Abstract
Replication through a diverse array of DNA lesions occurs by the sequential action of two translesion synthesis (TLS) DNA polymerases (Pols), in which one inserts the nucleotide opposite the lesion and the other carries out the subsequent extension. By extending from the nucleotide inserted by another Pol, Polζ plays an indispensable role in mediating lesion bypass. Polζ comprises the Rev3 catalytic and Rev7 accessory subunits. Pol32, a subunit of the replicative polymerase Polδ, is also required for Polζ-dependent TLS, but how this Polδ subunit contributes to Polζ function in TLS has remained unknown. Here we show that yeast Polζ is a four-subunit enzyme containing Rev3, Rev7, Pol31, and Pol32; in this complex, association with Pol31/Pol32 is mediated via binding of the Rev3 C terminus to Pol31. The functional requirement of this complex is supported by evidence that mutational inactivation of Rev3’s ability to bind Pol31 abrogates Polζ’s role in TLS in yeast cells. These findings identify an unexpected role of Pol31 and Pol32 as two essential subunits of Polζ, and clarify why these proteins are required for Polζ-dependent TLS, but not for TLS mediated by Polη in yeast cells. To distinguish the four-subunit complex from the two-subunit Polζ, we designate the four-subunit enzyme “Polζ-d,” where “-d” denotes the Pol31/Pol32 subunits of Polδ.
Keywords: mutagenesis, DNA repair
Specialized translesion synthesis (TLS) DNA polymerases (Pols) promote replication through DNA lesions and, depending on whether they function in an error-free or mutagenic manner, act as inhibitors or promoters of carcinogenesis. Polζ functions primarily in TLS mediated by the consecutive action of two Pols, where it extends from the nucleotide inserted opposite a lesion site by another Pol (1–3). Because Polζ can extend from both correct and incorrect nucleotides, it contributes to mutagenic TLS; thus, in the absence of Polζ, the frequency of UV-induced mutations is greatly reduced in eukaryotes. Polζ is a B family Pol composed of the Rev3 catalytic and Rev7 accessory subunits (3). The high-fidelity B-family Pol, Polδ, plays an indispensable role in replication of the yeast genome. It is composed of the catalytic Pol3 and the accessory Pol31 and Pol32 subunits, wherein Pol31 binds to both Pol3 and Pol32 (4). The Pol32 subunit is also required for DNA damage-induced mutagenesis in yeast cells, and as indicated from genetic studies in yeast, it functions in the Polζ-dependent TLS pathway (5–7). The role of this Polδ subunit in Polζ-dependent TLS has remained unknown, however.
The N-terminal ∼90 residues of Pol32 are required for binding Pol31 (7), and deletion of this N-terminal region of Pol32 also inactivates Polζ-dependent DNA damage-induced mutagenesis. The requirement for binding of Pol32 to Pol31 has been attributed to the role of Polδ in Polζ-mediated TLS (7); however, based on genetic studies indicating that Polδ replicates the lagging strand (8), the requirement for Pol31/Pol32 in Polδ for Polζ-dependent mutagenesis would limit Polζ function in TLS to only the lagging strand, which fails to account for how TLS would occur on the leading strand, which, as inferred from genetic studies, is replicated by Polε (9). In this study, we examined whether Pol31 and Pol32 are integral subunits of Polζ, which would explain the requirement of Polζ for lesion bypass on both the DNA strands (10). We purified a highly stable and stoichiometric four-subunit complex of Polζ containing Rev3, Rev7, Pol31, and Pol32, and determined from genetic studies that mutations that confer defects in the binding of Rev3 to Pol31 inactivate Polζ function in TLS in yeast cells. Thus, in its subunit composition and structural complexity, Polζ resembles the high-fidelity replicative Pols. We discuss the implications of these observations for the various cellular roles of Polζ in both yeast and human cells.
Results
Homology of Rev3 with Pol3.
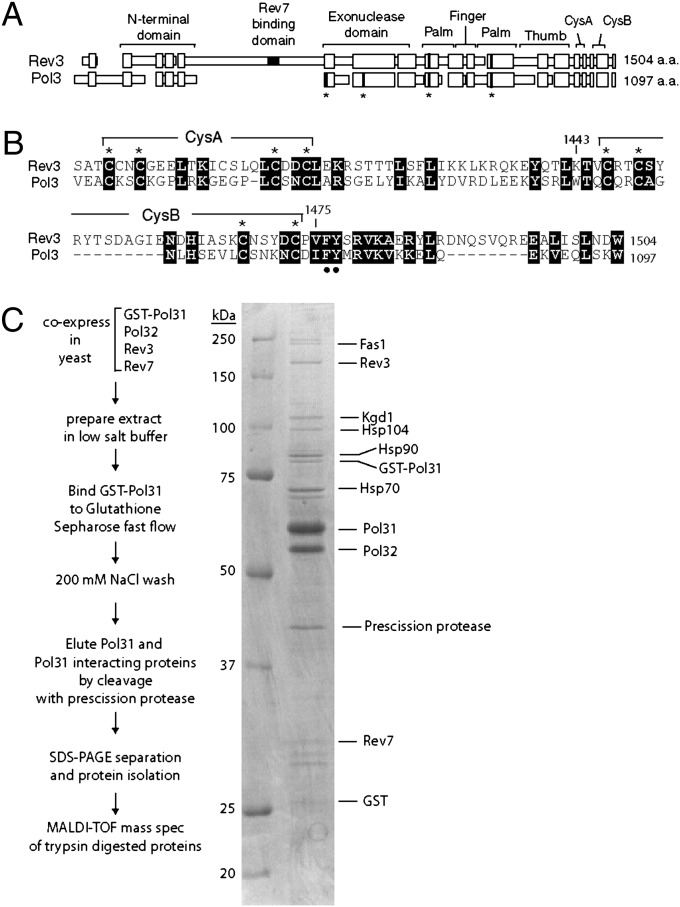
The interaction of the catalytic Pol3 subunit of Polδ with its B-subunit, Pol31, is mediated via the C terminus of Pol3 and requires the ∼70 C-terminal residues (4) that include the second metal-binding domain (11). In fact, each eukaryotic replicative B-family DNA polymerase (δ, ε, and α) harbors a cognate B-subunit (Pol31, Dpb2, and Pol12, respectively) that binds to the C-terminal region of the catalytic subunit, but such a B-subunit has not yet been identified for Rev3. Rev3 shares a high degree of homology with Pol3 in the N terminus and the conserved DNA polymerase domains (Fig. 1A). Interestingly, Rev3 and Pol3 also exhibit considerable homology in their C-termini encompassing the two cysteine-containing metal-binding domains, CysA and CysB (Fig. 1B). Despite this homology, however, previous two-hybrid studies have failed to identify an interaction between the C-terminal portion of Rev3 with Pol31 in either Saccharomyces cerevisiae or Schizosaccharomyces pombe (4), and in pull-down experiments with the purified Rev3–Rev7 complex, we found no interaction of Pol31 with Rev3. However, we have observed that the C terminus of Pol3 is susceptible to cleavage when Pol3 is expressed without its B-subunit Pol31. We hypothesized that this might be the case for Rev3 as well, and that interactions between Rev3 and Pol31 have not been observed due to the proteolytic cleavage or other disruptions of the Rev3 C terminus when the Rev3/Rev7 proteins are expressed in yeast cells in the absence of a cognate B subunit. Thus, we examined whether Pol31/Pol32 could be purified with Rev3/Rev7 in yeast cells.
Fig. 1.
Homology between Rev3 and Pol3, and copurification of Rev3/Rev7 with the Pol31/Pol32 complex. (A) Schematic alignment of amino acid sequences. Boxes represent protein sequences that share similar residues, and spaces indicate gaps introduced into the sequence for alignment. The positions of the conserved N-terminal domain, exonuclease domain, and DNA polymerase domains identified in the crystal structure of Pol3 are shown. Asterisks below the black boxes indicate the positions of the catalytic residues present in the exonuclease domain of Pol3, which are absent in Rev3, and in the polymerase domains in the palm region of Pol3 and Rev3. The Rev7-binding region in Rev3 and the positions of the C-terminal metal-binding motifs, CysA and CysB, are indicated. (B) C-terminal regions of Rev3 and Pol3. Identical and highly conserved residues are indicated by white on black lettering. Cysteine residues in the metal-binding domains, CysA and CysB, are indicated by asterisks, and the conserved F1476 and Y1477 residues of Rev3 that were changed to alanines in this study are indicated by solid circles. (C) Copurification of Rev3 and Rev7 with Pol31/Pol32. The purification scheme is shown on the left. Proteins were resolved by 10% SDS/PAGE and stained with Coomassie blue. The positions of molecular weight markers and the identities of various proteins are indicated.
Pol31 and Pol32 Copurify with Rev3/Rev7.
Because Polζ is a very low-abundance enzyme whose purification requires coexpression of the Rev3/Rev7 proteins in yeast cells (12), we examined whether the Rev3/Rev7 subunits of Polζ would form a complex with Pol31/Pol32 when all of these proteins were coexpressed in yeast cells. To express all four proteins together, we used a Pol31/Pol32 expression plasmid in which the Pol31 subunit was tagged with GST, as was used previously for purification of Polδ (13), and cotransformed it with a plasmid expressing the native Rev3 and Rev7 (Polζ) proteins into yeast cells. Yeast cells (∼6 g) were disrupted under low-salt conditions [200 mM NaCl, 175 mM (NH4)2SO4], and soluble extract was incubated with glutathione Sepharose. Bound proteins were eluted with prescission protease, which releases untagged Pol31 and its interacting proteins from the glutathione Sepharose. SDS/PAGE analysis revealed that a number of proteins copurified with Pol31/Pol32 under these mild conditions, but interestingly, two proteins were observed, albeit in limited amounts, that migrate at positions identical to those of purified Rev3 and Rev7 (Fig. 1C). The identity of the Rev3 and Rev7, as well as of the Pol31 and Pol32 proteins, was confirmed through MALDI-TOF mass spectrometry of the protein bands excised from the SDS/PAGE gel. Although several other proteins that are not expected to specifically interact with Pol31 or Pol32, including heat shock proteins Hsp104, Hsp90, and Hsp70, were also identified by MALDI-TOF mass spectrometry (Fig. 1C), the presence of Rev3 and Rev7 proteins together with Pol31/Pol32 raised the possibility that all four of these proteins together form a complex.
Purification of a Stoichiometric Four-Subunit Polζ Complex.
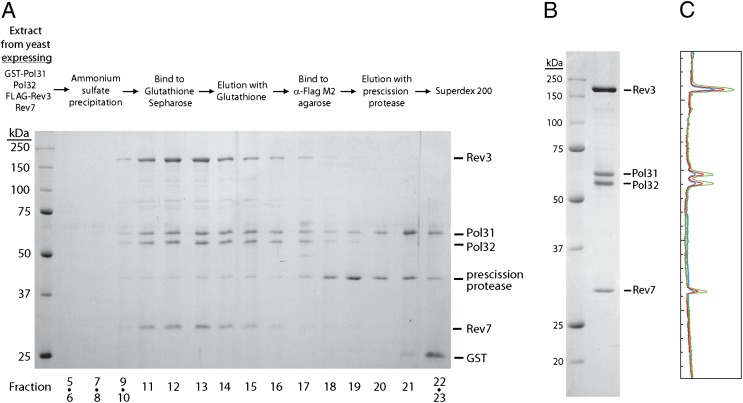
To purify the four-subunit Polζ complex that contained Rev3, Rev7, Pol31, and Pol32 and was devoid of any other contaminating proteins, we used a similar protein expression system, but with a precission protease-cleavable FLAG tag introduced onto the N terminus of Rev3. Yeast cells (80 g) were lysed, and after an initial ammonium sulfate precipitation step, proteins were resuspended in low-salt buffer and bound to glutathione Sepharose. Proteins were then eluted by treatment with glutathione, releasing the proteins with the tags intact, and subsequently bound to anti-FLAG agarose, followed by elution with prescission protease. The protein eluate was concentrated and then run on a Superdex-200 Gel Filtration column (GE Healthcare). The Rev3/Rev7 proteins comigrated with Pol31/Pol32 such that the four proteins were present in stoichiometric amounts in fractions 11–13 (Fig. 2A). Peak fractions were pooled and incubated with glutathione Sepharose to remove residual precission protease and any remaining GST-Pol31. The resulting enzyme preparation was then concentrated. SDS/PAGE analyses indicated a high degree of purity, and gel scanning confirmed an almost 1:1:1:1 ratio of each subunit (Fig. 2 B and C and Table S1). The interaction of Rev3/Rev7 with Pol31/Pol32 was resistant to salt concentrations up to 1 M NaCl (Fig. S1), suggesting that this interaction is highly specific and highly stable. Also, in contrast to the two-subunit Polζ (Rev3/Rev7), which is prone to precipitation and has a maximal solubility of ∼0.15 mg/mL, the four-subunit Polζ complex is stable even up to 1 mg/mL and does not readily precipitate from solution, indicating that binding of Pol31/Pol32 stabilizes the Polζ (Rev3/Rev7) structure. We designate the four-subunit complex “Polζ-d,” where “-d” indicates the Pol31/Pol32 subunits of Polδ and distinguishes the two-subunit Polζ containing only the Rev3/Rev7 subunits from the four-subunit complex.
Fig. 2.
Purification of the Rev3/Rev7/Pol31/Pol32-containing Polζ-d complex. (A) Pol31 and Pol32 comigrate with Rev3 and Rev7 during gel filtration. An abbreviated purification scheme is shown at the top. Each fraction (2.5 μL) from the Superdex 200 step was resolved by 10% SDS/PAGE. Molecular weight markers and protein identities are indicated. (B) Polζ-d complex. Peak fractions from the Superdex 200 column were pooled, incubated with glutathione Sepharose, and concentrated. The purified Polζ-d (2 μL) was resolved by 10% SDS/PAGE and stained with Coomassie blue. (C) Subunit stoichiometry of the Polζ-d complex. The protein gel in B was analyzed using ImageQuant software, and protein band intensity was plotted along the length of the gel. The results shown are from three independent lanes, each containing increasing amounts of Polζ-d (0.5 μL, 1 μL, and 2 μL), in which the green line is the plot of the lane shown in B. The relative amount of each subunit was quantified, and the resulting respective stoichiometry of the Rev3, Pol31, Pol32, and Rev7 subunits was determined to be 0.93:0.93:1.15:1.20 (Table S1).
Lesion Bypass by the Four-Subunit Polζ.
Polζ functions in lesion bypass primarily at the extension step of TLS, in which Polζ carries out the subsequent extension after insertion of a nucleotide opposite the lesion site by another Pol (1–3, 14). Thus, opposite a cis-syn TT dimer or a (6–4) TT photoproduct, Polζ does not insert a nucleotide opposite the 3′T of either lesion, but extends from an inserted nucleotide. Because of its ability to extend from a mispair opposite a lesion, Polζ contributes to mutagenic TLS (1, 2). To determine whether Pol31/Pol32 proteins affect the lesion bypass properties of Polζ, we compared the proficiency of Polζ and Polζ-d for nucleotide insertion opposite the 3′T of a TT dimer or a (6–4) TT photoproduct. However, Polζ-d remained as ineffective at inserting a nucleotide opposite both these DNA lesions as Polζ (Fig. S2). Both Pols carried out the extension reaction from an A or a G opposite from the 3′T of the lesion (Fig. S3), and, as determined from steady-state kinetic analyses, Polζ-d exhibited twofold to fivefold greater catalytic efficiency for extension compared with Polζ (Table S2). As is the case for Polζ (1, 14), Polζ-d is inhibited from inserting a nucleotide opposite an abasic site; for this lesion as well, Polζ-d exhibits approximately threefold to fivefold greater efficiency for extension compared with Polζ (Table S2). The increased efficiencies result primarily from an increase in kcat value, which reflects a more rapid turnover of substrate by the enzyme.
Binding of the Rev3 C Terminus to Pol31 Mediates the Association of Rev3/Rev7 with Pol31/Pol32.
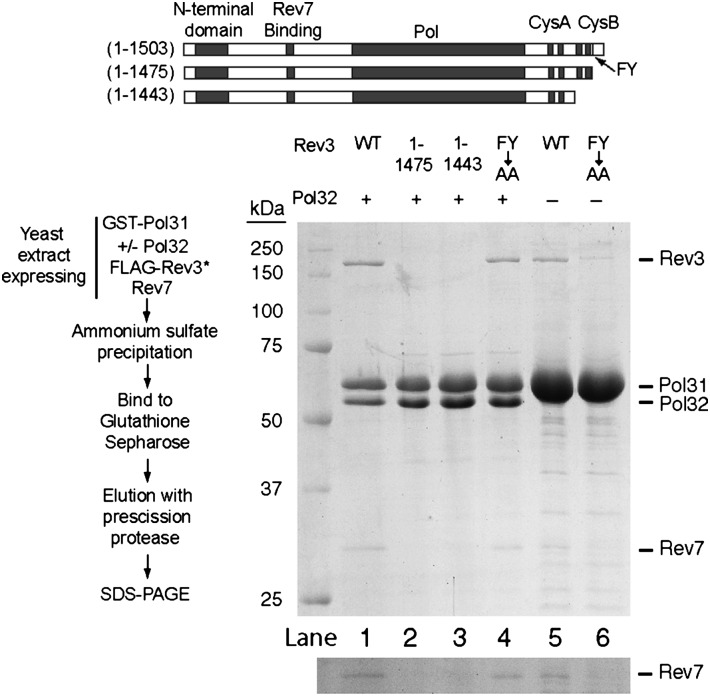
To determine whether the interaction of the Pol31/Pol32 complex with Rev3/Rev7 is mediated through the Rev3 C-terminal region and the Pol31 subunit, we first made two C-terminal truncations of the Rev3 subunit, Rev3(1-1443) and Rev3(1-1475), and examined their ability to interact with Pol31/Pol32. The GST Pol31/Pol32 and FLAG Rev3/Rev7 proteins were coexpressed in yeast cells, affinity-purified by binding to glutathione Sepharose, and eluted by treatment with prescission protease. Neither Rev3(1-1443) or Rev3(1-1475) copurified with the Pol31–Pol32 complex (Fig. 3; compare lanes 2 and 3 with lane 1). The Rev3(1-1475) protein retains both metal-binding domains and deletes only the C-terminal tail, yet it fails to interact with Pol31/Pol32. In Pol3 also, deletion of the C-terminal tail (residues 1085–1097) abrogates Pol3’s ability to interact with Pol31 (4). Thus, in both Rev3 and Pol3, the C-terminal region lying beyond the second metal-binding domain is indispensable for their binding to Pol31. We next examined whether the Pol32 subunit is required for the interaction of Polζ with Pol31. When Pol32 was not coexpressed with Pol31, Rev3 could still interact with Pol31 (Fig. 3, lane 5). Based on these observations, we conclude that the association of Rev3/Rev7 and Pol31/Pol32 proteins is mediated via the binding of Rev3 C terminus to Pol31. Moreover, our data show that the cognate B-subunit of Polζ is Pol31, which is also the B-subunit of Polδ.
Fig. 3.
Effects of mutations in Rev3 C terminus on Pol31 binding. GST-tagged Pol31 was expressed in yeast cells with or without Pol32 (indicated by +/−) and together with FLAG-tagged WT or mutant Rev3 protein (indicated by Rev3*) and Rev7. After the initial ammonium sulfate precipitation step, pellets were resuspended in buffer and dialyzed, and proteins were affinity-purified by gluthathione Sepharose, followed by elution with prescission protease. Pull-down experiments were performed on extracts from yeast cells expressing proteins, as follows: lane 1, GST-Pol31/Pol32/Rev3/Rev7; lane 2, GST-Pol31/Pol32/Rev3(1-1475)/Rev7; lane 3, GST-Pol31/Pol32/Rev3(1-1443)/Rev7; lane 4, GST-Pol31/Pol32/Rev3(F1476A,Y1477A)/Rev7; lane 5, GST-Pol31/Rev3/Rev7; lane 6, GST-Pol31/Rev3(F1476A, Y1477A)/Rev7. FY→AA indicates the F1476A Y1477A mutation. The lower panel provides an enhanced view of the region of the Rev7 protein band.
In the sequence immediately following the second metal-binding domain, Rev3 shares a number of conserved residues with Pol3 (Fig. 1B). Among these, mutations of F1077 Y1078 residues to AA in Pol3 diminish Pol3’s ability to bind Pol31 (4). To determine whether mutations of the corresponding F1476 Y1477 residues in Rev3 to AA impair its interaction with Pol31, we first examined whether the F1476A Y1477A mutant Rev3 protein copurifies with the Pol31–Pol32 protein complex. The binding of Rev3/Rev7 with the Pol31–Pol32 complex was not affected by these FY→AA mutations in Rev3 (Fig. 3, lane 4); however, recovery of Rev3/Rev7 proteins was greatly reduced when these proteins were purified from yeast cells in which Pol32 was not coexpressed with Pol31 (Fig. 3, lane 6). These findings suggest that the ability of the F1476A Y1477A mutant Rev3 protein to bind Pol31 is weakened, and that Pol32 helps stabilize it.
Defects in Ability of Rev3 to Bind Pol31 Impair Polζ Function in UV Mutagenesis.
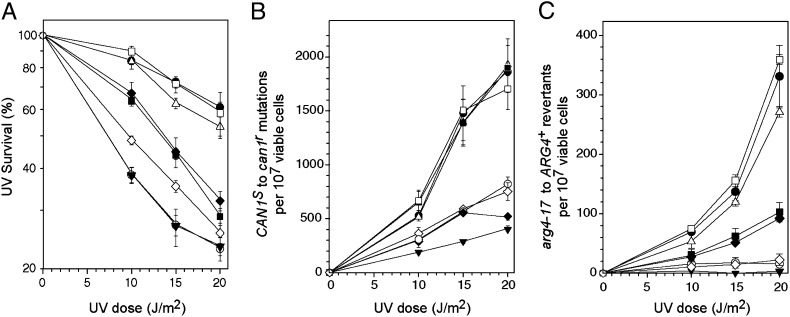
To evaluate the biological significance of the ability of Rev3 to bind Pol31, we examined the effects of mutations in Rev3 that impair its interaction with Pol31 on Polζ TLS function in yeast cells. Low-copy number centromeric plasmids carrying the WT REV3 gene or its mutant alleles, expressed from the native REV3 promoter, were introduced into rev3Δ cells, and their effects on UV survival and UV mutagenesis were analyzed. Compared with rev3Δ cells harboring the WT REV3 gene, the catalytically inactive D975A rev3 exhibited increased UV sensitivity, and the frequency of UV-induced CAN1S to can1r forward mutations was greatly reduced (Fig. 4 A and B). UV- induced mutations at arg4-17 show a very strong dependence on Polζ, and in cells carrying the catalytically inactive rev3 gene, UV-induced reversion of arg4-17 to ARG4+ was completely eliminated (Fig. 4C). The rev3 (1–1475) mutant, which expresses the C-terminally deleted Rev3 protein that fails to interact with Pol31 (Fig. 3), conferred an increase in UV sensitivity (Fig. 4A) and a defect in UV mutagenesis (Fig. 4 B and C) similar to that observed for the catalytically inactive mutant. Because this deletion of Rev3 C-terminal region has no effect on the DNA synthetic activity of the purified Rev3/Rev7 enzyme, these results imply that Rev3’s ability to bind Pol31 is necessary for Polζ-d function in TLS in yeast cells.
Fig. 4.
Effects of mutations in the Rev3 C terminus and of the pol31 K358E (sdp5-15) mutation on UV survival and UV-induced mutagenesis. (A) UV survival of yeast cells. The rev3Δ strain or the rev3Δ strain harboring the pol31 K358E mutation in the genomic POL31 (rev3Δ pol31 K358E) was transformed with plasmid carrying the WT REV3 gene or mutant rev3 alleles as indicated. Yeast cells were grown to mid-logathmic phase, washed with water, and sonicated to disperse clumps. Appropriate dilutions were plated on synthetic complete medium lacking leucine (SC-leu) and irradiated with UV light at a dose rate of 1 J/m2/s. Plates were incubated in the dark for 3–4 d. (B) UV-induced CAN1S to can1r forward mutation. Cells were grown as described above and plated on SC medium lacking arginine but containing canavanine (SC-arg+can) and UV- irradiated. (C) UV-induced reversion of arg4-17 to ARG4+. Cells were grown as described above, plated on SC-arg and UV-irradiated. The UV survival and UV-induced mutation curves represent the mean ± SEM of at least three independent experiments. Symbols denote the rev3Δ strain carrying the following: ●, WT Rev3; △, F1476A Y1477A Rev3; ■, C1468S Rev3; ○, (1-1475) Rev3; ▼, D975A Rev3; rev3Δ pol31 K358E strain carrying the following: □, WT Rev3; ◆, F1476A, Y1477A Rev3; and ◇, C1468S Rev3.
The pol3-11 mutation in the catalytic subunit of Polδ, in which the third cysteine of CysB, C1069 (Fig. 1B), is replaced by serine, confers temperature-sensitive growth; and mutant cells, although viable, exhibit slow growth at the permissive temperature of 30 °C (15, 16). These phenotypes of the pol3-11 mutant are suppressed by dominant mutations in the POL31 gene (originally designated SDP5, suppressor of DNA polymerase). The suppressor mutation sdp5-15 harbors a change of the lysine (K) 358 residue to glutamic acid (E) in Pol31 (15, 16). To determine whether the pol31 K358E mutation has a similar suppressive effect on the rev3 C1468S mutation, which corresponds to the pol3-11 mutation in Polδ, we examined UV mutagenesis in rev3Δ cells expressing these mutant proteins. We found that UV survival as well as the frequency of UV-induced CAN1S to can1r forward mutations and arg4-17 to ARG4+ reversions were not affected by the pol31 K358E mutation alone (Fig. 4). The rev3 C1468S mutation conferred an increase in UV sensitivity but did not affect the frequency of UV induction of can1r mutations, whereas it reduced the frequency of arg4-17 reversion considerably (Fig. 4). Thus, similar to the deleterious effect of the C1069S mutation in pol3-11, the analogous cysteine-to-serine mutation in REV3 also resulted in a limited loss of function. The DNA synthetic activity of Rev3/Rev7 was not affected by this mutation, however. Interestingly, compared with either single mutant, the rev3 C1468S pol31 K385E double mutant exhibited a further decline in UV survival, and the frequency of UV-induced mutations for both the CAN1S and arg4-17 genes was greatly reduced (Fig. 4). Thus, rather than a suppressive effect, the pol31 K358E mutation exerted an inhibitory effect on the TLS function of the four-subunit Polζ-d enzyme containing a C1468S mutation in Rev3.
Because of the weakened interaction of F1476A Y1477A mutant Rev3 protein with Pol31 in the absence of Pol32 (Fig. 3, lane 6), we next examined whether this mutation alone or in combination with pol31 K358E affects the TLS function of Polζ-d. UV survival and UV mutagenesis were not affected by the F1476A Y1477A mutations, as might be expected given the lack of any perceptible effect of these mutations on the binding of the Pol31–Pol32 complex (Fig. 3, lane 4). In striking contrast, UV sensitivity was enhanced and UV mutagenesis was greatly reduced in the rev3 F1476A Y1477A, pol31 K358E double mutant (Fig. 4). Thus, although Polζ-d carrying either of these mutations in Rev3 or Pol31 could promote WT levels of TLS, the simultaneous presence of both these mutations greatly reduced its ability to carry out TLS (Fig. 4). Taken together, these data show that Pol31 functions in Polζ-mediated TLS, and that this role is manifested through its binding to the Rev3 C terminus. Furthermore, our observation that the pol31 K358E mutation had a deleterious effect rather than a suppressive effect on TLS in the presence of Rev3 C-terminal mutations indicates that although Pol31 binds to Pol3 and to Rev3 via their C-terminal regions, there are differences in the manner in which they associate.
Discussion
The results presented here show that Pol31 and Pol32 associate in a stable complex with the Rev3 and Rev7 subunits of Polζ. The demonstration that Pol31 and Pol32 are also Polζ subunits was made possible by our ability to purify a four-subunit stoichiometric complex. From the severe reduction in UV survival and UV-induced mutagenesis of C-terminal truncations in Rev3, which eliminate its ability to bind Pol31, we could establish that the Rev3 C terminus is required for Polζ function in TLS. Based on these observations, our genetic evidence for functional interaction of Rev3 with Pol31, and the requirement of Pol32 for damage-induced Polζ-dependent mutagenesis, we conclude that Pol31 and Pol32 are essential subunits of Polζ. These findings raise the possibility that some of the phenotypes associated with mutations of Pol31 or Pol32 may derive from their role in Polζ-mediated functions rather than from their role as Polδ subunits.
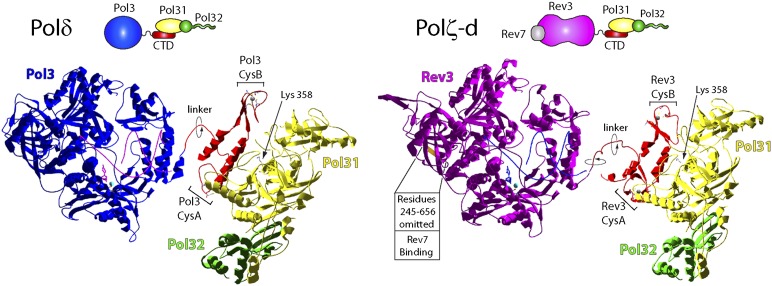
The presence of the Pol31 and Pol32 subunits in both Polδ and Polζ-d, along with the requirement of the C-terminal domains of Pol3 and Rev3 for their binding to Pol31, suggests structural similarity of the two Pols. To generate a 3D model of Polδ and Polζ-d (Fig. 5), we derived the structures of the various subunits using the data available for several B-family DNA polymerases, which include a ternary structure of the catalytic core of yeast Pol3 (PDB ID: 3IAY) (17), the structure of the human p50/p66(N-terminal domain) Polδ accessory subunits (PDB ID: 3E0J) (18), and a structure of yeast Pol1 (Polα) C-terminal domain (CTD) bound to its cognate B-subunit, Pol12 (PDB ID: 3FLO) (19). We first generated models of the Pol31/Pol32(1-104) complex and of the C-terminal regions of both Pol3 and Rev3. We then aligned these structures to the Pol1 CTD/Pol12 structure to model the binding of the Pol3 and Rev3 CTDs to the Pol31 subunit. Finally, we modeled an ∼14-aa linker based on a similar region found in Pfu Pol (PDB ID: 3A2F) (20) to join the CTD-bound Pol31/Pol32 structures to their respective catalytic subunits (Fig. 5). The model of the Rev3 catalytic subunit was generated based on the Pol3 catalytic core, excluding the large insertion within Rev3 that lacks homology to Pol3 and contains the Rev7-binding region (Fig. 1A). In both the Polδ and Polζ-d models, the α helix between CysA and CysB (helix 1) mediates the interactions of Pol3 and Rev3 with Pol31. Our modeling predicts that the ∼30 C-terminal residues of Rev3, which are required for Pol31 binding (Fig. 3), form a second helix that likely stabilizes the overall structure of the C terminus, such that helix 1 can mediate binding to Pol31 (Fig. 5). Although shown here in a random, static position, the CTD/Pol31/Pol32 region could occupy various positions, through rotation and bending of the linker region. Such a scenario is consistent with small-angle X-ray scattering analysis of the yeast Polδ holoenzyme, which indicates that the Pol31/Pol32 subunits exhibit high conformational variability with respect to the catalytic domain (21). Based on the biochemical and genetic data provided here, and the placement of accessory subunits in the models, we suggest that Polδ and Polζ-d will share architectural similarity (Fig. 5), but because of their association with other proteins that relate to their distinct biological roles, they will differ in many other respects.
Fig. 5.
A model of Polδ (Pol3-Pol31-Pol32) and Polζ-d (Rev3-Rev7-Pol31-Pol32) complexes. (Upper) Schematic representation of subunit interactions in Polδ and Polζ-d. CTD refers to the C-terminal domain of the Pol3 and Rev3 catalytic subunits. The C terminus of Pol32 is shown as an extended tail, but this region is not depicted in the models shown below. (Lower) The crystal structure of the catalytic core of Pol3 (blue; PDB ID: 3IAY), shown for Polδ, was used to generate the model of the Rev3 DNA polymerase domain (magenta). For this purpose, the intervening region of Rev3 from residues 245–656 that lacks homology to Pol3 and contains the Rev7- binding domain (Fig. 1A) was omitted from the analysis. The predicted location of this large region in the Rev3 structure is indicated. The modeled Pol31 and the N terminus of Pol32 (residues 1–104) are shown in yellow and green, respectively. The C-terminal regions of Pol3 (residues 998–1097) and Rev3 (residues 1384–1504), including their modeled linker regions, are shown in red. Conformational flexibility of the CTD/Pol31/Pol32 region relative to the catalytic Pol3 or Rev3 domain is indicated by rotational arrows around the linker regions. The incoming dNTP and Mg2+ are shown in the active site of each Pol. The DNA in the Pol3 structure (magenta helix) was used to model the DNA in Rev3 (blue helix). The position of the lysine 358 residue in Pol 31 is indicated. The positions of metals in CysA and CysB of Pol3 and Rev3 are shown as spheres.
It was recently reported that a C-terminal peptide of human Rev3 encompassing the two metal-binding regions shows physical interaction with the P50 and P66N proteins (the latter of which contains the N-terminal 141 residues), which are the human counterparts of yeast Pol31 and Pol32, respectively, and that mutations of all four cysteines to alanines in the second metal-binding region of yeast Rev3 affect UV mutability (22). Based on this finding, the authors concluded that the binding of P50 and P66 proteins to Polζ represents a polymerase-switch mechanism. In particular, they proposed that when Polδ encounters a DNA lesion, the Pol3 catalytic subunit dissociates from P50/P66, and Polζ then gains access to the replication fork by its binding to the P50/P66 heterodimer. This conclusion implies that Polζ could function in TLS only in conjunction with Polδ; it fails to account for how Polζ would access Polɛ, which, according to genetic studies in yeast, replicates the leading strand (9). Even if Polδ replicates both DNA strands, the idea that the Pol3 subunit would disassociate from the Pol31/Pol32 (P50/P66) complex seems quite untenable, given that the three-subunit Polδ holoenzyme is a highly stable entity. Also, our observation that the C-terminal region of both Pol3 and Rev3 is very labile unless bound to Pol31/Pol32 argues against the proposed exchange mechanism. The lability of the Pol3 C terminus has been noted by others as well (11). Moreover, how the Pol31/Pol32 proteins would disassociate from Rev3/Rev7 and reengage with Pol3 once Polζ has completed its role in TLS is not clear. Furthermore, the proposal allows only for the exchange of Polζ with Polδ, and fails to account for how TLS Pols, such as Polη, would access Polδ at the site of stalled replication, given that genetic studies have indicated that Pol31/Pol32 function in TLS in conjunction only with Polζ, not with Polη (5–7).
Our ability to purify a highly stable and stoichiometric four-subunit complex, together with other biochemical and genetic data that we have presented, provides strong evidence that a distinct four-subunit Polζ holoenzyme, Polζ-d, comprising Rev3, Rev7, Pol31, and Pol32 proteins, functions independent of Polδ or any other replicative Pol in carrying out its role in TLS and other cellular processes. Such a complex negates the need to postulate complex exchange mechanisms that differ for different TLS Pols. Although the exchange mechanism remains to be defined, genetic studies in yeast have provided strong evidence for a key role of Rad6-Rad18–mediated proliferating cell nuclear antigen ubiquitylation in this process (23–25).
The four-subunit Polζ-d complex exhibits greatly enhanced stability compared with the two-subunit Polζ. In the absence of Pol31/Pol32, the Rev3/Rev7 enzyme tends to aggregate, and complex formation with Pol31/Pol32 greatly enhances the solubility of Polζ. The catalytic efficiency for nucleotide incorporation is increased between threefold and fivefold in Polζ-d compared with Polζ, but the lesion bypass characteristics, such as a defect in the ability to insert a nucleotide opposite DNA lesions, are similar in both Pols. Complex formation with Pol31/Pol32 also stimulates the DNA synthetic activity of Pol3 (11, 13) and enhances its solubility; thus, the effects of these subunits on Rev3/Rev7 are similar to their effects on Pol3.
The indispensability of Pol31 and Pol32 for Polζ function in TLS in yeast cells could derive not only from their role in stabilizing the Rev3/Rev7 structure, but also from their role in providing sites for interaction with Rev1 and Rad5, which are also required for Polζ-dependent mutagenic TLS (26–28). Rev1 has been shown to interact physically with Rev3 as well as with Pol32, and Rev1 also binds to Rad5 (26, 28, 29), raising the possibility that a higher-order complex comprising Polζ-d, Rev1, and Rad5 functions in TLS.
In addition to its role in TLS, Rev3 is required for spontaneous mutagenesis (30, 31) and for double-strand break (DSB) repair-induced mutagenesis in yeast cells (32, 33). Polζ localizes with HO-induced DSBs, and genetic analyses in yeast have suggested a role for Polζ in Ku-dependent nonhomologous end-joining as well (34). Furthermore, disruption of mouse Rev3 (35–37) but not of other TLS Pols, such as Pols η, ι, or κ, causes embryonic lethality, and Rev3-deficient mouse B cells exhibit reduced efficiency of class switch recombination and increased frequency of DNA breaks in the Ig H locus (38). These observations have suggested a role for mammalian Polζ in class switch recombination-associated DSB repair by nonhomologous end-joining. The potential ability of multiple subunits to interact with a diverse set of protein factors could allow Polζ-d to function in its many biological roles.
Materials and Methods
Details of protein expression and purification, enzymatic assays, and genetic studies are provided in SI Materials and Methods. All proteins were expressed in yeast cells and purified using glutathione Sepharose and anti-FLAG agarose affinity chromatography, and Superdex-200 gel filtration. For UV survival and mutagenesis studies, yeast cells harboring the genomic rev3Δ mutation and either the WT POL31 gene or the pol31 K358E (sdp5-15) mutation were transformed with low-copy number plasmids containing the WT REV3 gene or its various mutants.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant CA107650.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206052109/-/DCSupplemental.
See Commentary on page 12268.
References
- 1.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol Cell Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA. The C-terminal zinc finger of the catalytic subunit of DNA polymerase δ is responsible for direct interaction with the B-subunit. Nucleic Acids Res. 2004;32:3005–3016. doi: 10.1093/nar/gkh623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 6.Huang M-E, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 7.Johansson E, Garg P, Burgers PMJ. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 8.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pursell ZF, Isoz I, Lundström E-B, Johansson E, Kunkel TA. Yeast DNA polymerase ɛ participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagès V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci USA. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Netz DJA, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RE, Prakash L, Prakash S. Yeast and human translesion DNA synthesis polymerases: Expression, purification, and biochemical characterization. Methods Enzymol. 2006;408:390–407. doi: 10.1016/S0076-6879(06)08024-4. [DOI] [PubMed] [Google Scholar]
- 13.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc Natl Acad Sci USA. 2011;108:17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haracska L, et al. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giot L, Simon M, Dubois C, Faye G. Suppressors of thermosensitive mutations in the DNA polymerase δ gene of Saccharomyces cerevisiae. Mol Gen Genet. 1995;246:212–222. doi: 10.1007/BF00294684. [DOI] [PubMed] [Google Scholar]
- 17.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baranovskiy AG, et al. X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle. 2008;7:3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klinge S, Núñez-Ramírez R, Llorca O, Pellegrini L. 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. EMBO J. 2009;28:1978–1987. doi: 10.1038/emboj.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida H, et al. Structural determinant for switching between the polymerase and exonuclease modes in the PCNA-replicative DNA polymerase complex. Proc Natl Acad Sci USA. 2009;106:20693–20698. doi: 10.1073/pnas.0907780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain R, et al. Structural insights into yeast DNA polymerase δ by small angle X-ray scattering. J Mol Biol. 2009;394:377–382. doi: 10.1016/j.jmb.2009.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baranovskiy AG, et al. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 25.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 26.Acharya N, Johnson RE, Prakash S, Prakash L. Complex formation with Rev1 enhances the proficiency of yeast DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26:9555–9563. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagès V, et al. Requirement of Rad5 for DNA polymerase ζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2008;180:73–82. doi: 10.1534/genetics.108.091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acharya N, Johnson RE, Pagès V, Prakash L, Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc Natl Acad Sci USA. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunz BA, Ramachandran K, Vonarx EJ. DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae. Genetics. 1998;148:1491–1505. doi: 10.1093/genetics/148.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roche H, Gietz RD, Kunz BA. Specificity of the yeast rev3Δ. antimutator and REV3 dependency of the mutator resulting from a defect (rad1Δ) in nucleotide excision repair. Genetics. 1994;137:637–646. doi: 10.1093/genetics/137.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand break repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase ζ-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- 36.Esposito G, et al. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 37.Wittschieben J, et al. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 38.Schenten D, et al. Pol ζ ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009;206:477–490. doi: 10.1084/jem.20080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.