Abstract
Phenotypic heterogeneity displayed by a clonal bacterial population permits a small fraction of cells to survive prolonged exposure to antibiotics. Although first described over 60 y ago, the molecular mechanisms underlying this behavior, termed persistence, remain largely unknown. To systematically explore the genetic basis of persistence, we selected a library of transposon-mutagenized Escherichia coli cells for survival to multiple rounds of lethal ampicillin exposure. Application of microarray-based genetic footprinting revealed a large number of loci that drastically elevate persistence frequency through null mutations and domain disruptions. In one case, the C-terminal disruption of methionyl-tRNA synthetase (MetG) results in a 10,000-fold higher persistence frequency than wild type. We discovered a mechanism by which null mutations in transketolase A (tktA) and glycerol-3-phosphate (G3P) dehydrogenase (glpD) increase persistence through metabolic flux alterations that increase intracellular levels of the growth-inhibitory metabolite methylglyoxal. Systematic double-mutant analyses revealed the genetic network context in which such persistent mutants function. Our findings reveal a large mutational target size for increasing persistence frequency, which has fundamental implications for the emergence of antibiotic tolerance in the clinical setting.
Keywords: genetic interactions, evolution, adaptation, systems biology
In a classic study conducted in 1944, Joseph W. Bigger discovered that a subpopulation of cells within a genetically homogeneous culture of Staphylococcus aureus was consistently able to survive prolonged exposure to bactericidal concentrations of the then-novel antibiotic penicillin (1). The behavior of these cells, called persisters, was not due to a mutation because the initial survivors, once regrown, displayed the same level of sensitivity to penicillin. Instead, persistence has been attributed to noninherited antibiotic resistance (2) and was shown to be a feature of a subpopulation of bacteria exhibiting phenotypic heterogeneity (3). It is thought that this heterogeneity has evolved to ensure the longevity of a population threatened with a potentially catastrophic event such as lethal antibiotic exposure (4). Recent years have witnessed renewed interest in persistence due to its potential role in chronic and recalcitrant infections. Understanding the biology of persistence is thus central to achieving effective antibiotic treatment.
Many years after Bigger’s description, a substantial contribution to our understanding of persistence was made by the identification of high persistence (hip) mutations in an Escherichia coli operon (5). One of these, hipA7, increases persistence frequency by 10,000-fold. Using these mutants and microfluidic devices, Balaban and colleagues provided a rich phenotypic description by showing that the persistent subpopulation exists in a slow-growing state in the presence of the β-lactam drug, ampicillin; because ampicillin is a bactericidal agent that targets cell wall biosynthesis, nondividing persister cells are immune to the lethal effects of this drug (6). Although the dormant state of persisters is accompanied by a cessation of most cellular processes, translation continues at a reduced rate (7). In a recent study, Allison and colleagues take advantage of this observation and show that metabolic stimulation of persisters makes them vulnerable to killing by aminoglycosides (8), which are bactericidal antibiotics that target the ribosome. The enhanced killing of persisters by the addition of metabolites was found to be specific to aminoglycosides, as metabolic stimulation failed to reduce the number of persisters when exposed to quinolones or β-lactam antibiotics (8), which target DNA replication and cell division, respectively.
Despite efforts to identify the genetic determinants contributing to persister cell formation, we still have a limited understanding of the molecular basis of this phenotype. In this study, we applied a highly sensitive experimental strategy for large-scale characterization of the genetic basis of persistence. After a series of selections using a high-density library of transposon-insertional mutants, we combined competitive selection and microarray-based genetic footprinting to measure the contribution of every genetic locus to persistence. Our results implicate a large number of genes that increase persistence by varying degrees and through diverse mechanisms. We used the same framework to carry out global epistasis analyses, which revealed the genetic network context in which these loci function to affect persistence.
Results and Discussion
Selection Strategy to Identify Genetic Determinants for Persistence.
We designed and implemented a unique selection strategy to identify the genetic loci contributing to persistence. Because of its widespread occurrence, we hypothesized that persistence is an evolutionary adaptation with a discrete set of genetic determinants. We sought to identify these determinants by exposing a library of high-density transposon-insertional mutants of E. coli to several rounds of selection in the presence of lethal antibiotic exposure.
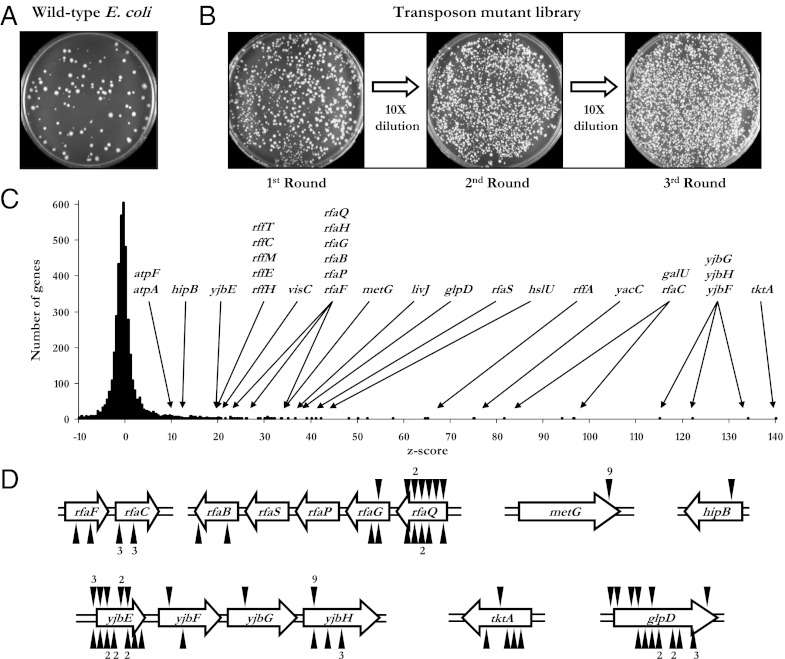
Because persisters are commonly observed in biofilms or associated with a solid surface (9), we expected to observe a more robust persistence phenotype if we used a solid growth medium as a substrate for adhesion. To enrich for mutants exhibiting an increasing propensity for persistence, a culture of transposon-insertional mutants was grown to stationary phase, plated on LB agar containing ampicillin, and incubated at 37 °C for 24 h. The plates were then sprayed with penicillinase to inactivate the ampicillin followed by an additional incubation to permit the growth of surviving colonies. This procedure constituted one round of persister cell enrichment, and we reasoned that multiple cycles of selection would enrich for mutants with a high rate of persistence. Colonies were eluted from the agar plates and used to inoculate fresh media, which was then taken through the enrichment cycle again. We performed three rounds of enrichment and, to ensure a large diversity of mutants, we used 50 plates (each containing ∼1,500 independent colonies) for each of the three rounds.
We were encouraged by our strategy after observing that the transposon mutant library displayed a persistence frequency ∼10-fold higher than the wild type at the initial round of enrichment (Fig. 1A). As can be seen from a representative set of plates in Fig. 1B, the fraction of persister cells in the mutant library substantially increases with each cycle of enrichment. To quantify the contribution of every genetic locus to increased persistence, we applied our microarray-based genetic footprinting (10) to the entire population of enriched cells. The hybridization value for each ORF on the array is compared with five hybridizations of the unselected library, yielding a positive z score for favorable mutations and a negative z score for deleterious insertions (10). Roughly 50 loci had extremely high z scores (Fig. 1C). These genes belong to a variety of categories such as metabolism, cell structure, and genes of unknown function.
Fig. 1.
Enrichment for persister mutants. (A) A 100-μL sample of an undiluted stationary phase culture was spread on the surface of LB agar containing ampicillin. The ampicillin was inactivated with penicillinase after 24 h of incubation at 37 °C. The plate was imaged following 24 h of additional incubation. (B) The library of mutants was enriched for persister mutants and cultivated in identical growth conditions as the wild type. After three enrichment cycles, the persister frequency of the enriched population was 1,000-fold higher than the starting population. (C) Distribution of z scores calculated from the mean and SD of five unselected library replicates. (D) Transposon insertions from multiple mutants were mapped to these genes. Triangles above or below the genes denote transposon insertions with the T7 promoter oriented to the right or left, respectively. The exact locations of the insertions and a full list of mutants identified using this procedure can be found in Table S2.
Isolation of Mutants with High-Level Persistence.
To isolate individual mutants for further analysis, a small aliquot from the eluted pool of mutants was diluted and plated. Over 100 of these colonies were selected at random and cultured separately. Genetic footprinting was applied to each of the mutants to PCR amplify and sequence the region of DNA adjacent to the transposon insertion. The sequencing results revealed that each mutant contained a single transposon insertion, and that many of insertions were mapped to different locations within a defined set of genes (Fig. 1D and Table S1). Among the genes that contained multiple insertions were metG, tktA, glpD, and members of the rfa and yjb operons. The sequencing results corroborate the hybridization data and suggest that a distinct set of genetic determinants contribute to increased persistence.
To confirm the sequencing and hybridization results, we performed assays to measure the persistence frequency of mutants we isolated. We performed the persistence assays by replicating the conditions used for selection. We cultured individual isolates to stationary phase, diluted and plated on LB agar with ampicillin, and incubated the plates for 24 h before and after spraying with penicillinase. Persistence was calculated as the ratio of survival fraction of the mutant to the wild type. The mutants showed a wide range of persistence frequencies, from approximately 3 to 10,000 times higher than the wild type (Table S1).
We expected that the z score of a gene would be congruent with the survival ratio of the corresponding mutant. This concordance was largely the case except for mutants with insertions in two genes, metG and hipB. Given the parameters of the experiment, the z scores reflect not only the capacity to survive exposure to antibiotic, but also the ability to grow in the absence of the antibiotic. With this understanding, we hypothesized that the metG::Tn and hipB::Tn insertional mutants would exhibit reduced growth rates relative to the wild type. We observed that these mutants have only a slightly reduced growth rate, but exhibited a substantial lag phase (Fig. S1). Because bacterial growth rate has been shown to be proportional to antibiotic susceptibility (11), the extended lag period displayed by these mutants may be a contributing factor; however, the lag difference is small in comparison with the time scale of persistence assays and insufficient to explain the three-log increase in persistence frequency. Additional kill curves in the presence of fluoroquinolones (Fig. S2), which are capable of killing nongrowing cells but much less effective in clearing persisters (12), clearly show that these mutants are true hyperpersisters. Growth rates for other mutants isolated in this study were similar to wild type.
Our selection strategy may have given rise to mutants that are tolerant to ampicillin through a resistance mechanism rather than persistence. Using two complementary methods of determining the minimal inhibitory concentration (MIC) of these strains to ampicillin, we showed that the persistence phenotype exhibited by the strains listed in Table S1 was not a result of genetically acquired antibiotic resistance (Fig. S3).
Quantitative Determination of Persistence Frequency.
Kill curves provide a quantitative measure of persistence frequency by representing the survival fraction of cells as a function of the length of exposure to antibiotics (6). To generate a kill curve, a culture in stationary phase is diluted and plated on LB agar containing ampicillin. At designated time points, penicillinase is sprayed onto the surface of the agar to inactivate the ampicillin. Colonies that form after a subsequent incubation period are counted, and the survival fraction at each time point is calculated relative to the initial number of cells. For each kill curve, the hipA7 mutant and the wild type are used as strains that represent high- and low-persistence frequency, respectively. To make the comparisons meaningful, the hipA7 allele was transferred to the same genetic background as the wild type. To transfer this allele, we took advantage of a selectable marker closely linked to hipA7 and a cold sensitivity phenotype conferred by the allele (13) to transduce hipA7 from TH1269 (14) to our wild-type E. coli MG1655 (Fig. S4). We created quantitative kill curves for a subset of the mutants identified in this study (discussed below).
Insertions in metG Create a Hyper-Persistence Phenotype.
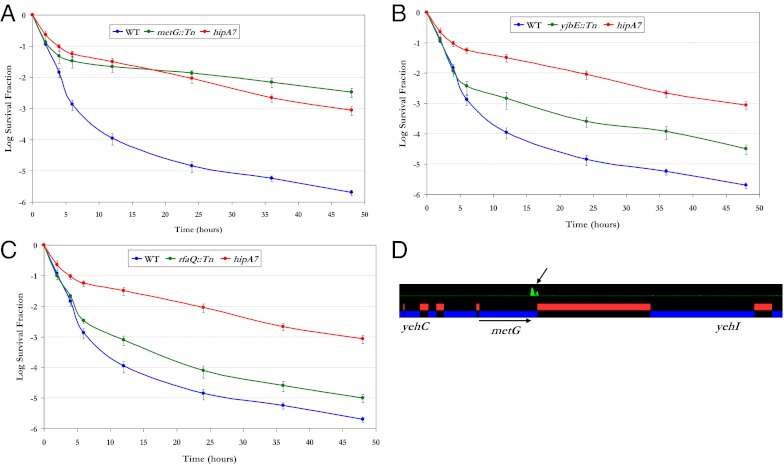
We isolated several mutants with insertions in the 3′ region of metG (Fig. 1D). The persistence frequency of one of these mutants was evaluated by generating a kill curve, and remarkably, this metG mutant showed a level of survival that is very similar to the hipA7 mutant (Fig. 2A). To ensure that the observed phenotype was due to the insertion in metG and not a secondary mutation, we used P1 transduction to move the insertion back into the wild-type parent strain. This strain showed a level of persistence indistinguishable from the original isolate (Fig. S5). The sequencing results revealed that the transposon insertion occurred at the 3′ end of metG, presumably disrupting the C-terminal domain and preserving the essential function of the gene product. This observation was confirmed through a high-resolution mapping of transposon-insertion sites using a high-density tiling array (Fig. 2D).
Fig. 2.
Phenotypic characterization of insertional mutants. (A–C) Kill curves generated for mutants with insertions in metG (A), yjbE (B), and rfaQ (C) are shown in green. For comparison, kill curves were also generated for the wild type (blue) and the hipA7 strain (red). Error bars represent SDs of three independent measurements. (D) A high-resolution tilling array covering the entire E. coli genome with overlapping 25-mer oligos at four base pair resolution was used to display the abundance of insertions in a population of transposon mutants that have been enriched for persistence. This profile of the metG locus shows a strong transposon-insertion signal limited to the 3′ end of metG. Peaks in green represent insertion sites.
The metG gene encodes methionyl-tRNA synthetase (MetRS), an enzyme essential for viability that functions to covalently link methionine amino acids to their cognate tRNA molecules. MetRS is a large protein with multiple domains, and the distal C-terminal domain is involved in dimerization (15). C-terminal truncations of MetRS, leading to the monomeric form of the enzyme, have been reported to have minimal effect on the activity of the enzyme (16). It has been found that monomeric MetRS has reduced binding affinity to the methionine tRNA (tRNAMet) (17). The reduced affinity to tRNAMet exhibited by the metG mutants may be the key factor in this mutant's persistence phenotype. Inefficiencies in translation may lead to a heterogeneous population in which a minority of cells in the population experiences a temporary cessation of growth. The similarities in the kill kinetics of this mutant to the hipA7 mutant (Fig. 2A) suggest that both mutants exhibit the persistence phenotype by the common mechanism of stochastic switching from the growing to the nongrowing state (6).
Increased Persistence Frequency Through Activation of the Rcs-Signaling Pathway.
Some of the mutants we identified had insertions in genes encoding cellular structures. For example, many of the constituents of the rfa operon, which are associated with LPS synthesis, exhibited high z scores. Mutations in genes comprising the rfa operon have been found to activate the Rcs-signaling pathway (18), and activation of this pathway has been linked to increased tolerance to β-lactam antibiotics (19). Because activation of the Rcs pathway by LPS mutations has been found to induce colanic acid capsular polysaccharide synthesis (18), we hypothesized that the increased polysaccharide synthesis displayed by LPS mutants serves as a barrier that reduces antibiotic permeability. We confirmed this hypothesis by disrupting the Rcs pathway in our LPS mutants and observing a persistence frequency comparable to the wild type.
Another operon with multiple insertions contributing to persistence was the yjb operon. The genes in this operon have no assigned function; however, the yjbEFGH locus has been found to be part of the Rcs regulon (20). This locus is also implicated in exopolysaccharide production (21). As with the rfa mutants, introducing mutations that block signaling through the Rcs pathway abolished the observed elevation in persistence of yjb insertional mutants. Therefore, the persistence phenotype may be more related to the generation of a drug permeability barrier rather than an intracellular state that is impervious to antibiotic effect.
Most of the genes that comprise the yjb and rfa operons showed high z scores in our selection (Fig. 1C), and we isolated a disproportionately large number of mutants with insertions in yjbE and rfaQ (Fig. 1D). Quantitative kill curves were generated using yjbE::Tn (Fig. 2B) and rfaQ::Tn (Fig. 2C), which revealed an intermediate level of persistence, below the frequency for hipA7 yet 10-fold higher than the wild type.
Increased Persistence Through the Accumulation of the Intracellular Metabolite Methylglyoxal.
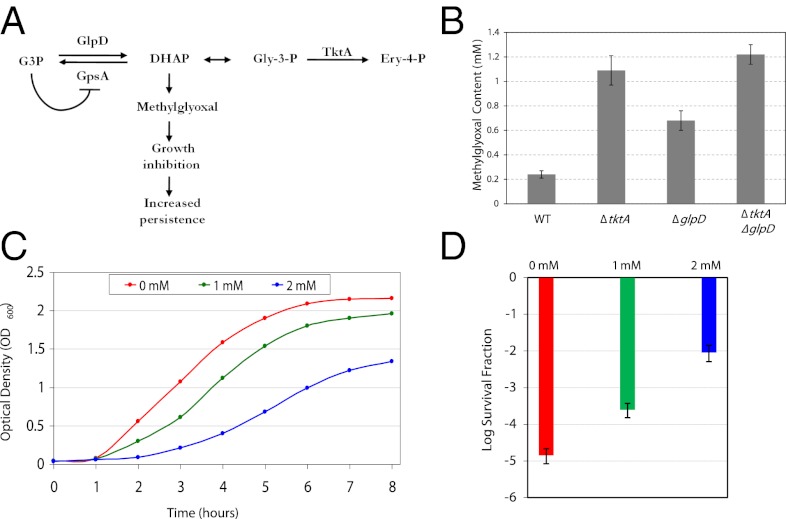
Loss of function mutations in two genes associated with metabolism, G3P dehydrogenase (glpD) and transketolase A (tktA), gave rise to increased persistence frequency. Glycerol-3-phosphate dehydrogenase is encoded by glpD, and tktA codes for transketolase in E. coli. The gene products of glpD and tktA function at the juncture between respiration and glycolysis. During aerobic respiration, glycerol-3-phospate (G3P) is converted to dihydroxyacetone phosphate (DHAP) by GlpD. The reverse reaction—conversion of G3P from DHAP—is catalyzed by G3P synthase (GpsA). In a glpD mutant, the accumulation of G3P inhibits GpsA by a negative feedback mechanism, which leads to the accumulation of DHAP (22) (Fig. 3A). Likewise, mutations in TktA prevent the conversion of glyceraldehyde-3-phosphate (Gly-3-P) to erythrose-4-phosphate (Ery-4-P), which also leads to an accumulation of DHAP (Fig. 3A). Elevated levels of DHAP have been shown to produce methylglyoxal (23), which causes growth arrest (24) and could be responsible for the high-persistence frequency observed by glpD and tktA mutants.
Fig. 3.
Role of methylglyoxal in the persistence phenotype of glpD and tktA mutants. (A) A model illustrating the metabolic consequences of glpD and tktA disruptions. G3P is converted to DHAP by GlpD during aerobic respiration, and the reverse reaction is catalyzed by GpsA. DHAP is a precursor to Gly-3-P, which is converted to Ery-4-P by TktA. Mutations in glpD cause elevated levels of G3P, which inhibits the function of GpsA and leads to increased amounts of DHAP. DHAP also accumulates as a consequence of TktA malfunction and leads to production of methylglyoxal, which causes growth cessation and increased persistence. (B) Methylglyoxal concentration was determined by ESI/LC/MS. Error bars represent SDs of three independent measurements. (C) Dose-response curves were used to monitor sensitivity to methylglyoxal. LB media containing the indicated methylglyoxal concentrations was inoculated with an overnight culture of the wild type. Growth was monitored using OD600 spectrophotometric readings. (D) Survival fractions of the wild type exposed to various concentrations of methylglyoxal were determined. Aliquots were taken three hours postinoculation from the dose–response curves shown in C and plated on LB agar with ampicillin. Plates were sprayed with penicillinase after 24-h incubation at 37 °C and incubated again for colony formation. Error bars represent SDs of three independent measurements.
To determine whether the glpD and tktA mutants are producing elevated levels of methylglyoxal, we measured the intracellular levels of this metabolite in these mutants. Both the glpD and tktA mutants showed statistically significant elevated levels of methylglyoxal (Fig. 3B). We created a glpD–tktA double mutant and found even higher levels of methylglyoxal in the double mutant than each of the individual mutants (Fig. 3B). This double mutant displayed persistence frequency of 18.2-fold higher than the wild type (Table S1), which is slightly higher than each of the individual mutants.
Next we sought to determine whether exogenous addition of methylglyoxal causally influences persistence rate. We created dose–response curves to monitor sensitivity to various concentration of methylglyoxal, and found an inverse relationship between metabolite concentration and growth rate (Fig. 3C). Next, we measured persistence frequency of the wild type when exposed to these concentrations, and observed that exposure to methylglyoxal increases persistence in a dose-dependent manner (Fig. 3D). These findings confirm our hypothesis that glpD and tktA loss of function lead to elevated persistence through increased intracellular concentration of methylglyoxal.
Uncharacterized Genes Contributing to Persistence.
Genes with unknown function that were found to be associated with persistence are visC, yacC, and yiiS (Table S1). VisC is a predicted oxidoreductase with an FAD/NAD(P)-binding domain; however, deletion of visC has been found to have no effect on aerobic respiration (25). The gene products of yacC and yiiS have no assigned function, although, yiiS has been identified as a member of the sigma E regulon (26). Further experimentation is required to characterize the role of these genes in persistence.
Insertions in hipB Increase Persistence Frequency.
We identified an insertion in hipB (Table S1), whose product is known to suppress persistence by its physical interaction with HipA (27). Expression of hipA causes an increased rate of persistence through growth cessation (28), and the two substitution mutations in HipA7—G22S and D291A—confer a high-persistence phenotype in hipA7 strains through a weakened HipA–HipB interaction (29). These observations suggest that the inability of HipB to sequester HipA is likely the mechanism by which the hipB insertional mutant is able to exhibit a 10,000-fold increase in persistence frequency.
Characterization of Persistence Through Genetic Interaction Mapping.
In this study, we have shown that single-gene disruptions can produce substantial increases in persistence frequency. However, as with other complex phenotypes, persistence is a manifestation of multigene networks. The microarray-based genetic footprinting approach used in the study is well suited for exploring genetic interaction networks. We used this experimental framework to identify genes that interact with hipA7 and metG::Tn, each of which confers a persistence frequency 10,000-fold higher than the wild type. The first step in this process involved creating double-mutant libraries in each of these genetic backgrounds (10). However, because metG::Tn already contained a transposon with the same selectable marker used to create libraries, we created a mutant by site-directed recombination to replace 21 nucleotides at the 3′ end of metG with a selectable marker (30) that was removed using Flippase recombination enzyme (31), resulting in a gene product missing seven amino acids at the C-terminal. The resulting mutant strain (referred to as metG2) showed a level of persistence identical to metG::Tn (Fig. S5).
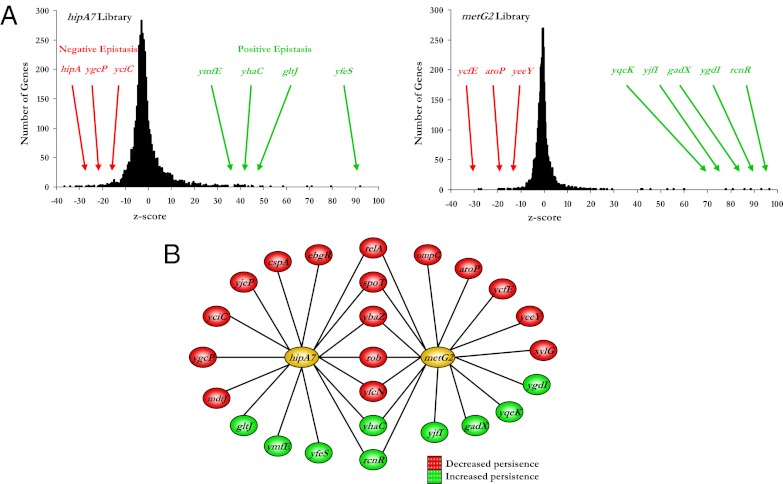
Saturated transposon libraries were generated in the hipA7 and metG2 strains and were exposed to three rounds of persistence selection. Genetic footprinting was applied to the cells after the third stage of selection and the resulting PCR products were hybridized to DNA microarrays. Hybridization values were converted to z scores using three hybridizations for each of the hipA7 and metG2 unselected libraries as references. We obtained many genes with large positive or negative z scores, reflecting genetic interactions that increase or decrease, respectively, the persistence frequency of the parent strains (Fig. 4A).
Fig. 4.
Epistatic interaction map of hipA7 and metG2. (A) Z-score distribution of the hipA7 and metG2 libraries after selection. Positive or negative epistasis refers to secondary mutations that increase or decrease persistence, respectively. (B) Genetic interaction network for the hipA7 and metG2 loci. The map was constructed with genes that exhibited a positive or negative interaction with the hipA7 or metG2 alleles. Genes are color-coded according to the nature of their interaction; that is, whether they decreased (red) or increased (green) the persistence frequency of the hipA7 or metG2 strains.
To validate the microarray results, double mutants were created and evaluated individually for persistence. Using this strategy, we identified many genes that, when mutated, either increased or decreased the persistence frequency of the hipA7 or metG2 mutants in agreement with the microarray data (Fig. S6). Null mutations found to alter persistence in one of these strains were transduced and tested in the other strain. A systematic application of this approach allowed us to create a genetic interaction map for the hipA7 and metG2 loci (Fig. 4B).
We identified a number of genes that interact, positively and negatively, with either hipA7 or metG alone. Among these genes are those that encode transcriptional regulators (ebgR, cspA, and yeeY), which is consistent with the results of a previous large-scale screen for persister alleles (32). We identified three alleles previously associated with drug tolerance: disruption of mdtJ reduced persistence of the hipA7 strain presumably because this gene is involved in multidrug export (33); disruption of gadX, which is involved with multidrug resistance (34), increased the persistence rate of the metG2 strain; and we observed reduced persistence of hipA7 by deleting cspA, whose expression has been shown to be induced by antibiotic treatment (35).
Two of the genes interacting with metG2, aroP and xylG, encode amino acid and sugar transporters, respectively. In addition, gltJ, which encodes an amino acid transporter, interacts with hipA7. Furthermore, ebgR, which encodes a negative regulator of lactose metabolism, interacts with hipA7. These findings suggest a relationship between persistence and the metabolism of sugars and amino acids.
We identified seven interactions shared by both hipA7 and metG2, suggesting the involvement of common pathways. We were encouraged by finding that relA and spoT were among the genes that reduce persistence in both backgrounds, because mutations in these have previously been shown to abolish the persistence phenotype of the hipA7 strain (14). We also found that insertions in rob reduce persistence in both strains. Overexpression of rob has been shown to increase antibiotic tolerance (36), which suggests that this mutation may be modulating antibiotic sensitivity. In addition, we identified genes encoding products involved with DNA mismatch repair (ybaZ), transcriptional regulation (rcnR), and conserved proteins of unknown function (yfcN and yhaC).
Conclusions
We used a library of transposon-insertional mutants and a genomewide mapping strategy to identify the genetic determinants contributing to bacterial persistence. Although previous attempts to identify persister mutants using transposon mutagenesis were met with limited success (37), we were able to substantially increase the number of genes associated with this phenotype. Our favorable outcome can be attributed to three experimental innovations used in this study: (i) we used a high-density library of insertional mutants to perform selections en masse; (ii) we performed our selections on solid growth media rather than planktonic cultures because the former better approximates the environment found in biofilms; and (iii) we combined genetic footprinting with microarray hybridization to obtain a quantitative readout of population dynamics during the selection process.
The results of this study represent an initial genomewide genetic map of persistence from the perspective of persister mutants. A major observation of our study is the modulation of persistence through many chromosomal mutations. In particular, persistence frequency that can increase through acquired disruptions has fundamental implications for emergence of antibiotic tolerance in the clinical setting. Such persister mutants will not be detected in standard assays that test for minimal inhibitory concentration. However, high-persistence rates may be a major source of recolonization in a wide variety of chronic infections that may be initially cleared by a seemingly successful round of antibiotics. In addition, the large number of persister bacteria constitutes a potentially important reservoir for the emergence of high-level antibiotic tolerance through the acquisition of mobile genetic elements. A better molecular understanding of persistence may lead to the development of drugs that reduce persistence rate to levels low enough to allow successful eradication of the entire population by the application of existing antibiotics. Historically, the genetic basis of persistence has been difficult to study. However, the identification of dozens of loci that substantially alter persistence frequency should aid in future experimental discovery and analysis of persistence pathways.
Materials and Methods
Bacterial Strains and Microbiological and Molecular Techniques.
All strains in this study are isogenic derivatives of E. coli MG1655 and are listed in Table S2. Bacteriophage and plasmids are listed in Table S3, and oligonucleotide sequences can be found in Table S4. Transposon mutant libraries were constructed using hyperactive Tn5 EZ::TN transposase (1.0 U/μL; Epicentre Technologies) according to the manufacturer's guidelines. Chromosomal markers were transferred by transduction with P1vir. Ampicillin (Sigma) was used in LB agar plates at a concentration of 100 μg/mL Penicillinase (Sigma) was diluted to 2,500 U/mL in penicillinase buffer consisting of 50 mM of Tris-Cl (pH 7.5), 50mM NaCl, 1 mM EDTA (pH 7.5), 1 mM DTT, and 20% (vol/vol) glycerol, and stored at −80 °C. Before use, penicillinase was diluted to 50 U/mL in LB and filter sterilized. Genetic footprinting and subsequent hybridization to DNA spotted arrays were performed as described previously (10).
Enrichment for Increased Persistence.
Erlenmeyer flasks containing 50 mL of LB were inoculated with 500 μL of 109 transposon-insertional mutants and cultivated at 37 °C with aeration to early stationary phase; 100 μL of each culture was plated onto each of 50 LB plates with 100 μg/mL of ampicillin and incubated at 37 °C for 24 h. Plates were then sprayed with a fine mist of penicillinase solution to destroy the ampicillin in the media. After another incubation period, colonies were eluted from the agar surface, pooled, and 500 μL of the eluent was used to inoculate 50 mL of fresh LB, which was incubated at 37 °C to stationary phase. This procedure constitutes one cycle of enrichment, which was repeated for three rounds.
Generating Kill Curves.
Kill curves were performed as described previously (6). Briefly, a clonal population was cultivated for 6 h at 37 °C with aeration, diluted, and plated. At specified time points, plates were sprayed with penicillinase and incubated again to permit colony formation. Average survival fractions and SDs were calculated from triplicate sets of plates at each time point.
Quantification of Methylglyoxal.
Methylglyoxal concentrations were determined by electrospray ionization-liquid chromatography-mass spectrometry as previously described (38) with some modifications. Log-phase cultures were grown in LB at 37 °C, washed twice in deionized H2O at 4 °C, followed by sonication at 3 × 5 s 10-W bursts; 5 M hydrochloric acid was added at 0.1 volume and the mixture was centrifuged at 16,000 × g for 10 min to remove cell debris and precipitated proteins. The supernatant samples were derivatized at 4 °C for 4 h with 500 nmol of O-phenylenediamine (Sigma Chemical), loaded with 2.5 nmol 2,3-hexanedione (Sigma Chemical) as an internal standard, and quantified by ESI/LC/MS.
Supplementary Material
Acknowledgments
S.T. was supported National Institute of Allergy and Infectious Diseases Award 5R01AI077562 and National Institutes of Health Director's Pioneer Award 1DP10D003787.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205124109/-/DCSupplemental.
References
- 1.Bigger JW. Treatment of staphylococcal infections with penicillin. Lancet. 1944;ii:497–500. [Google Scholar]
- 2.Levin BR. Noninherited resistance to antibiotics. Science. 2004;305:1578–1579. doi: 10.1126/science.1103077. [DOI] [PubMed] [Google Scholar]
- 3.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 4.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 7.Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc Natl Acad Sci USA. 2008;105:6145–6149. doi: 10.1073/pnas.0711712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol. 1986;132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 12.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 13.Scherrer R, Moyed HS. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J Bacteriol. 1988;170:3321–3326. doi: 10.1128/jb.170.8.3321-3326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 15.Cassio D, Waller JP. Modification of methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1971;20:283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 16.Mellot P, Mechulam Y, Le Corre D, Blanquet S, Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989;208:429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- 17.Crepin T, Schmitt E, Blanquet S, Mechulam Y. Structure and function of the C-terminal domain of methionyl-tRNA synthetase. Biochemistry. 2002;41:13003–13011. doi: 10.1021/bi026343m. [DOI] [PubMed] [Google Scholar]
- 18.Parker CT, et al. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrières L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol. 2003;50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- 21.Ferrières L, Aslam SN, Cooper RM, Clarke DJ. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology. 2007;153:1070–1080. doi: 10.1099/mic.0.2006/002907-0. [DOI] [PubMed] [Google Scholar]
- 22.Clark D, et al. Regulation of phospholipid biosynthesis in Escherichia coli. Cloning of the structural gene for the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J Biol Chem. 1980;255:714–717. [PubMed] [Google Scholar]
- 23.Freedberg WB, Kistler WS, Lin EC. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J Bacteriol. 1971;108:137–144. doi: 10.1128/jb.108.1.137-144.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackerman RS, Cozzarelli NR, Epstein W. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J Bacteriol. 1974;119:357–362. doi: 10.1128/jb.119.2.357-362.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakahigashi K, Miyamoto K, Nishimura K, Inokuchi H. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J Bacteriol. 1992;174:7352–7359. doi: 10.1128/jb.174.22.7352-7359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezuchova B, Miticka H, Homerova D, Roberts M, Kormanec J. New members of the Escherichia coli sigmaE regulon identified by a two-plasmid system. FEMS Microbiol Lett. 2003;225:1–7. doi: 10.1016/S0378-1097(03)00480-4. [DOI] [PubMed] [Google Scholar]
- 27.Black DS, Irwin B, Moyed HS. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falla TJ, Chopra I. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob Agents Chemother. 1998;42:3282–3284. doi: 10.1128/aac.42.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher MA, et al. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 32.Hansen S, Lewis K, Vulić M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higashi K, et al. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J Bacteriol. 2008;190:872–878. doi: 10.1128/JB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishino K, Yamaguchi A. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J Bacteriol. 2004;186:1423–1429. doi: 10.1128/JB.186.5.1423-1429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, Jones P, Inouye M. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J Bacteriol. 1993;175:5824–5828. doi: 10.1128/jb.175.18.5824-5828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ariza RR, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, Coates AR. Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol Lett. 2005;243:117–124. doi: 10.1016/j.femsle.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 38.Randell EW, Vasdev S, Gill V. Measurement of methylglyoxal in rat tissues by electrospray ionization mass spectrometry and liquid chromatography. J Pharmacol Toxicol Methods. 2005;51:153–157. doi: 10.1016/j.vascn.2004.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.