Abstract
Aneuploidy, or an aberrant karyotype, results in developmental disabilities and has been implicated in tumorigenesis. However, the causes of aneuploidy-induced phenotypes and the consequences of aneuploidy on cell physiology remain poorly understood. We have performed a metaanalysis on gene expression data from aneuploid cells in diverse organisms, including yeast, plants, mice, and humans. We found highly related gene expression patterns that are conserved between species: genes that were involved in the response to stress were consistently upregulated, and genes associated with the cell cycle and cell proliferation were downregulated in aneuploid cells. Within species, different aneuploidies induced similar changes in gene expression, independent of the specific chromosomal aberrations. Taken together, our results demonstrate that aneuploidies of different chromosomes and in different organisms impact similar cellular pathways and cause a stereotypical antiproliferative response that must be overcome before transformation.
Keywords: stress response, trisomy, chromosomal instability
Eukaryotic organisms have evolved elaborate mechanisms that ensure that chromosomes are partitioned equally during cell division (1). Aberrant segregation events can result in aneuploidy, a condition in which cells acquire a karyotype that is not a whole-number multiple of the haploid complement. In humans, aneuploidy is the leading cause of spontaneous abortions and developmental disabilities, and aneuploid karyotypes are observed in greater than 90% of solid tumors (2–4). Thus, understanding the consequences of aneuploidy has broad relevance for the study of mammalian development and cancer.
The cause of aneuploidy-induced syndromes remains an open question. For instance, it has been hypothesized that the phenotypes associated with Down syndrome (trisomy 21) are caused by the triplication of a small set of genes that lie within a 5.4-Mb “Down syndrome critical region” on chromosome 21 (5, 6). However, evidence from mouse models suggest that this region is not sufficient to recapitulate Down syndrome–like phenotypes, and genetic mapping of partially trisomic individuals has revealed that numerous regions of chromosome 21 affect clinical presentation (7–9). Alternately, changes in gene dosage across an entire chromosome might have additive effects on organismal development (10). It has been observed that the three human trisomies that survive until birth (trisomy 13, 18, and 21) have the fewest protein-coding genes on them, implying that these karyotypes can be tolerated in utero because they have the lowest net dosage imbalances (11). However, the consequences of copy number variation range from benign to extremely deleterious, demonstrating that different genes exhibit varying levels of dosage sensitivity (12).
To examine the consequences of aneuploidy, we have previously constructed and analyzed a series of haploid budding yeast strains and mouse embryonic fibroblasts (MEFs) that carry single extra chromosomes (13–17). These aneuploid cells display defects in cell proliferation as well as sensitivity to conditions that interfere with protein folding and turnover. Microarray expression analyses revealed that haploid yeast strains carrying an additional chromosome (henceforth disomes) exhibit a common transcriptional signature, dubbed the environmental stress response (ESR). The ESR consists of ∼300 genes that are upregulated and ∼600 genes that are downregulated by various exogenous stresses, including heat shock or oxidative stress (18). Most of these genes also vary in expression in response to growth rate; inducing slow proliferation by nutrient limitation mimics the ESR (19, 20). No such common signature has been reported among aneuploid cells in any other organism. Additionally, it has been suggested (21) that the phenotypes detected in the disomic strains may be a unique consequence of the method that was used to generate aneuploidy, in which genetic markers were used to select for rare chromosome transfer events between nuclei (13). To further our understanding of aneuploidy, we examined gene expression data from aneuploid cells from diverse organisms. We detected a conserved transcriptional response that was associated with stress and decreased cell proliferation that was apparent in aneuploid yeast, plants, mice, and humans. These data suggest that aneuploidy in various species is detrimental to cell fitness, and that many consequences of aneuploidy are a common response to chromosomewide dosage imbalances.
Results
Aneuploid Strains of Budding Yeast Share a Chromosome-Independent Stress Response.
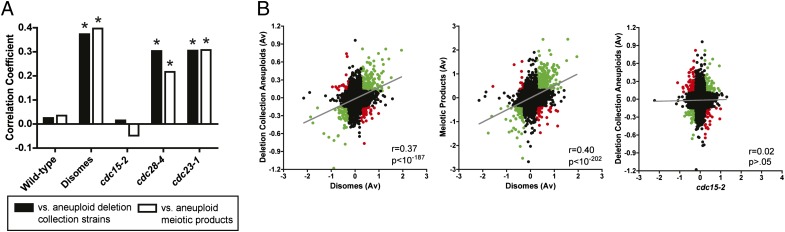
We previously reported that disomic yeast produced via chromosome transfer exhibit an ESR, in which genes related to RNA processing and the ribosome are downregulated, and genes involved in protein folding, detoxification of reactive oxidative species, and various other processes are upregulated (13). We sought to determine whether aneuploid cells generated in other ways also exhibited a stress and/or slow-growth response similar to that observed in the disomes. For all subsequent analyses, aneuploid chromosomes were excluded from consideration so as to avoid artifacts due to normalization. We first compared the disomes to 23 aneuploid strains generated during the construction of the yeast deletion collection (22). These aneuploidies include multiple chromosome gains and losses, and thus display more complex karyotypes than those present in the disomes. We found that the Pearson correlation coefficient (PCC) between the mean expression levels in these strains and in the disomic yeast was 0.37 (P < 10−187), while the PCC between these strains and a wild-type strain was 0.02 (P > 0.05), which demonstrated highly significant transcriptional similarity between these aneuploid populations (Fig. 1 A and B).
Fig. 1.
Transcriptional similarities among aneuploid strains of S. cerevisiae. (A) The correlation coefficients between either aneuploid strains from the yeast deletion collection (black bars) or aneuploid products of triploid meiosis (white bars) and the indicated strains of S. cerevisiae are displayed. Asterisks denote statistically significant correlations (P < 0.05). (B) Scatter plots comparing gene expression values between the indicated yeast strains. Points in green represent genes that are expressed ±1 SD from the mean codirectionally, and points in red represent genes that are expressed ±1 SD in opposite directions. Gray lines are linear regressions plotted against the data.
We hypothesized that the correlation between strains was due to a shared underlying stress or slow-growth response. To test this, we compared the aneuploid strains obtained from the deletion collection to gene expression data from three cdc mutant strains: cdc28-4 and cdc23-1, which divide slowly and exhibit a significant ESR at the permissive temperature, and cdc15-2, which proliferates at a wild-type rate at the permissive temperature and does not display an ESR (13). Both cdc28-4 and cdc23-1 exhibited significant correlations with aneuploid strains from the deletion collection (r = 0.30, P < 10−114, and r = 0.31, P < 10−116, respectively), while cdc15-2 was uncorrelated with the aneuploid strains (Fig. 1 A and B; r = 0.02, P > 0.05). Next, we quantified the intensity of the stress response in each disomic strain by averaging the expression levels of genes annotated to the ESR (Fig. S1). We found that 15 out of 16 disomes exhibited significant pairwise correlations with the average expression level in the aneuploid strains obtained from the deletion collection, and the strength of the correlation increased with the intensity of the stress response in the disomes (Figs. S2A and S2B; r = 0.64, P < 0.01). Disomes that did not display an ESR (e.g., disome I) exhibited minimal correlations with the deletion collection aneuploid strains, and disomes that displayed significant ESRs (e.g., disome IV) tended to exhibit stronger correlations. These results are consistent with our hypothesis that a shared transcriptional stress response underlies the similarity between different aneuploid populations.
We note, however, that the various deleted genes within the deletion collection are likely to be at least partially responsible for the stress phenotype. Two observations suggest that aneuploidy is also a relevant cause of the similarity with the disomic transcriptomes. First, among individual deletion strains, there was a significant positive correlation between the percent of the genome that was aneuploid and the PCC with the disomes (Fig. S3A; r = 0.54 P < 0.01). Second, we found that the stress response gained intensity as the number of genes on aneuploid chromosomes increased (Fig. S3B; r = 0.49, P < 0.05). This relationship was true for both aneuploid strains from the deletion collection and disomic strains that we constructed (r = 0.58, P < 0.0005). Thus, it is likely that aneuploidy contributes to the similarities in gene expression between these sets of strains.
We next sought to identify aneuploidy-responsive genes in yeast. We sorted the disomes and aneuploid deletion strains according to the number of genes present on aneuploid chromosomes, and then calculated correlation coefficients between the expression levels of each gene and the degree of aneuploidy across the panel of strains. There were 446 genes identified whose expression levels were significantly correlated or anticorrelated with the degree of aneuploidy across all strains (Fig. S4; PCC > 0.5 or PCC < −0.5; P < 0.002). Among transcripts that were positively correlated with increasing aneuploidy, Gene Ontology (GO) term analysis revealed an enrichment of genes related to the response to oxidative stress (P < 10−6) and protein refolding (Table S1; P < 10−3). Transcripts that decreased with increasing aneuploidy were enriched for noncoding RNA processing (P < 10−11) and ribosome biogenesis genes (P < 10−6). Importantly, there was highly significant overlap among aneuploidy-responsive genes and the ESR (P < 10−13, hypergeometric test). Furthermore, among the 446 genes, 414 of them exhibited a codirectional change in the slow-growing cdc mutants. Discordant transcripts (i.e., those that increased with aneuploidy but were expressed at less than wild-type levels in the cdc strains, and vice versa) were not significantly enriched for any GO terms. Taken together, these data indicate that most (but not all) transcriptional changes caused by aneuploidy are related to stress and/or slow growth, and that increasing degrees of aneuploidy generally exerts increasing degrees of stress on cell homeostasis.
Next, we obtained gene expression data from seven aneuploid strains that were derived via triploid meiosis (21). The average correlation between these strains and the disomes and five out of seven pairwise correlations with the disomes were highly significant (Fig. 1A and Fig. S3C). There was a mild anticorrelation between the aneuploid products of triploid meiosis and cdc15-2 (r = −0.05, P < 0.001), but a significant positive correlation with the ESR-exhibiting mutants cdc28-4 and cdc23-1 (r = 0.21, P < 10−55, and r = 0.30, P < 10−113, respectively). Among the five aneuploid strains that were correlated with the disomes, four showed a significant stress/slow growth response relative to a euploid strain (Fig. S3D). We conclude that a shared transcriptional response is a common although not obligate consequence of aneuploidy in yeast, and this response is independent of the mechanism by which aneuploidy is generated.
Aneuploidy Causes a Stress Response in Fission Yeast.
We next sought to determine whether aneuploidy causes a stress response in other organisms. We averaged gene expression data from two aneuploid strains of the fission yeast Schizosaccharomyces pombe, then identified upregulated and downregulated genes using a ±1.3-fold change (FC) cutoff (23). GO term analysis of upregulated genes revealed that the most enriched functional category was the response to stress (Table S2; P < 10−25). Downregulated genes included many terms associated with the ribosome, including ribosome biogenesis (P < 10−13) and the nucleolus (P < 10−12). Similar GO term enrichments were obtained using rank products, a cutoff-independent method of identifying differentially expressed genes [Table S3 (24)]. We noted that these GO terms are a hallmark of the budding yeast ESR, suggesting that aneuploidy in different yeasts causes a similar stress-related transcriptional response. Indeed, an environmental stress response has also been described in S. pombe (25), and out of 236 genes that constitute the fission yeast ESR, 203 genes exhibited codirectional transcriptional changes in the aneuploid S. pombe (Fig. S5).
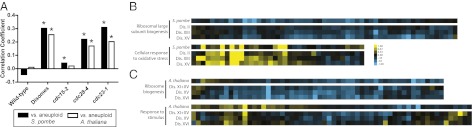
To determine whether aneuploidy caused genomewide similarities in gene expression in different species, we identified one-to-one orthologs between Saccharomyces cerevisiae and S. pombe and then calculated the PCC between the averaged aneuploid strains in each organism. The correlation coefficient between disomic budding and fission yeast strains was highly significant (Fig. 2A; r = 0.30, P < 10−47). In addition, there was a weak correlation between aneuploid fission yeast and cdc15-2 (r = 0.04, P < 0.05), but stronger correlations with cdc28-4 and cdc23-1 (r = 0.22, P < 10−26, and r = 0.31, P < 10−52). We found that 14 out of 16 individual disomes also exhibited significant pairwise correlations with S. pombe, and these transcriptional similarities were particularly striking when genes annotated to GO terms affected by aneuploidy were compared (Fig. 2B and Fig. S2C). Moreover, the PCC between individual disomes and S. pombe tended to increase based on the intensity of the stress response in each disomic strain (r = 0.60, P < 0.02; Fig. S2D). Last, we sought to determine whether specific groups of genes exhibited coordinate changes in expression in both species. Orthologous genes that were upregulated in both organisms were significantly enriched for those involved in the response to oxidative stress (P < 10−6) and the response to heat (P < 10−3), and downregulated genes were enriched for ribosome biogenesis factors (P < 10−6) and those associated with the nucleolus (Table S4; P < 10−3). We conclude that aneuploidy in different fungal species induces a highly related stress response.
Fig. 2.
Aneuploidy causes a stress response in S. pombe and A. thaliana. (A) The correlation coefficients between aneuploid strains of either S. pombe (black bars) or A. thaliana (white bars) and the indicated strains of S. cerevisiae are displayed. (B and C) Heat maps of orthologous genes annotated to aneuploidy-related GO terms from aneuploid strains of (B) S. pombe or (C) A. thaliana and the indicated disomes are displayed.
Aneuploidy Causes a Stress Response in Arabidopsis thaliana.
Based on the conserved transcriptional response to aneuploidy among different fungi, we hypothesized that aneuploidy in higher organisms could also result in a stress/decreased proliferation response similar to that seen in aneuploid yeast. To test this, we analyzed gene expression data from Arabidopsis thaliana plants that were trisomic for chromosome 5 (26). GO term enrichment analysis revealed that many of the same pathways were perturbed by aneuploidy in plants as in yeast (Tables S5 and S6). “Response to chemical stimulus” and “response to stress” were among the most upregulated GO terms (P < 10−24 and P < 10−19, respectively), and the cytosolic ribosome and ribosome biogenesis were highly enriched among downregulated genes (P < 10−11 and P < 10−8, respectively). Furthermore, we identified one-to-one orthologs between budding yeast and A. thaliana, and found that trisomic plants and disomic yeast exhibited a significant genomewide transcriptional correlation (Fig. 2A; r = 0.26, P < 10−5). There was no correlation between trisomic plants and cdc15-2 (r = 0.02, P > 0.05), but significant correlations with cdc28-4 and cdc23-1 (r = 0.17, P < 0.002 and r = 0.20, P < 0.0002, respectively) as well as 11 out of 16 individual disomes (Fig. 2C and Fig. S2E). As with S. pombe, the correlation coefficient between the yeast disomes and trisomic A. thaliana increased with the intensity of the stress response in the budding yeast strains (Fig. S2F; r = 0.69, P < 0.003). We conclude that a shared stress response underlies significant transcriptional similarity between aneuploid budding yeast and A. thaliana.
Aneuploid Mouse and Human Cells Share Slow Growth-Related Changes in Gene Expression.
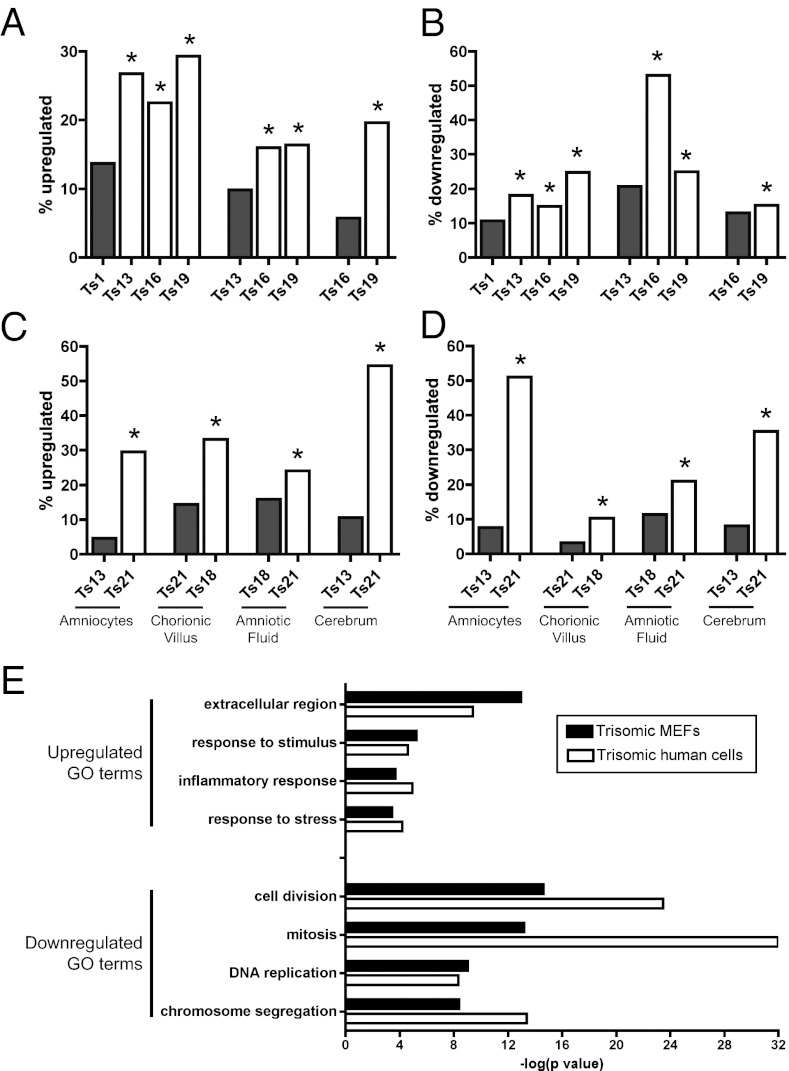
We next analyzed expression data from mouse embryonic fibroblasts trisomic for one of four chromosomes (chromosome 1, 13, 16, and 19) that were normalized to MEFs obtained from their euploid littermates (15). We first sought to determine whether different trisomies caused similar changes in gene expression. We found highly significant overlap among differentially expressed genes across the trisomies: in 12 out of 12 pairwise comparisons, a gene that was up- or downregulated in one trisomic cell line was significantly more likely to exhibit a similar change in expression in a different trisomy (Fig. 3 A and B). For instance, ∼6% of all genes on euploid chromosomes in trisomy 16 were upregulated at a 1.5-FC cutoff, but among genes that were upregulated in trisomy 19, 20% were also upregulated in trisomy 16 (P < 10−25, hypergeometric test). Significant similarities were also observed when differentially expressed genes were identified using t tests or more stringent FC cutoffs (Fig. S6 A–D). To determine whether the same genes were affected across the trisomic cell lines, we applied a permutation test, in which gene expression values were randomized within each trisomy. Although 78 genes were upregulated and 168 genes were downregulated in three or more trisomies, no more than 37 and 92 genes were up- or downregulated, respectively, among 100,000 random permutations of the expression data (Fig. S7 A and B). GO terms enriched among upregulated genes were highly variable and reflected perturbations in many aspects of cell physiology (Tables S7 and S8). Of note, we observed that many upregulated terms were related to stress and inflammation, including the response to wounding (P < 10−4), the acute inflammatory response (P < 10−3), and the response to stress (P < 10−3). The most enriched GO term among upregulated genes was the extracellular region (P < 10−12), which reflected increased transcript levels of cytokines as well as various matrix-related genes. Downregulated GO terms were more specific: the most downregulated term was cell division (P < 10−14), and nearly all affected GO terms were directly related to progression through the cell cycle, including mitosis (P < 10−13), DNA replication (P < 10−9), and chromosome condensation (P < 10−5). This is consistent with our previous finding that trisomic MEFs exhibit poor proliferative capacity relative to euploid cells (15). We conclude that trisomic MEFs display some chromosome-independent transcriptional similarities that are indicative of slow growth and cellular stress.
Fig. 3.
Aneuploidy causes similar transcriptional changes in primary mouse and human cells. (A and B) Genes up- or downregulated in one trisomic MEF line are significantly more likely to exhibit a similar change in another trisomy. Gray bars indicate the percentage of all genes up- or downregulated in the indicated cell line at a 1.5-FC cutoff. White bars indicate the percentage of genes up- or downregulated in that trisomy that are also up- or downregulated in the trisomy represented with a gray bar. Asterisks indicate statistically significant overlap (P < 0.05, hypergeometric test). (C and D) Genes up- or downregulated in one trisomic human type are significantly more likely to exhibit a similar change in another trisomy from the same tissue of origin. Data from cultured amniocytes and chorionic villi are from (44). Data from fetal cerebra are from (45). Data from cell-free amniotic fluid are from (31) and (46). (E) GO terms that are enriched among up- and downregulated genes in trisomic MEFs and human cells. Complete lists are presented in Tables S7 and S9.
Does a chromosome-independent response to aneuploidy exist in humans as well? To test this, we examined gene expression data from four datasets that included trisomy 13, 18, and 21. Within each sample, we asked whether genes that are upregulated or downregulated in one trisomy were more likely to be up- or downregulated in another trisomy. In eight out of eight pairwise comparisons, the overlap between different trisomies was significantly more than expected by chance (Fig. 3 C and D and Fig. S6 E–H). To determine whether similar genes were affected across datasets, we performed a permutation test. Although 94 and 137 genes were up-or downregulated, respectively, in four or more trisomic samples, no more than 59 and 49 genes were up- or downregulated, respectively, among 100,000 random permutations of the expression data (Fig. S7 C and D). Surprisingly, dysregulated genes in human trisomies were enriched for many of the same GO terms as were found in the trisomic MEFs (Fig. 3E; Tables S9 and S10). The extracellular region (P < 10−9), inflammatory response (P < 10−4), and response to stress (P < 10−4) were significantly enriched among upregulated genes, while mitosis (P < 10−13), DNA replication (P < 10−8), and chromosome condensation (P < 10−6) were enriched among downregulated genes. Out of 97 GO terms enriched among downregulated genes in trisomic MEFs, 64 were also enriched among downregulated genes in trisomic human cells, including all 29 of the most enriched terms (Table S7).
Analysis of individual datasets further clarified the origins of the transcriptional changes caused by aneuploidy in humans. First, trisomy 13 and trisomy 18 were primarily responsible for the observed enrichment of stress-related and cell-cycle-related GO terms (cf. Tables S11 and S12). As these chromosomes are both larger than chromosome 21, this is consistent with our hypothesis that the degree of aneuploidy determines the severity of the transcriptional response. Second, the origin and/or culturing of trisomic tissue affected the penetrance of the stress signature. Cultured trisomy 13, trisomy 18, and, to a lesser extent, trisomy 21 samples displayed enrichments of stress-related and cell-cycle-related GO terms, and those GO terms were not enriched among differentially expressed genes when only data from fetal cerebra and amniotic fluid were considered (Table S13). Natural limits on cell division that exist in utero may partially mask the different proliferative capacities of trisomic and euploid cells, while unconstrained growth in culture highlights this disparity. For this reason, we used gene expression data from cultured trisomic human cells for subsequent comparisons.
The similarity between enriched GO terms in trisomic human and mouse cells suggested that aneuploidy causes a conserved transcriptional response across mammals. To test this, we identified one-to-one orthologs between humans and mice, then calculated the correlation between the average gene expression values from trisomic MEFs and cultured trisomic human cells. The PCC across all genes was moderate but highly significant (r = 0.11, P < 10−20). In addition, we noted significant overlap between the sets of differentially expressed genes in trisomic mouse and human cells (P < 10−17, hypergeometric test), which was particularly evident among cell-cycle transcripts (Table S14). Thus, aneuploidy induces a similar gene expression pattern indicative of slow growth and/or cellular stress in both mouse and human cells.
Stress-Related Transcriptional Similarities across all Aneuploid Cell Types.
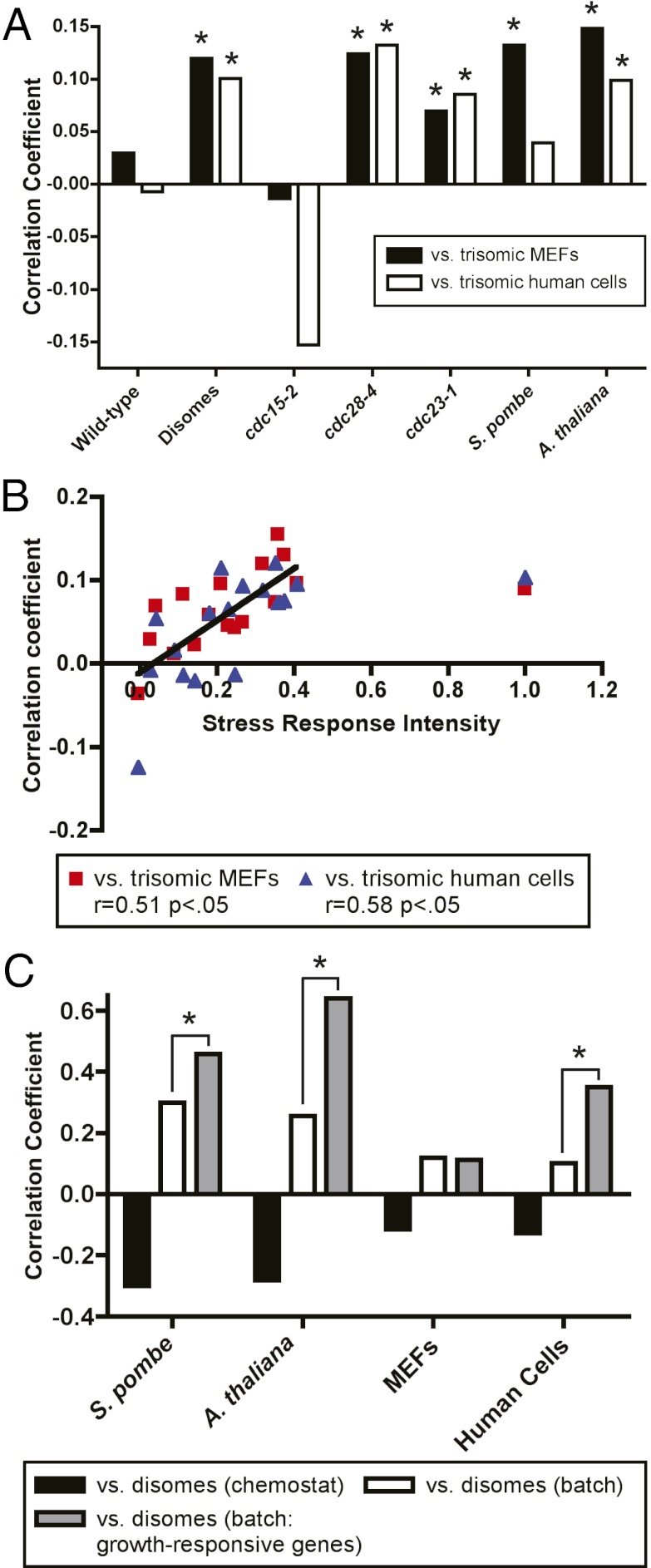
The common stress response in aneuploid cells of highly divergent species raised the possibility that yeast and mammalian cells share a transcriptional response to aneuploidy. To test this, we identified one-to-one orthologs between yeast, plants, mice, and humans. We found that disomic yeast exhibited a small but statistically significant correlation with the averaged expression values of trisomic MEFs and of cultured trisomic human cells (Fig. 4A; r = 0.12, P < 10−4, and r = 0.10, P < 0.002, respectively). The significance of these correlations was also confirmed by permutation testing (Fig. S7 E–H). A majority of individual disomes also exhibited significant pairwise correlations with the trisomic mammalian cells (Fig. S2G). Furthermore, we observed significant correlations between the aneuploid strains of S. pombe and A. thaliana and the trisomic MEFs, as well as between A. thaliana and the trisomic human cells (Fig. 4A).
Fig. 4.
Transcriptional similarities among all aneuploid cell types. (A) Correlation coefficients between the indicated cell type and either trisomic MEFs (black bars) or cultured trisomic human cells (white bars) are displayed. Asterisks indicate a statistically significant correlation (P < 0.05). (B) The stress-response intensity of the disomic strains is plotted against the pairwise correlations with the trisomic mouse and human cells. The black line is a linear regression plotted against the data excluding disome IV (SRI∼1). (C) Correlation coefficients between the indicated aneuploid cell types and chemostat-grown disomes (black bars), batch-grown disomes (white bars), and 500 growth-responsive genes in batch-grown disomes (gray bars) are displayed.
We hypothesized that the similarities in gene expression between the aneuploid transcriptomes were a consequence of the slow growth/stress response that was common among aneuploid cells of all species. Consistent with this hypothesis, there was a significant positive correlation between the stress-response intensities in each disome and their PCC with the trisomic mammalian cells (Fig. 4B). Moreover, cdc28-4 and cdc23-1 exhibited significant positive correlations with both trisomic mammalian cell types, and cdc15-2 did not (Fig. 4A). Taken together, these findings suggested that conserved aspects of the transcriptional response to stress and/or slow growth, rather than aneuploidy per se, drives the expression correlations between aneuploid cell types. To test this, we compared gene expression data from aneuploid cells to chemostat-grown yeast disomes. In chemostats, the doubling times between disomic and euploid cells were equalized by nutrient titration, thereby masking the slow growth/stress response. Indeed, for each interspecies comparison, aneuploid cells were anticorrelated with chemostat-grown disomes (Fig. 4C). Next, we examined the set of yeast genes whose expression levels directly vary according to the rate of cell division (19). In batch culture, the correlation coefficients between the disomes and other species were significantly increased in three out of four comparisons when only orthologs of the 500 strongest growth-responsive genes in yeast were compared. These results demonstrate that conserved elements of a transcriptional stress or slow growth program in eukaryotes underlie the significant transcriptional similarities between aneuploid yeast, plant, and mammalian cells. The lesser predictive value of the growth-responsive yeast genes in comparisons with mammalian cells likely reflects the fact that ribosome synthesis is strongly downregulated by slow proliferation in yeast, while the most striking transcriptional changes among aneuploid mammalian cells were the downregulation of cell cycle transcripts (Tables S14 and S15 and Fig. S8).
A second prediction derived from these results is that aneuploid cells should exhibit similar gene expression changes to euploid cells experiencing exogenous stresses. Aneuploidy in budding and fission yeast affects known stress-response genes in those organisms (Figs. S1 and S5); therefore we examined stress-induced transcription in A. thaliana and mammalian cells. We analyzed gene expression data from A. thaliana grown under salt, reactive oxygen species (ROS), or drought stress (27), and from MEFs treated with salt, the protein synthesis inhibitor anisomycin, or TNF-α (28). The transcriptomes of stressed plants and MEFs were significantly correlated with the corresponding trisomic tissue (Fig. S9; r = 0.40, P < 10−300, and r = 0.11, P < 10−16, respectively). Furthermore, the PCC values were significantly higher when we examined only genes that changed codirectionally by a certain threshold under all stress conditions. As expected, many GO terms that were enriched among upregulated genes in both stressed and trisomic tissue were stress related, including the response to chemical stimulus (P < 10−26) and the defense response (P < 10−12) in Arabidopsis and the inflammatory response (P < 10−4) and cytokine activity in MEFs (Tables S16 and S17; P < 10−4).
Last, we sought to identify conserved changes in expression in response to aneuploidy among single-ortholog genes across species. We used relatively permissive criteria, and asked which genes changed codirectionally in aneuploid budding yeast, fission yeast, mouse, and human cells. We found that 254 single-ortholog genes changed codirectionally in all four species (240 down and 14 up), significantly more than expected by chance (P < 10−5 and P < 0.007, respectively, Fig. S7 I and J). The most affected GO terms among those genes were the nucleus (P < 10−17) and nucleic acid metabolism (Table S18; P < 10−14). These terms reflected the downregulation of ribosomal genes (primarily in fungi) and cell-cycle-related genes (primarily in mammalian cells; Fig. S8 and Table S18). Out of the 254 genes that exhibited codirectional changes across species, 230 of the genes exhibited a similar change in the yeast cdc mutants. Among discordant genes, no significant GO term enrichments were observed. We conclude that aneuploidy in different cell types induces a conserved transcriptional program that is also elicited by exogenous stress and/or slow growth.
Discussion
We have identified a stress/slow-growth-related transcriptional signature that is present in aneuploid cells of diverse organisms and is largely independent of the identity of the extra chromosome(s). Previous analyses have described an oxidative stress response and the downregulation of proliferation-related genes in human samples and mouse models of Down syndrome (29–32). The data presented here suggest that these phenotypes and others found in aneuploid cells may be a common consequence of aneuploidy, as eukaryotic cells appear to exhibit a stereotypical transcriptional response to chromosomewide gene dosage changes.
Why is aneuploidy associated with a stress response? First, aneuploidy increases a cell’s energy needs. This may result from the wasteful transcription, translation, and degradation of proteins encoded by extra chromosomes, and is evidenced by the decreased efficiency at which aneuploid cells convert nutrients into biomass (13, 15). Altered metabolism may also increase the production of reactive oxygen species (33), and ROS-related GO terms were commonly upregulated in aneuploid cells. Second, protein folding and turnover pathways are burdened by aneuploidy. Overexpression of certain proteins may saturate key chaperones, prohibiting them from folding client proteins whose functions are required for viability. Proteins that escape proper folding or degradation may also form cytotoxic aggregates (34, 35). Last, although many aneuploidy-induced phenotypes appear to be independent of the identity of the extra chromosome, copy number changes of a few particularly dosage-sensitive genes may have direct consequences. For instance, budding yeast cells are exquisitely sensitive to tubulin levels, and a single extra copy of beta-tubulin causes the lethality of disome VI (13, 36, 37). We posit that these factors contribute to limit the proliferative capacity of aneuploid cells, thereby resulting in the common downregulation of cell cycle and ribosomal genes.
It is interesting to note that euploid yeast strains that displayed a basal stress response (cdc23-1 and cdc28-4) exhibited significant correlations with aneuploid cells in every organism, and the intensity of the ESR in disomic yeast predicted the strength of their transcriptional similarity with aneuploid cells in other organisms. The ESR was first described as a common transcriptional signature in yeast cells treated with multiple independent stresses, although later research demonstrated that it could also result from a slowed rate of cell division (18–20). Whether the ESR-like transcriptional changes observed in aneuploid higher eukaryotes result from stress, or whether they are also a byproduct of differences in growth rate, remains to be tested.
Not every aneuploid strain that we examined displayed a significant stress response. In many cases, this can be explained by threshold effects of dosage imbalance, as the degree of aneuploidy is proportional to the intensity of the transcriptional response (Fig. S3B). Still, some outliers fail to follow this overall trend: although chromosome XII contains the third most ORFs of any yeast chromosome, disome XII displays the third lowest stress response of any disomic strain (Fig. S1). An extra copy of chromosome XII is also found in strains A2 and A3, the two products of triploid meioses that were uncorrelated with the gene expression pattern in the disomic yeast strains. It is interesting to note that strains A2 and A3 do exhibit significant PCCs with disome XII (r = 0.30, P < 10−92, and r = 0.24, P < 10−59, respectively), but they do not exhibit a PCC > 0.2 with any other disome. Thus, a gene or genes present on chromosome XII may modulate the effects of aneuploidy on transcription. Discovering what underlies the differences between aneuploid cells that do and do not exhibit stress responses may shed further light on the interplay between gene dosage alterations and cellular phenotype.
Among higher eukaryotes, the shared transcriptional response of primary cells to aneuploidy has relevance for the study of cancer. Although 90% of solid tumors display whole-chromosome aneuploidy, it is not clear what role aneuploidy plays in transformation and cancer progression. In mouse models, chromosomal instability (CIN) can both instigate and inhibit tumorigenesis (33, 38–41). In human patients, CIN in tumor cells is generally associated with aggressive disease (42), although in some contexts high levels of CIN actually correlate with improved prognosis (43). We have found that single-chromosome aneuploidy causes a transcriptional response indicative of increased cellular stress and decreased proliferative capacity. These results present aneuploidy as a complex phenomenon with potentially antitumorigenic properties. Although aneuploidy can contribute to transformation by altering the dosage of oncogenes and tumor suppressors, the stresses induced by aneuploidy on metabolism and protein folding limit growth potential. Yeast can adapt to aneuploidy by developing mutations, which improve their proliferative capacity (14), and it may be the case that cancer cells must develop similar genetic changes that allow them to tolerate aneuploidy and acquire robust proliferative capacity as well.
Materials and Methods
RNA collection, hybridization, and analysis of strains derived via triploid meiosis were performed as previously described (13). Other datasets were acquired from the Gene Expression Omnibus or were downloaded from the relevant publication. Further experimental details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Joan Clara Smith and Emily Mitchell for technical assistance, Rong Li for providing strains used in this analysis, and Mike Laub, Aviv Regev, and members of the Amon and Gilbert labs for reading this manuscript. This work was supported by the National Institutes of Health (GM056800 to A.A. and GM094306 to M.J.D.) and an NSF Predoctoral Fellowship (to J.M.S.). M.J.D. is a Rita Allen Foundation Scholar. A.A. is an HHMI investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray data has been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE35853).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209227109/-/DCSupplemental.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Hassold T, et al. Human aneuploidy: Incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Brown S. Miscarriage and its associations. Semin Reprod Med. 2008;26:391–400. doi: 10.1055/s-0028-1087105. [DOI] [PubMed] [Google Scholar]
- 5.Delabar JM, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 6.Korenberg JR, et al. Down syndrome phenotypes: The consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson LE, et al. Trisomy for the Down syndrome ‘critical region’ is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet. 2007;16:774–782. doi: 10.1093/hmg/ddm022. [DOI] [PubMed] [Google Scholar]
- 9.Korbel JO, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci USA. 2009;106:12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roper RJ, Reeves RH. Understanding the basis for Down syndrome phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres EM, Williams BR, Amon A. Aneuploidy: Cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 14.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regenberg B, et al. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 23.Chikashige Y, et al. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct Funct. 2007;32:149–161. doi: 10.1247/csf.07036. [DOI] [PubMed] [Google Scholar]
- 24.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 25.Chen D, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huettel B, Kreil DP, Matzke M, Matzke AJM. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000226. doi: 10.1371/journal.pgen.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilian J, et al. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 28.Ferreiro I, et al. Whole genome analysis of p38 SAPK-mediated gene expression upon stress. BMC Genomics. 2010;11:144. doi: 10.1186/1471-2164-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moldrich RX, et al. Proliferation deficits and gene expression dysregulation in Down’s syndrome (Ts1Cje) neural progenitor cells cultured from neurospheres. J Neurosci Res. 2009;87:3143–3152. doi: 10.1002/jnr.22131. [DOI] [PubMed] [Google Scholar]
- 30.Contestabile A, Fila T, Bartesaghi R, Ciani E. Cell cycle elongation impairs proliferation of cerebellar granule cell precursors in the Ts65Dn mouse, an animal model for Down syndrome. Brain Pathol. 2009;19:224–237. doi: 10.1111/j.1750-3639.2008.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slonim DK, et al. Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc Natl Acad Sci USA. 2009;106:9425–9429. doi: 10.1073/pnas.0903909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jovanovic SV, Clements D, MacLeod K. Biomarkers of oxidative stress are significantly elevated in Down syndrome. Free Radic Biol Med. 1998;25:1044–1048. doi: 10.1016/s0891-5849(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 33.Li M, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci USA. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiler-Samerotte KA, et al. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci USA. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefani M. Protein misfolding and aggregation: New examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: Consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders KR, et al. A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC Genet. 2009;10:36. doi: 10.1186/1471-2156-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remeseiro S, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sotillo R, Schvartzman J-M, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: A meta-analysis. Gut. 2008;57:941–950. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 43.Birkbak NJ, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altug-Teber O, et al. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007;119:171–184. doi: 10.1159/000112058. [DOI] [PubMed] [Google Scholar]
- 45.Mao R, et al. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koide K, et al. Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18. Hum Genet. 2011;129:295–305. doi: 10.1007/s00439-010-0923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.