Opin Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okada S, et al. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: Functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 2009;50:1049–1061. doi: 10.1093/pcp/pcp056. [DOI] [PubMed] [Google Scholar]
- 44.Daniel X, Lacomme C, Morel JB, Roby D. A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 1999;20:57–66. doi: 10.1046/j.1365-313x.1999.00578.x. [DOI] [PubMed] [Google Scholar]
- 45.Lee J, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 46.Catala R, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin JB, Hasegawa PM. Flowering time regulation by the SUMO E3 ligase SIZ1. Plant Signal Behav. 2008;3:891–892. doi: 10.4161/psb.3.10.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.