Abstract
A cell membrane can be considered a liquid-phase plane in which lipids and proteins theoretically are free to diffuse. Numerous reports, however, describe retarded diffusion of membrane proteins in animal cells. This anomalous diffusion results from a combination of structuring factors including protein–protein interactions, cytoskeleton corralling, and lipid organization into microdomains. In plant cells, plasma-membrane (PM) proteins have been described as relatively immobile, but the control mechanisms that structure the PM have not been studied. Here, we use fluorescence recovery after photobleaching to estimate mobility of a set of minimal PM proteins. These proteins consist only of a PM-anchoring domain fused to a fluorescent protein, but their mobilities remained limited, as is the case for many full-length proteins. Neither the cytoskeleton nor membrane microdomain structure was involved in constraining the diffusion of these proteins. The cell wall, however, was shown to have a crucial role in immobilizing PM proteins. In addition, by single-molecule fluorescence imaging we confirmed that the pattern of cellulose deposition in the cell wall affects the trajectory and speed of PM protein diffusion. Regulation of PM protein dynamics by the plant cell wall can be interpreted as a mechanism for regulating protein interactions in processes such as trafficking and signal transduction.
Proteins within membranes play significant roles in signal perception and transduction, solute partitioning, and secretion. Accordingly, more than 25% of the proteome of higher plants is predicted to be membrane-associated proteins (1, 2).
Proteins diffuse within the plane of a membrane through thermal agitation. Each protein diffusing freely (3) has a diffusion constant that is dependent on the protein’s hydrodynamic radius and the viscosity of the membrane and surrounding medium (4). In a hypothetical uniform membrane, proteins would be distributed randomly. However, biological membranes are spatially complex, with regions of protein and lipid concentration. Numerous reports describe retarded diffusion of membrane proteins (5–8) because of structuring factors such as protein–protein interactions (9), cytoskeleton corralling (10), and lipid organization into nanodomains (11). Membrane nanostructuring is crucial for protein–protein interactions and can either segregate or colocalize membrane proteins, thus optimizing protein interactions in processes such as trafficking and signal transduction (12).
Like yeast and animal cells, plant cells have a subcompartmentalized plasma membrane (PM). Membrane rafts (reviewed in ref. 13) have been demonstrated in plant PMs by proteomics on detergent-insoluble membranes (DIMs). DIMs are enriched in signaling, stress response, cellular trafficking, and cell-wall metabolism proteins (14–16). The Chlorella kessleri hexon-proton symporter HUP1 and Solanum tuberosum remorin StREM1.3 have been visualized in clusters within the PM (17, 18), and the clustering localization pattern of HUP1 is disrupted in mutant yeast lines lacking typical ergosterol and sphingolipid microdomains (17). The physiological role of plant PM substructuring has been demonstrated in several studies. For instance, in the sterol mutant cyclopropylsterol isomerase1-1 (cpi1.1) of Arabidopsis thaliana, the asymmetric localization of PIN2 and, hence, polar auxin transport are perturbed (19). In polarized cells such as pollen tubes, perfusion with the sterol-binding toxin filipin not only perturbs membrane microdomain structure but also alters calcium gradients, production of reactive oxygen species, and normal cell elongation (20).
The relationship between membrane subcompartmentalization and protein diffusion has not been studied in detail, however, and only a few reports have quantified protein diffusion in plant-cell membranes. In the case of PM proteins, such as KAT1, PMA2 H+ATPase, PIN2, PIP2;1, BOR1, NIP5;1, and AtFH1, only a small fraction of the protein pool is mobile (19, 21–23). In contrast, endoplasmic reticulum-associated proteins, nuclear membrane proteins, and tonoplast-associated proteins diffuse more freely within the membrane (24–26). Consequently, we are led to believe that the plant cell has specific properties that constrain PM protein diffusion.
Here, we studied PM protein diffusion in plant cells to understand better PM structure and function. Protein mobility was quantified using fluorescence recovery after photobleaching (FRAP) for a set of 13 plant PM proteins fused to fluorescent proteins. Then we developed a set of modified PM proteins that we term “minimal” because only the membrane-interacting or -spanning domains are present. Minimal PM proteins were designed to reduce the effect of protein interactions on diffusion and showed that DIM association and cytoskeleton have very little effect on protein mobility. However, PM proteins that normally are almost immobile become mobile when the cell wall is absent or when the distance between PM and cell wall is increased. Then cell-wall interaction with PM proteins was confirmed by single-molecule tracking using total internal reflection fluorescence (TIRF) microscopy. Our results show that the cell wall constrains protein diffusion, especially for proteins with larger extracellular domains, even in the absence of binding interactions between proteins and cell-wall components.
Results
PM Proteins Are Relatively Immobile.
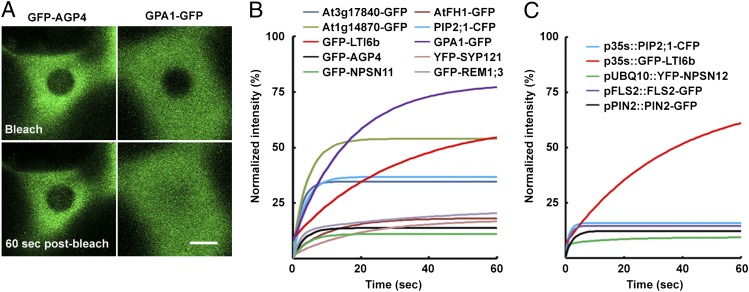
Sutter et al. (27) demonstrated low lateral mobility of the potassium channel KAT1 and the H+-ATPase PMA2. These observations have been supported recently by similar results on other PM proteins (19, 21–23, 28, but systematic study of PM protein dynamics is lacking. We selected 10 Arabidopsis PM proteins, including examples of several different membrane-association types: transmembrane domains, lipid modifications, and peripheral membrane proteins (Table 1). These proteins were fused to fluorescent protein and transiently expressed in Nicotiana tabacum leaves (29). All 10 constructs marked the cell PM. We used FRAP experiments to quantify protein mobility. GFP fluorescence was bleached in a small region of PM, and fluorescence recovery within the region was monitored (Fig. 1A; see Fig. S1 for technique). Using a nonlinear curve-fitting approach, we determined the relative fraction of the protein free to diffuse within the bleached region during 60 s postbleaching (I60s). We observed large differences between constructs in this characteristic. For instance, the bleaching area of GFP-AGP4 remains visible much longer than that of GPA1-GFP (Fig. 1A). The implication is that I60s is lower for GFP-AGP4 than for these other proteins. This difference becomes clear when data are plotted (Fig. 1B). Indeed, I60s is highly variable among constructs (Table 1), indicating that a proportion of the protein potentially does not diffuse but is fixed in place, probably through interaction with other cellular components. Monitoring of this relatively short fluorescence-recovery phase was intended to exclude artifacts that would result from endo- or exocytotic removal or insertion of protein from the membrane (30). Relatively low levels of fluorescence that occur in the cytoplasm for some constructs (e.g., GFP-NPSN11 or PIP2;1-CFP) result in an apparent two-phase recovery process in which rapid initial recovery during the first few seconds is followed by a flat line indicating no subsequent recovery (Fig. 1B). The bleaching laser bleaches not only the PM but also the underlying cytoplasm, which recovers within seconds because of cytoplasmic streaming. Any subsequent change in fluorescence intensity within the bleached spot is the result of lateral diffusion within the PM.
Table 1.
Fluorescence recovery after photobleaching for different PM proteins fused to GFP
| Gene (from Arabidopsis) | Type of anchoring* | No. of amino acids | Expressed in | Promoter | I60 s ± SEM (%) | R2 for curve fit |
| GFP-NPSN11 | 1 TM (type 2) | 265 | Tobacco | p35S | 10.4 ± 0.8 | 0.75 |
| PIP2;1-GFP | 6TM (type 4) | 287 | Arabidopsis | p35S | 10.8 ± 1.1 | 0.71 |
| YFP-NPSN12 | 1 TM (type 2) | 265 | Arabidopsis | pUBQ10 | 11.8 ± 1.7 | 0.34 |
| PIN2-GFP | 9TM (type 4) | 647 | Arabidopsis | pPIN2 | 13.3 ± 0.8 | 0.42 |
| AtFH1-GFP | 1 TM (type 1) | 1051 | Tobacco | p35S | 18.1 ± 0.5 | 0.76 |
| FLS2-GFP | 1 TM (type 1) | 1173 | Arabidopsis | pFLS2 | 19.0 ± 2.0 | 0.43 |
| GFP-AGP4 | GPI | 135 | Tobacco | p35S | 20.0 ± 2.2 | 0.52 |
| GFP-REM;3.1† | Extrinsic - inner leaflet | 198 | Tobacco | p35S | 23.4 ± 1.2 | 0.46 |
| YFP-SYP121 | 1 TM (type 2) | 346 | Tobacco | p35S | 23.7 ± 3.4 | 0.69 |
| At1g14870-GFP | 1 TM (type 2) | 152 | Tobacco | p35S | 36.2 ± 2.7 | 0.74 |
| PIP2;1-CFP | 6TM (type 4) | 287 | Tobacco | p35S | 43.7 ± 1.7 | 0.41 |
| At3g17840-GFP | 1 TM (type 1) | 647 | Tobacco | p35S | 58.9 ± 3.8 | 0.73 |
| GFP-LTI6b | 2 TM (type 4) | 54 | Tobacco | p35S | 72.4 ± 2.7 | 0.84 |
| GPA1-GFP | Myristoylated and palmitoylated | 383 | Tobacco | p35S | 79.8 ± 3.9 | 0.90 |
| GFP-LTI6b | 2 TM (type 4) | 54 | Arabidopsis | p35S | 91.4 ± 3.6 | 0.99 |
Maximum recovery (I60s) of prebleaching fluorescence intensity during 133 s.
*TM, transmembrane domain. ”Type” refers to membrane-anchoring topology.
†Gene from Solanum tuberosum.
Fig. 1.
Plant PM proteins are relatively immobile. (A) FRAP examples illustrating slow (GFP-AGP4) and fast (GPA1-GFP) membrane lateral diffusion. (Scale bar: 5 μm.) (B and C) FRAP curves of PM protein GFP fusions. (B) GFP fusions expressed in N. tabacum leaf cells. GFP-LTI6b and GPA1-GFP have relatively high mobility; others experience an initial rapid (2–10 s postbleaching) recovery of fluorescence that results from diffusion within the cytoplasmic fraction followed by almost no recovery during the subsequent 50 s, showing that they are highly immobile when anchored within the PM. (C) GFP fusions expressed in A. thaliana. p35S::GFP-LTI6b behaves with relatively high mobility, as in tobacco leaf cells, but all others tested (including pFLS2::FLS2-GFP and pPIN2::PIN2-GFP, both of which were expressed under native promoters) were very immobile.
Protein Crowding Within the PM Has a Limited Effect on Protein Diffusion.
Protein crowding within membranes should reduce protein lateral mobility, because collision between molecules restricts diffusion (31). Consequently, we quantified diffusion of the overexpressed proteins p35S::GFP-LTI6b, p35S::PIP2;1-GFP, and pUBQ10::YFP-NPSN12 in hypocotyl cells of stably transformed Arabidopsis. Again we observed two typical types of fluorescence-recovery curves: one-phase diffusion for GFP-LTI6b and two-phase with little diffusion for PIP2;1-GFP and YFP-NPSN12 (Fig. 1C). In addition, we tested two constructs under their endogenous promoters, pFLS2::FLS2-GFP and pPIN2::PIN2-GFP, and again found that a large proportion of each protein remains immobile. To test whether the relative amount of a protein within a membrane was related to the extent of fluorescence recovery in FRAP experiments, we plotted mean prebleach intensity within a bleaching region vs. maximum recovery (I60s). We also tested whether the relative amount of protein within the PM was associated with observed FRAP levels, but no relationship was observed (R2 = 0.03) (Fig. S2).
Relative Immobility of Many PM Proteins Is Not Caused by Protein Interactions.
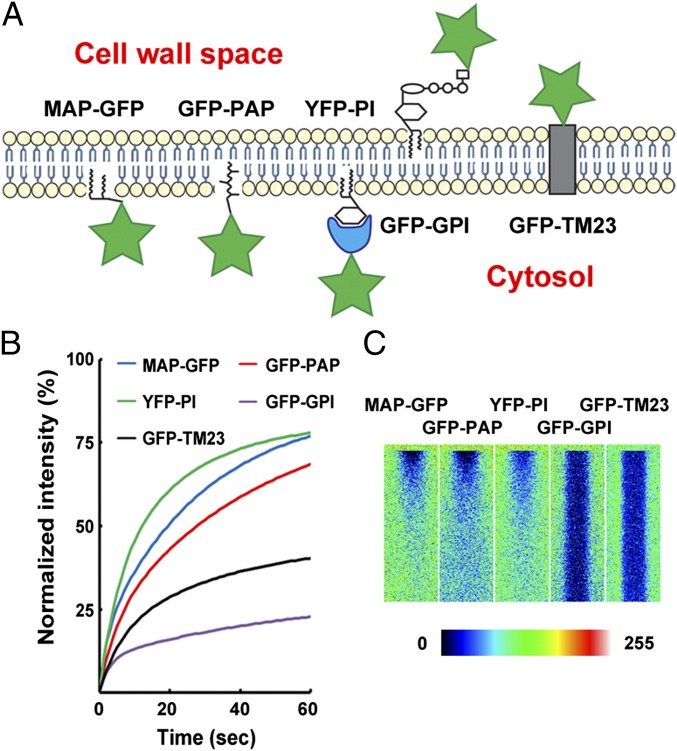
Eleven of the 13 full-length PM protein constructs tested were shown to be relatively immobile (Table 1). Proteins can form larger complexes within membranes by self-association or through protein–protein interactions with other endogenous proteins. Complex formation could limit diffusion, so we designed a set of modified PM proteins as fluorescent protein fusions. These minimal PM proteins consist of only the membrane-anchoring residues, and we predicted that they would have no ability to interact with other cellular constituents. Several different membrane-anchoring types of minimal construct were generated, including a myristoylated and palmitoylated GFP (MAP-GFP), a prenylated GFP (GFP-PAP), a glycosylphosphatidylinositol-anchored GFP (GFP-GPI), a phosphatidylinositol-4-phosphate–binding protein YFP (PI-YFP) (32), and the LAMP1 transmembrane domain fused to GFP (GFP-TM23) (33) (Fig. S3A). We verified the correct targeting of all constructs to the PM in leaves (Fig. S3B). Minimal FP constructs in which the FP moiety was intracellular and anchored by a single transmembrane domain were tested but lost their targeting to the PM, presumably because of the removal of a C-terminal targeting signal (Fig. S4). As described previously (33), TM23 accumulates mainly in the PM but also is present in Golgi bodies and within the endoplasmic reticulum membrane. Then, we confirmed the predicted topology of these constructs (Fig. 2A) by incubating tobacco mesophyll protoplasts with an antibody against GFP or YFP. As expected, only GFP-GPI and GFP-TM23 showed labeling (Fig. S5). This result confirms that in two constructs the GFP is outside the cell in the apoplastic space, whereas in MAP-GFP, GFP-PAP, and YFP-PI the fluorescent protein remains within the symplastic space. Finally, Arabidopsis transgenic lines were generated for each minimal construct.
Fig. 2.
Minimal FP constructs targeted to the PM have different diffusion dynamics in A. thaliana. (A) Schematic of the topology of minimal FPs. Green stars represent the fluorescent protein. (B) FRAP curves for minimal FPs. MAP-GFP, GFP-PAP, and YFP-PI are highly mobile; GFP-TM23 and GFP-GPI are relatively less mobile. (C) Kymograms of protein diffusion within the PM. MAP-GFP, GFP-PAP, and YFP-PI show a centripetal movement of fluorescence. No recovery is observable for GFP-GPI and GFP-TM23. Color scale indicates pixel intensity from 0 (black) to 255 (brightest intensity possible).
FRAP experiments on constructs in which the GFP projects into the cell (i.e., MAP-GFP, GFP-PAP, and YFP-PI) showed the one-phase type of gradual but continuous recovery during 60 s postbleaching, with PM proteins finally recovering to relatively high levels [MAP-GFP I60s = 90.4 ± 3.0% (R2 = 0.80); GFP-PAP I60s = 83.4 ± 3.2% (R2 = 0.86); YFP-PI I60s = 80.3 ± 2.5% (R2 = 0.78)] (Fig. 2B). In addition, for MAP-GFP, GFP-PAP, and YFP-PI, kymographic analysis clearly shows that recovery of fluorescence is centripetal and therefore results from lateral diffusion (Fig. 2C). The constructs in which GFP projects out of the cell (i.e., GFP-GPI and GFP-TM23) recover to significantly lower levels [GFP-GPI I60s = 26.6 ± 3.1% (R2 = 0.54); GFP-TM23 I60s = 45.8 ± 2.9% (R2 = 0.66)] (Fig. 2B). The shapes of their recovery curves are very similar to the ones that we describe for full-length proteins in the previous section. GFP-GPI and GFP-TM23 are relatively immobile, and kymographic analysis of their fluorescence recovery shows that recovery does not proceed from the margins of the bleached area (Fig. 2C). These results show that, even for minimal PM protein constructs, protein mobility can be limited and suggest that secondary protein association and interactions are not for the cause of limited diffusion within the PM.
Lipid Domain Organization and the Cytoskeleton Have Little Effect on Protein Diffusion.
Lipid organization resulting in inhomogeneity within the membrane could induce sequestration of proteins and influence their diffusion. We used detergent fractionation to isolate DIM proteins from solubilized membrane proteins and analyzed fractions to test for the presence of minimal PM protein constructs. As a control, GFP-REM, but not PMA2-GFP, appears in DIM fractions 1, 2, and 3, confirming its previously published DIM localization (Fig. S6A) (18). Similar to GFP-REM, MAP-GFP and GFP-GPI co-occurred in DIM and non-DIM fractions. Myristoylated proteins (MAPs) and glycosylphosphatidylinositol-anchored proteins (GPIs) often are abundant in DIM fractions (15). Finally, GFP-PAP occurred mostly in non-DIM fractions, and YFP-PI and GFP-TM23 occurred only in non-DIM fractions (Fig. S6A). Surprisingly, however, minimal PM proteins that occurred in membrane microdomains (DIM fractions) had contrasting mobile fractions (I60s MAP-GFP = 90.4 ± 3.0%; I60s GFP-GPI = 26.6 ± 3.2%), indicating that there is no direct relationship between membrane subcompartmentation and mobility. Filipin III is a 3-β-hydroxysterol–binding antibiotic (34) that induces changes in cholesterol organization (35). In FRAP experiments on Arabidopsis treated with 100 μM filipin III, the mobile fraction of minimal PM protein constructs did not fluctuate (control vs. filipin, P = 0.87, two-way ANOVA) (Fig. S6B).
In animal cells the cytoskeleton has been shown to corral membrane proteins within subregions (36) by forming “fences” that limit protein diffusion. To test if actin or microtubule cytoskeletons might exert a similar influence on membrane-protein mobility in plant cells, we incubated minimal GFP-expressing Arabidopsis seedlings with either cytochalasin D or oryzalin to depolymerize actin microfilaments or microtubules, respectively. No increase in mobile fraction for these constructs was observed (Fig. S6C). Consequently, we believe that the cytoskeleton is not responsible for the relative immobility of GFP-GPI or GFP-TM23.
Plant Cell Walls Have a Major Effect on Protein Lateral Mobility.
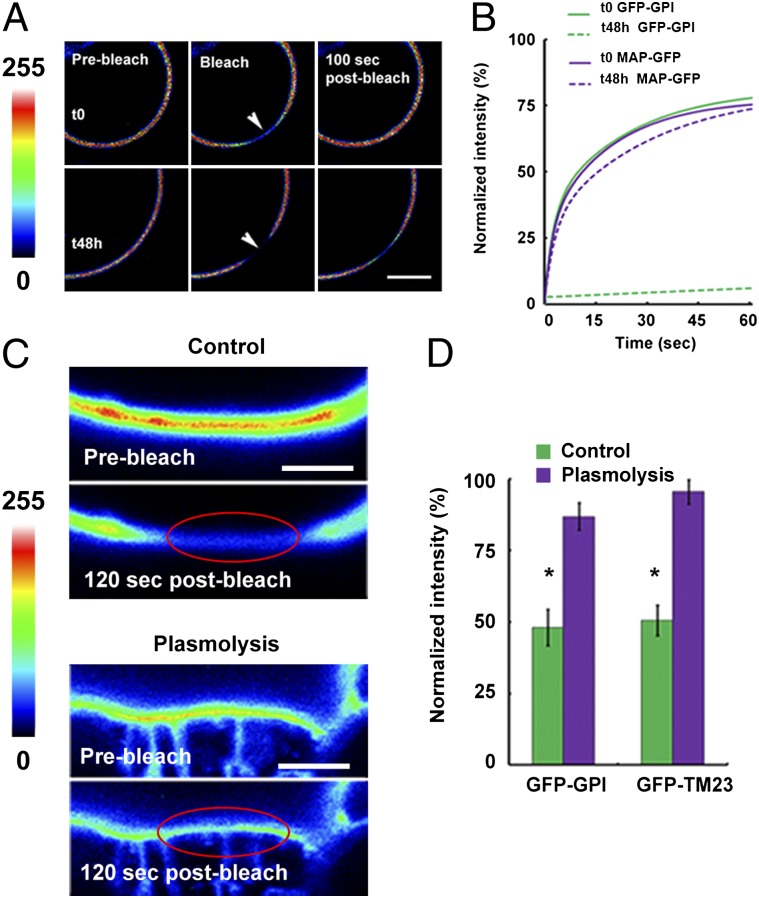
The two least mobile of our minimal PM protein constructs, GFP-GPI and GFP-TM23 (Fig. 2A), orient in the PM with GFP projecting into the cell-wall space. The remaining three minimal proteins orient in the PM so that the fluorescent protein projects into the cytoplasm. We decided to investigate whether the cell wall influences protein diffusion within the PM. To do so, cell walls either were removed by protoplasting or were separated from the PM by plasmolysis. First, we studied protein mobility during neosynthesis of protoplast cell walls (Fig. S7). FRAP experiments were carried out both on freshly prepared protoplasts (cell-wall absent) and on protoplasts in which the cell wall had been regenerated for 24 or 48 h. GFP-GPI has a high mobile fraction in fresh protoplasts that decreases by a factor of more than 20 after cell-wall regrowth (GFP-GPI I60s t0 = 79.9 ± 3.2%; I60s t48h = 2.6 ± 0.6%; P < 0.001, t test) (Fig. 3 A and B). In contrast, the mobile fraction of MAP-GFP, in which GFP projects into the cell, did not differ between fresh protoplasts and those with regrown cell walls (I60s t0 = 76.6 ± 1.8%; I60s t48h = 75.6 ± 1.73%; P = 0.71, t test). This result suggests that the cell wall plays a role in immobilization of PM proteins that project into the cell-wall space.
Fig. 3.
The cell wall limits lateral mobility of plant PM proteins. (A) Protoplasts expressing GFP-GPI. Fluorescence recovers quickly in freshly prepared protoplasts, but the protein is much less mobile once the cell wall has regrown for 48 h. Arrowheads indicate the bleached region. Color scale indicates pixel intensity from 0 (black) to 255 (brightest intensity possible). (Scale bar: 5 μm.) (B) Mobile fraction of GFP-GPI and MAP-GFP in freshly prepared protoplasts (t0) and after cell-wall regeneration (t48h). The mobile fraction (I60s) of GFP-GPI decreased significantly as the cell wall was neosynthesized (I60s GFP-GPI t0 = 79.9 ± 3.2%; t48h = 2.6 ± 0.6%; P < 0.001, t test), but that of MAP-GFP did not change. (C) GFP-GPI is relatively immobile in FRAP experiments in control cells but becomes mobile in cells plasmolyzed with 0.5 M mannitol. Color scale indicates pixel intensity from 0 (black) to 255 (brightest intensity possible). Red ellipse marks the bleached region. (Scale bars: 2 μm.) (D) Plasmolysis induces a highly significant increase in fluorescence recovery (*P < 0.001 for both I60s GFP-GPI control vs. plasmolysis and I60s GFP-TM23 control vs. plasmolysis; t test).
These results were verified by incubating tissue in a hyperosmotic buffer to induce plasmolysis, a shrinkage of the protoplast that results in the creation of a space between the cell wall and the PM. FRAP experiments were carried out on control and plasmolyzed cells expressing GFP-GPI (Fig. 3C) and GFP-TM23 and showed a significant increase in mobility of the proteins’ I60s in plasmolyzed cells (I60s GFP-GPI control vs. plasmolysis, P < 0.001, t test; I60s GFP-TM23 control vs. plasmolysis, P < 0.001, t test) (Fig. 3D).
Single-Molecule Tracking Reveals an Effect of Cellulose Deposition on paGFP-LTI6b Diffusion.
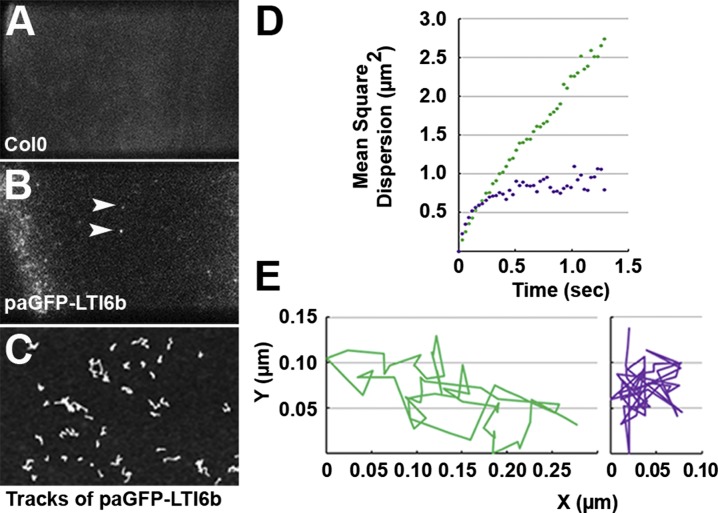
Single-molecule imaging by TIRF microscopy has been used previously to detect and track individual proteins in plant samples (37, 38). We used this technique to observe the PM of plants expressing photoactivable GFP (paGFP) (24, 39) fused to LTI6b. Before photoactivation, a small fraction of paGFP molecules occur naturally in the activated state, i.e., they behave as GFP without the requirement for photoactivation. This pool of autoactivated paGFP is of sufficiently low density to be useful for single-molecule tracking studies even when overexpressed. In these circumstances normal GFP is too bright, but paGFP-LTI6b has a very good signal-to-noise ratio and appears as discrete spots that are trackable over time (Fig. 4 A–C and Movie S1). paGFP molecules show a typical blinking behavior (Fig. S8) with a short time of residence (1.28 ± 0.09 s) at the PM before bleaching. Mean squared displacement (MSD) describes the average motion in a population of diffusing molecules as a function of time and is a useful means of characterizing the type of molecular motion that occurs. LTI6b is a relatively mobile PM protein (Fig. 1B) with only two residues predicted to be in the extracellular space. Its MSD in control cells has a linear dependence on time (Fig. 4D), as predicted for molecules that diffuse freely (40). When seedlings were incubated with 20 μM isoxaben for 1 h to disrupt cell-wall structure, we observed greatly restricted molecular movement and sublinear MSD, indicating constrained diffusion of paGFP-LTI6b in this condition (Fig. 4E).
Fig. 4.
Single-molecule tracking observations of paGFP-LTI6b molecules in the PM of A. thaliana. (A) Wild-type PM (Col0) did not show spots. (B) PM in plants expressing nonphotoactivated paGFP-LTI6b. A small population of paGFP molecules fluoresces without requiring photoactivation (arrowheads), and this density of emitting fluorochrome produced a signal-to-noise ratio sufficient to generate single-molecule tracking data sets. (Scale bar: 2 μm in A and B.). (C) Tracks of single molecules during a 2.5-s time series. (Scale bar: 1 μm.) (D) MSD of single paGFP-LTI6b molecules as a function of time. In control cells, movement of paGFP-LTI6b has a linear dependence on time (green). When seedlings were incubated with 20 μM isoxaben for 1 h to disrupt cell-wall structure, MSD of paGFP-LTI6b became sublinear (purple). (E) Single-molecule diffusion was more constrained in cells treated with isoxaben for 1 h (purple) than in control cells (green).
Discussion
Measurement of protein mobility has been used in this report to get a better understanding of control mechanisms that structure the plant cell PM.
PM Proteins Have Relatively Low Lateral Mobility.
Eleven of the 13 PM proteins fused with GFP were relatively immobile. This result is consistent with previous studies (19, 21, 23, 27). No doubt a subset of PM proteins is immobilized through specific physical interaction with cell-wall components. For instance, the potassium channel KAT1 is known to be strongly immobile at the PM (28) and forms dot patterns in the PM which coalign with cellulose microfibrils (41). The cell wall also anchors AtFH1 (23). We have demonstrated, however, that other low-mobility PM proteins have no physical interaction with cell-wall components and have tested for PM/cell-wall proximity effects that might mediate protein immobilization.
The two full-length proteins with the highest mobility were GFP-LTI6b and GPA1-GFP, which are predicted to have only two amino acids in the apoplast or to be inserted in the inner leaflet of the PM, respectively. Full-length proteins inserted only in the outer leaflet of the PM, such as GFP-GPI in which the GFP projects into the cell-wall space, were strongly immobilized by the cell wall.
A set of minimal PM proteins that retain PM targeting ability but have been modified to prevent any protein–protein interaction were used to rule out protein interactions as a diffusion constraint. Lateral mobility of minimal PM proteins within the plane of the PM was constrained, as it was for many full length proteins, so we tried to identify other cellular factors that might limit protein mobility. Protein association, membrane microdomain structure, and cytoskeletons were shown to have little effect on protein dynamics. The cell wall, however, was shown to be involved in controlling protein mobility in the PM of plants. Plasmolysis to separate the PM and cell wall physically resulted in increased protein mobility, and, conversely, cell-wall regeneration in protoplasts led to a decrease in PM protein mobility. These results show the importance of the cell wall in limiting lateral diffusion of PM proteins, but an interesting distinction was made based on the extent to which a protein projects into the cell-wall space.
Cell Wall Corrals PM Proteins.
Electron microscopy studies show that the distance between cell wall and PM must be smaller than a few tens of nanometers. This close association is highlighted by three factors. The first is the cellulose synthesis mechanism, which is mediated by the cellulose synthase complex (42). The cellulose synthase complex is a rosette protein complex inserted in the PM which secretes cellulose microfibrils directly to the apoplast, thus forming a direct link between the PM and one of the cell-wall components (43, 44). Second, some proteins, such as arabinogalactans (AGPs), wall-associated kinases, and formin1 (AtFH1), are known to form molecular bridges between the PM and cell wall (23). Third, plant cells have very high turgor pressure, which applies a pressure of 0.1–0.6 MPa on the PM in the direction of the cell wall (45). The cellulose microfibril meshwork and associated components of the cell wall such as pectins and hemicelluloses no doubt are appressed directly to the lipid bilayer. As a consequence, this meshwork might form bounded regions that serve to constrain the diffusion of PM proteins and phospholipids.
A cell-wall meshwork that acts as a constraint to PM protein lateral mobility is analogous with the animal cell anchored-protein picket model of Fujiwara et al. (36). Those authors used single-molecule tracking experiments to demonstrate that the actin cytoskeleton corrals proteins and limits protein and lipid lateral mobility in NRK cells. Our experiments show that actin and microtubule cytoskeletons do not perform this same function in plant cells but that a mechanism to stabilize proteins is conserved at the PM/cell-wall interface. Maintenance of protein localization and association within the PM is vital for cell signaling and transport and is a key component in the mechanism that maintains asymmetric distribution of PM proteins such as the PIN auxin efflux facilitators (22, 46).
The herbicide isoxaben inhibits cellulose synthases 2, 3, 5, and 6 (47) and therefore has a very detrimental effect on cell-wall structure. In single-molecule tracking experiments, paGFP-LTI6b, with only two residues in the apoplast, experienced a significant change to a more constrained movement pattern after very short (1 h) isoxaben treatment. Short-duration treatment of mature cells with isoxaben is unlikely to alter cell-wall structure dramatically, as occurs when seedlings are grown in the drug. In the short term, slight alteration to cellulose microfibril patterning might result from isoxaben treatment, but deposition of other cell-wall components such as pectins might be affected also. That the mobility of LTI6b is constrained by short-term isoxaben treatment shows not only that interactions at the PM/cell-wall interface have an effect on proteins with extracellular domains but also that cell-wall organization universally influences protein diffusion.
Materials and Methods
A full discussion of materials and methods can be found in SI Materials and Methods. FRAP and single-molecule tracking methods are presented briefly here.
FRAP Experiments.
The relative mobile fraction at time 60 s postbleaching (I60s) of different fluorescent proteins was assessed by FRAP following the technique of Martinière et al. (23) (Fig. S1). Circular regions of interest (ROIs) (radius 4.3 μm) were bleached in median optical sections of the fluorescent PM. Recovery of fluorescence was recorded during 60 or 120 s with a delay of 1.5 s between frames. Fluorescence intensity data were normalized using the equation:
where In is the normalized intensity, It is the intensity at any time t, Imin is the minimum intensity postphotobleaching, and Imax is the mean intensity before photobleaching.
Nonlinear regression was used to model the normalized FRAP data. In this case, a two-phase exponential association equation was used:
where Yt is normalized intensity, A, B, C, K1, and K2 are parameters of the curve, and t is time.
For each treatment, 10–20 cells were analyzed. The value of the fluorescence intensity recovery plateau was calculated for t = 60 and was used as an approximation of the relative mobile fraction (I60s).
TIRF Microscopy and Single-Molecule Tracking.
TIRF imaging was performed using a custom-built microscope equipped with a 100× objective (α-Plan-Fluar, NA = 1.45; Zeiss), 491-nm laser excitation (Cobolt), HQ525/50-nm emission filter (Chroma), and an electron-multiplication CCD (iXon; Andor) (48).
Cells overexpressing nonphotoactivated paGFP-LTI6b have a population of naturally fluorescent paGFP molecules that is at a suitable density for TIRF microscopy and single-particle tracking. Tracks were calculated as described in Rolfe et al. (49). We measured 939 spots in six repetitions. Mean time of PM residence and track length were recorded. The MSD was calculated for all molecules:
where |ri(t + Δt) − ri(t)| is the distance traveled by molecule i between time t and time t + Δt, and the expectation value is over all pairs of time points separated by Δt in each molecular track.
Supplementary Material
Acknowledgments
J.R. and A.M. were funded by Biotechnology and Biological Sciences Research Council Grant BB/F01407/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 12274.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202040109/-/DCSupplemental.
References
- 1.Schwacke R, et al. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 3.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 4.Saffman PG, Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson NL, Axelrod D. Reduced lateral mobility of a fluorescent lipid probe in cholesterol-depleted erythrocyte membrane. Biochim Biophys Acta. 1980;597:155–165. doi: 10.1016/0005-2736(80)90159-5. [DOI] [PubMed] [Google Scholar]
- 6.Yechiel E, Edidin M. Micrometer-scale domains in fibroblast plasma membranes. J Cell Biol. 1987;105:755–760. doi: 10.1083/jcb.105.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwik J, et al. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc Natl Acad Sci USA. 2003;100:13964–13969. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenne PF, et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 2006;25:3245–3256. doi: 10.1038/sj.emboj.7601214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieber JJ, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 10.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 11.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 12.Owen DM, Williamson D, Rentero C, Gaus K. Quantitative microscopy: Protein dynamics and membrane organisation. Traffic. 2009;10:962–971. doi: 10.1111/j.1600-0854.2009.00908.x. [DOI] [PubMed] [Google Scholar]
- 13.Mongrand S, Stanislas T, Bayer EM, Lherminier J, Simon-Plas F. Membrane rafts in plant cells. Trends Plant Sci. 2010;15:656–663. doi: 10.1016/j.tplants.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Borner GH, et al. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005;137:104–116. doi: 10.1104/pp.104.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morel J, et al. Proteomics of plant detergent-resistant membranes. Mol Cell Proteomics. 2006;5:1396–1411. doi: 10.1074/mcp.M600044-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre B, et al. Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 2007;144:402–418. doi: 10.1104/pp.106.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossmann G, Opekarova M, Novakova L, Stolz J, Tanner W. Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:945–953. doi: 10.1128/EC.00206-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raffaele S, et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Men S, et al. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol. 2008;10:237–244. doi: 10.1038/ncb1686. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, et al. Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 2009;60:303–313. doi: 10.1111/j.1365-313X.2009.03955.x. [DOI] [PubMed] [Google Scholar]
- 21.Takano J, et al. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feraru E, et al. PIN polarity maintenance by the cell wall in Arabidopsis. Curr Biol. 2011;21:338–343. doi: 10.1016/j.cub.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 23.Martinière A, Gayral P, Hawes C, Runions J. Building bridges: Formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J. 2011;66:354–365. doi: 10.1111/j.1365-313X.2011.04497.x. [DOI] [PubMed] [Google Scholar]
- 24.Runions J, Brach T, Kühner S, Hawes C. Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot. 2006;57:43–50. doi: 10.1093/jxb/eri289. [DOI] [PubMed] [Google Scholar]
- 25.Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2010;61:134–144. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 26.Sparkes I, Runions J, Hawes C, Griffing L. Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell. 2009;21:3937–3949. doi: 10.1105/tpc.109.072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutter JU, Campanoni P, Tyrrell M, Blatt MR. Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell. 2006;18:935–954. doi: 10.1105/tpc.105.038950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorieul M, Santoni V, Maurel C, Luu DT. Mechanisms and effects of retention of over-expressed aquaporin AtPIP2;1 in the endoplasmic reticulum. Traffic. 2011;12:473–482. doi: 10.1111/j.1600-0854.2010.01154.x. [DOI] [PubMed] [Google Scholar]
- 29.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- 30.Luu DT, et al. Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress. Plant J. 2012;69:894–905. doi: 10.1111/j.1365-313X.2011.04841.x. [DOI] [PubMed] [Google Scholar]
- 31.Umenishi F, Verbavatz JM, Verkman AS. cAMP regulated membrane diffusion of a green fluorescent protein-aquaporin 2 chimera. Biophys J. 2000;78:1024–1035. doi: 10.1016/S0006-3495(00)76661-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vermeer JE, et al. Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 2009;57:356–372. doi: 10.1111/j.1365-313X.2008.03679.x. [DOI] [PubMed] [Google Scholar]
- 33.Brandizzi F, et al. The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell. 2002;14:1077–1092. doi: 10.1105/tpc.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grebe M, et al. Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol. 2003;13:1378–1387. doi: 10.1016/s0960-9822(03)00538-4. [DOI] [PubMed] [Google Scholar]
- 35.Bonneau L, et al. Plasma membrane sterol complexation, generated by filipin, triggers signaling responses in tobacco cells. Biochim Biophys Acta. 2010;1798:2150–2159. doi: 10.1016/j.bbamem.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, et al. Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell. 2011;23:3780–3797. doi: 10.1105/tpc.111.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizcay-Barrena G, Webb SE, Martin-Fernandez ML, Wilson ZA. Subcellular and single-molecule imaging of plant fluorescent proteins using total internal reflection fluorescence microscopy (TIRFM) J Exp Bot. 2011;62:5419–5428. doi: 10.1093/jxb/err212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 40.Holtzer L, Schmidt T. The tracking of individual molecules in cells and tissues. In: Brauchle C, Lamb D, Michaelis J, editors. Single Particle Tracking and Single Molecule Energy Transfer. Weinheim, Germany: Wiley-VCH; 2010. [Google Scholar]
- 41.Homann U, Meckel T, Hewing J, Hütt MT, Hurst AC. Distinct fluorescent pattern of KAT1:GFP in the plasma membrane of Vicia faba guard cells. Eur J Cell Biol. 2007;86:489–500. doi: 10.1016/j.ejcb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Carpita NC. Update on mechanisms of plant cell wall biosynthesis: How plants make cellulose and other (1->4)-β-D-glycans. Plant Physiol. 2011;155:171–184. doi: 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giddings TH, Jr, Brower DL, Staehelin LA. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol. 1980;84:327–339. doi: 10.1083/jcb.84.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller SC, Brown RM., Jr Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol. 1980;84:315–326. doi: 10.1083/jcb.84.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Hukin D, Pritchard J, Thomas C. Comparison of plant cell turgor pressure measurement by pressure probe and micromanipulation. Biotechnol Lett. 2006;28:1147–1150. doi: 10.1007/s10529-006-9075-x. [DOI] [PubMed] [Google Scholar]
- 46.Kleine-Vehn J, et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol. 2011;7:540. doi: 10.1038/msb.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bischoff V, Cookson SJ, Wu S, Scheible WR. Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot. 2009;60:955–965. doi: 10.1093/jxb/ern344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke DT, et al. Optics clustered to output unique solutions: A multi-laser facility for combined single molecule and ensemble microscopy. Review of Scientific Instruments. 2011;82:093705. doi: 10.1063/1.3635536. [DOI] [PubMed] [Google Scholar]
- 49.Rolfe DJ, et al. Automated multidimensional single molecule fluorescence microscopy feature detection and tracking. Eur Biophys J. 2011;40:1167–1186. doi: 10.1007/s00249-011-0747-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.