Abstract
Acquired resistance to EGF receptor (EGFR) tyrosine kinase inhibitors (TKIs) is inevitable in metastatic EGFR-mutant lung cancers. Here, we modeled disease progression using EGFR-mutant human tumor cell lines. Although five of six models displayed alterations already found in humans, one harbored an unexpected secondary NRAS Q61K mutation; resistant cells were sensitive to concurrent EGFR and MEK inhibition but to neither alone. Prompted by this finding and because RAS/RAF/MEK mutations are known mediators of acquired resistance in other solid tumors (colon cancers, gastrointestinal stromal tumors, and melanomas) responsive to targeted therapies, we analyzed the frequency of secondary KRAS/NRAS/BRAF/MEK1 gene mutations in the largest collection to date of lung cancers with acquired resistance to EGFR TKIs. No recurrent NRAS, KRAS, or MEK1 mutations were found in 212, 195, or 146 patient samples, respectively, but 2 of 195 (1%) were found to have mutations in BRAF (G469A and V600E). Ectopic expression of mutant NRAS or BRAF in drug-sensitive EGFR-mutant cells conferred resistance to EGFR TKIs that was overcome by addition of a MEK inhibitor. Collectively, these positive and negative results provide deeper insight into mechanisms of acquired resistance to EGFR TKIs in lung cancer and inform ongoing clinical trials designed to overcome resistance. In the context of emerging knowledge about mechanisms of acquired resistance to targeted therapies in various cancers, our data highlight the notion that, even though solid tumors share common signaling cascades, mediators of acquired resistance must be elucidated for each disease separately in the context of treatment.
Keywords: NRAS mutation, gefitinib, erlotinib
Mutations in the gene encoding the EGF receptor (EGFR) are found in ∼10% of white and 30% of Asian non-small cell lung cancers, respectively (1–4). EGFR-mutant tumors with exon 19 deletions and L858R substitutions are highly sensitive initially to the EGFR tyrosine kinase inhibitors (TKIs), gefitinib or erlotinib (1–4). Unfortunately, tumor cells eventually acquire resistance, with progression of disease occurring in patients around 10–16 mo after starting treatment (5). Genetic mechanisms of resistance found in tumor samples from patients with acquired resistance include second-site EGFR mutations [>50% (6–9)], amplification of the gene encoding an alternative kinase, MET [(5–10% (9–12)], and mutations in the downstream signaling lipid kinase, PIK3CA [<5% (12)]. The majority of second-site mutations involve a threonine to methionine change at codon 790 of EGFR, which alters binding of drug to the ATP-binding pocket (6–9). A few percent seemingly undergo histologic transformation, displaying features of small cell lung cancer (12, 13) or epithelial–mesenchymal transition (EMT) (12). Up to 40% of cases of acquired resistance are mechanistically unexplained.
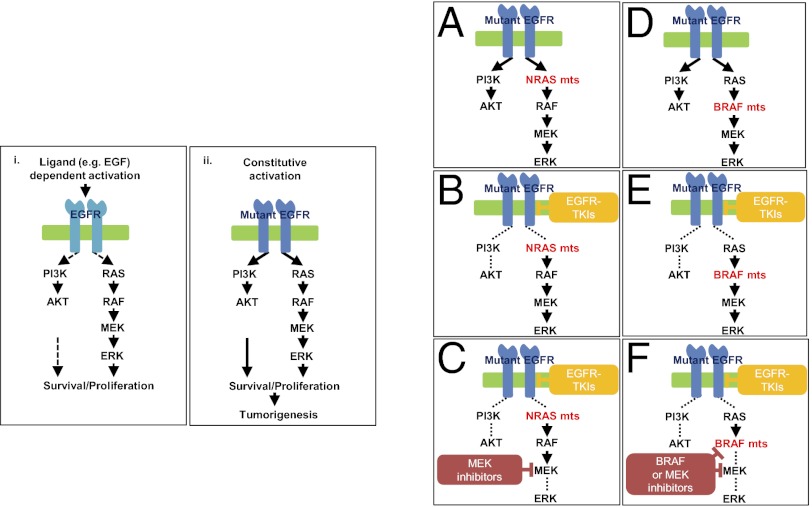
The RAS/RAF/MEK/MAPK signaling pathway downstream of EGFR plays a significant role in tumorigenesis, including lung cancers. Oncogenic recurrent driver mutations in KRAS, NRAS, BRAF, and MEK1 and are found in 15–30%, 1%, 3–5%, and 1% of non-small cell lung cancers, respectively (14–18). Unlike PIK3CA mutations, which occur concurrently with EGFR mutations in individual tumors, genetic alterations in KRAS, NRAS, BRAF, and MEK1 rarely occur in EGFR-mutant tumors (19–22). Mutations in these genes have been associated with primary resistance to targeted EGFR therapy, including EGFR TKIs in lung cancer (23) and anti-EGFR monoclonal antibodies in colon cancer (24). Recently, KRAS mutations also have been associated with acquired resistance to the anti-EGFR antibody cetuximab in colorectal cancers (25); NRAS and MEK1 mutations have been shown to mediate acquired resistance to the mutant BRAF kinase inhibitor, vemurafenib, in melanomas (26, 27); and BRAF mutations have been found in patients with acquired resistance to imatinib in KIT/PDGFRα-mutant gastrointestinal stromal tumors (GISTs) (28). Previous reports have shown that RAS mutations are not found in lung cancers from patients with acquired resistance to EGFR TKIs (7, 12, 29). However, the sample sizes were too small (n = 6, 37, and 14, respectively) to rule them out definitively as mediators of resistance. Only one study looked for BRAF mutations, and MEK1 status was not assessed.
Here, we modeled acquired resistance in vitro and, surprisingly, found that one cell-line model displayed a secondary NRAS mutation. Prompted by the data above and by the additional finding that a clinically relevant mouse lung tumor model of acquired resistance also has identified secondary Kras mutations (30), we systematically analyzed the frequency of secondary RAS pathway gene mutations in samples from patients with acquired resistance. The findings provide deeper insight into mechanisms of acquired resistance, inform ongoing clinical trials designed to overcome resistance, and suggest which mutations should be screened for routinely in samples from patients with acquired resistance.
Results
NRAS Q61K Mutation in an EGFR-Mutant Cell-Line Model of Acquired Resistance to Erlotinib.
To explore potential modes of drug resistance, we used well-established protocols (31) to develop cell models of acquired resistance to erlotinib. Including two lines previously reported (31), we developed a total of six resistant lines over 3–6 mo from five parental cells (PC-9, HCC827, HCC4006, HCC4011, and 11-18) with drug-sensitive EGFR mutations and known sensitivity to drug (Table 1). Using a sensitive assay that can detect mutations at an allele frequency of 5% (32), we found only two resistant lines (PC-9R and HCC827R1) that harbored the EGFR T790M second-site mutation (Table 1 and Fig. S1A). PC-9R cells displayed increased amplification of EGFR compared with parental cells (31), but HCC827R1 and R2 cells appeared to have no change in EGFR gene status. Two lines (HCC4006R and HCC4011R) appeared to lose copies of EGFR (Table 1). Two lines (HCC827R2 and HCC4011R) harbored MET amplification by fluorescent in situ hybridization (Fig. S2) and increased levels of MET protein by immunoblotting studies (Fig. S1B). Consistent with the loss of copies of EGFR, HCC4006R cells displayed loss of dependence on EGFR protein expression for cell growth. Instead, they showed features of EMT, i.e., increased expression of vimentin with loss of E-cadherin (Fig. S1 B and C). Similar results have been reported previously by us and/or others (10, 31, 33–36). No resistant cells displayed features of small cell lung cancer.
Table 1.
EGFR mutant cell-line models of acquired resistance to erlotinib
| Cell line | Primary EGFR mutation | Drug selection | Erlotinib IC50 (μM) | EGFR T790M | EGFR amp | MET amp | Other |
| PC-9 | Exon19 deletion | N/a | 0.01 | No | Yes* | No | N/a |
| PC-9R | Exon19 deletion | Erlotinib | >5 | Yes | Yes* | No | N/a |
| HCC827 | Exon19 deletion | 0.005 | No | Yes† | No | N/a | |
| HCC827R1 | Exon19 deletion | Erlotinib | >5 | Yes | Yes† | No | N/a |
| HCC827R2 | Exon19 deletion | Erlotinib | >5 | No | Yes† | Yes | N/a |
| HCC4006 | Exon19 deletion | N/a | 0.02 | No | Yes | No | N/a |
| HCC4006R | Exon19 deletion | Erlotinib | >5 | No | No | No | EMT |
| HCC4011 | L858R | N/a | 0.03 | No | Yes | No | N/a |
| HCC4011R | L858R | Erlotinib | >5 | No | No | Yes | N/a |
| 11-18 | L858R | N/a | 0.1 | No | No | No | N/a |
| 11-18R | L858R | Erlotinib | >5 | No | No | No | NRAS Q61K |
Six resistant cell lines were established from five parental cell populations. PC-9R and HCC827R1 cells harbored EGFR T790M. HCC827R2 and HCC4011R cells displayed MET amplification (amp) and increased levels of MET protein (Figs. S1A and S2). HCC4006R cells showed features of EMT (Fig. S1A). 11-18R cells harbored an NRAS Q61K mutation. N/a, not applicable.
*PC-9R cells showed further EGFR amplification.
†HCC827R1and HCC827R2 cells showed no further EGFR amplification.
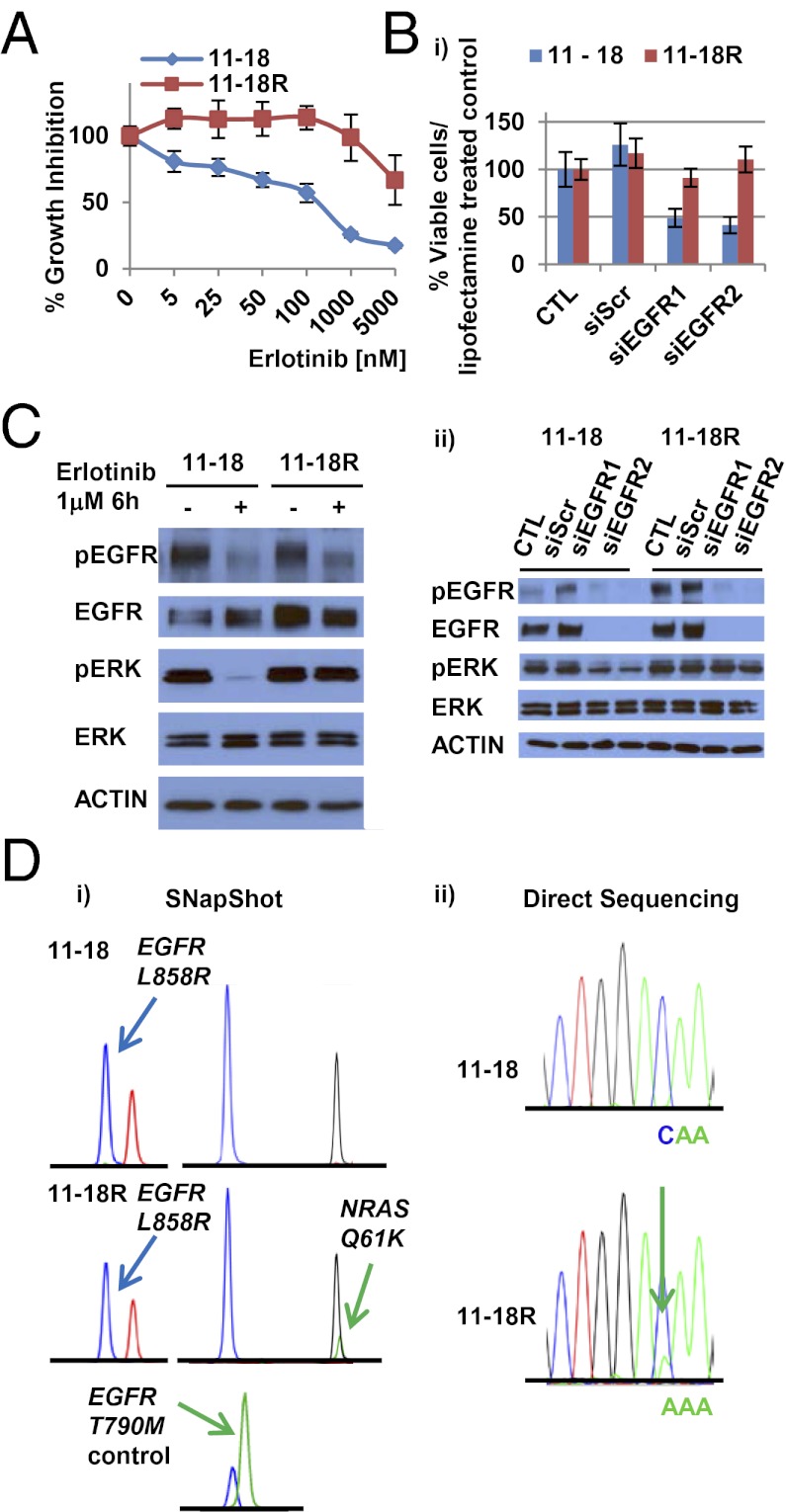
One EGFR T790M-negative line, 11-18R cells, had increased levels of EGFR and MET protein but lacked EGFR or MET amplification and did not display any morphologic changes (Fig. 1A and Figs. S1B, S2, and S3). Growth-inhibition studies after knockdown of EGFR protein using EGFR-specific siRNAs revealed that, unlike PC-9R cells (Fig. S1C), 11-18R cells were no longer dependent solely on EGFR for cell growth (Fig. 1B). Consistent with these findings, erlotinib treatment of 11-18R cells inhibited phosphorylation of EGFR but not the downstream signaling protein ERK (Fig. 1C). 11-18R cells also were resistant to growth inhibition by the more potent second-generation EGFR TKI afatinib (37), except at high doses (>1 μM) (Fig. S1D). These cells were insensitive to a MET TKI, either alone or in combination with erlotinib (Fig. S1E).
Fig. 1.
Characterization of 11-18R cells. (A) Cell growth-inhibition assays show the relative sensitivity of 11-18 and 11-18R cells to erlotinib. Data are expressed as percentage of viability compared with vehicle by the cell titer blue assay. Data shown are mean ± SD of three independent experiments performed in hextuplicate. (B) (i) EGFR knockdown experiments using siRNAs show that, compared with parental cells,11-18R cells are no longer dependent upon EGFR for survival. Data shown are mean ± SD of three independent experiments performed in hextuplicate. Scr, Scramble. Two different siRNAs against EGFR were used. (ii) Immunoblotting studies using the indicated antibodies show that downstream signaling is inhibited in parental but not resistant cells after knockdown of EGFR. (C) The effect of erlotinib on EGFR pathway signaling in 11-18/11-18R cells. (D) (i) SNaPshot assay reveals that, in addition to a baseline EGFR L858R mutation, 11-18R cells have acquired an NRAS Q61K mutation but not an EGFR T790M mutation. (ii) Direct sequencing confirms presence of the NRAS 181C > A (Q61K) mutation in 11-18R cells.
To determine possible mechanisms of resistance in 11-18R cells, we screened corresponding extracted cell DNA with a platform (a combination of SNaPshot and PCR-based sizing assays) that can detect more than 40 recurrent mutations in nine genes (AKT1, BRAF, EGFR, HER2, KRAS, MEK1, NRAS, PIK3CA, and PTEN) relevant to existing and emerging targeted therapies in lung cancer (32). Surprisingly, we found that, compared with parental cells, DNA from 11-18R cells harbored an acquired NRAS Q61K mutation in addition to the primary EGFR L858R substitution (Fig. 1D, Table 1, and Table S1). None of the other cell lines harbored any additional mutations. Consistent with these results, levels of phospho-ERK decreased after knockdown of EGFR in 11-18 parental cells but not in resistant cells (Fig. 1B). Ten individual single-cell clones derived from the resistant cells harbored both mutations (Table S2), suggesting that the NRAS and EGFR mutations were in the same cell. Because cell-line models of acquired resistance to date have harbored mechanisms of resistance found in human tumors, we performed additional characterization of 11-18R cells.
Functional Role of NRAS Q61K in 11-18R Cells.
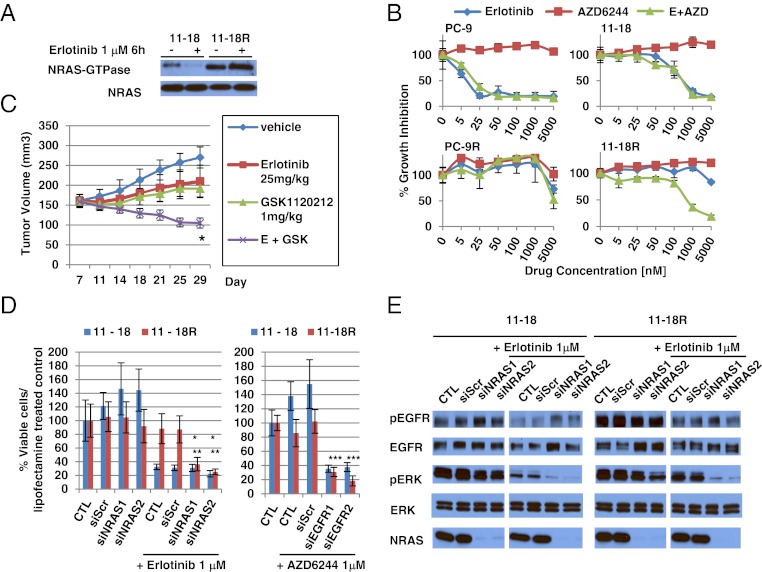
We assessed whether the acquired NRAS mutation plays a functional role in 11-18R cells. First, we validated the activation status of NRAS in 11-18R cells, using a Ras GTPase-specific pulldown assay. As expected, the expression of NRAS-GTPase was much higher in 11-18R cells than in 11-18 parental cells. Treatment with erlotinib had no effect on NRAS activation (Fig. 2A).
Fig. 2.
Functional role of NRAS Q61K in 11-18R cells. (A) RAS GTPase-specific pulldown assay shows increased activated NRAS in 11-18R cells compared with parental cells. Erlotinib has no effect on activated NRAS activity in 11-18R cells. (B) Cell growth-inhibition assays show the relative sensitivity of 11-18/11-18R and PC-9/PC-9R cells to erlotinib, the MEK inhibitor AZD6244, or the combination of erlotinib with AZD6244. Data shown are mean ± SD of three independent experiments performed in hextuplicate. (C) Athymic nude mice with 11-18R tumors were administered vehicle, erlotinib, MEK inhibitor, GSK1120212, or erlotinib plus GSK1120212. Tumor volume was determined at the indicated times after the onset of treatment. n = 5 mice per group. Error bars indicate SE. *P < 0.05 (Student’s t test) for the combination of erlotinib plus GSK1120212 versus either erlotinib or GSK1120212 alone. (D) siRNA-mediated knockdown of NRAS combined with erlotinib or siRNA-mediated knockdown of EGFR combined with AZD6244 inhibits growth of 11-18R cells. Scr, Scramble. Two different siRNAs against NRAS were used. Data shown are mean ± SD of three independent experiments performed in hextuplicate. *, **P < 0.05 (Student’s t test) for the combination of erlotinib plus siNRAS knockdown versus erlotinib or siNRAS alone in 11-18R cells. ***P < 0.05 (Student’s t test) for the combination of siEGFR plus AZD6244 knockdown versus AZD6244 alone in 11-18R cells. (E) Immunoblotting with the indicated antibodies demonstrates that siRNA-mediated knockdown of NRAS combined with erlotinib inhibits ERK activation in 11-18R cells. These samples were run on the same gel but were noncontiguous.
Next, we examined the effect of MEK inhibitors with and without erlotinib on 11-18R cell growth. The MEK serine-threonine kinase is downstream of RAS; therefore, its inhibition might be expected to affect cell viability in cells with activated RAS. The MEK inhibitor AZD6244 alone had no effect in either parental or resistant 11-18 cells. Addition of AZD6244 to erlotinib had no additive effect in parental cells, but the combination suppressed growth in the resistant cells (Fig. 2B). Similar results were obtained in vitro as well as in vivo with another MEK inhibitor, GSK1120212 (Fig. S4A and Fig. 2C). By contrast, addition of AZD6244 to erlotinib had no additive effect against PC-9 parental or resistant cells, the latter of which harbor EGFR T790M (Fig. 2B). Consistent with these findings, only the combination of erlotinib and AZD6244 strongly inhibited levels of phospho-ERK in 11-18R cells (Fig. S4B). Interestingly, the MEK inhibitor alone was able to inhibit the growth of H1299 cells, which harbor no drug-sensitive EGFR mutations but do have an NRAS Q61K mutation (Fig. S4C). This discrepancy suggests that signaling pathways in NRAS Q61K mutant cells are different in the context of wild-type and mutant EGFR. A selective PI3K inhibitor, GDC-0941, did not restore sensitivity of 11-18R cells to erlotinib (Fig. S4D). These data suggest that the MEK rather than the PI3K pathway is active downstream of mutant NRAS in these cells.
Third, we examined the effect of NRAS protein knockdown using siRNAs against NRAS. siRNA knockdown modestly inhibited the growth of 11-18R but not parental cells (Fig. 2D). When combined with erlotinib, NRAS knockdown led to an even greater growth reduction in resistant cells (Fig. 2D). Conversely, when combined with AZD6244, EGFR knockdown led to similar levels of growth inhibition (Fig. 2D). Consistent with these results, only the combinations of erlotinib plus siRNA knockdown of NRAS (Fig. 2E) or AZD6244 plus siRNA knockdown of EGFR (Fig. S4E) led to significant inhibition of phospho-ERK in resistant cells. Finally, to extend these results further, we performed growth-inhibition assays using siRNAs and kinase inhibitors as above with two separate 11-18R single-cell clones (C1 and C4) and observed essentially the same results (Fig. S4 F and G). Collectively, these data indicate that in 11-18R cells NRAS Q61K functions downstream of EGFR L858R and mediates resistance to erlotinib via the MEK signaling pathway.
RAS Signaling Pathway Gene Mutations in Tumors from Patients with Acquired Resistance.
Given the RAS-related findings in the murine (30) and cell-line models of acquired resistance and the reported data regarding KRAS/NRAS/BRAF/MEK1 mutations in other cancers that grow after responding to targeted therapies (25–28), we reexamined the frequency of mutations occurring in KRAS, NRAS, BRAF, and MEK1 in 195, 212, 195, and 146 tumor samples, respectively, from patients with acquired resistance to EGFR TKIs. Tumors were screened by a variety of methods including a SNapShot/sizing platform (32, 38), a Sequenom mass spectrometry-based assay (39), and/or direct sequencing. Although we did not detect any NRAS, KRAS, or MEK1 mutations, we did identify two BRAF mutations. One tumor harbored simultaneous EGFR exon19 deletion, EGFR T790M, and BRAF V600E mutations, and another tumor harbored EGFR exon 19 deletion and BRAF G469A (Table 2 and Fig. S5). Both patients were found at diagnosis to have tumors with an EGFR exon19 deletion that initially responded radiographically to erlotinib monotherapy. Lack of sufficient tissue precluded further analysis of the acquired-resistance specimens. A pretreatment tumor sample was unavailable for the first patient. This patient also had no history of melanoma, a disease in which BRAF V600E mutations are common. In the second case, the BRAF G469A mutation was confirmed to be absent before treatment. Collectively, these data demonstrate that RAS pathway gene mutations are rare, but BRAF mutations can occur in 1% of tumors with acquired resistance to EGFR TKIs.
Table 2.
RAS signaling pathway gene mutations in tumor samples from 212 patients with EGFR-mutant lung cancer and acquired resistance to EGFR TKIs
| Institution | EGFR T790M | NRAS | KRAS | BRAF | MEK1 |
| MSKCC* | 57/103 | 0/103 | 0/103 | 0/103 | 0/103 |
| Vanderbilt-Ingram Cancer Center† | 8/8 | 0/8 | 0/8 | 1/8 | 0/8 |
| Massachusetts General Hospital‡ | 42/84 | 0/84 | 0/84 | 1/84 | 0/35 |
| Others§ | n/d | 0/17 | n/d | n/d | n/d |
| Total | 107/195 (54.9%) | 0/212 | 0/195 | 2/195 (1.0%) | 0/146 |
n/d, no data.
*Sixty-eight MSKCC patient samples were assessed by a mass spectrometry-based (Sequenom) assay (ref. 39), and 35 samples were assessed using a SNaPshot-based assay (ref. 32).
†Eight samples were analyzed at Vanderbilt-Ingram Cancer Center by the SNaPshot assay.
‡Eighty-four samples were analyzed at Massachusetts General Hospital by SNapshot assay (ref. 38).
§Seventeen samples were analyzed for NRAS mutations by direct sequencing at other institutions (nine patients at Okayama University, Japan, five patients at National Taiwan University Hospital, and three patients at the Max Planck Institute, Cologne, Germany).
Ectopic Expression of NRAS Q61K, BRAF V600E, or BRAF G469A Confers Resistance to Cells Harboring Drug-Sensitive EGFR Mutations.
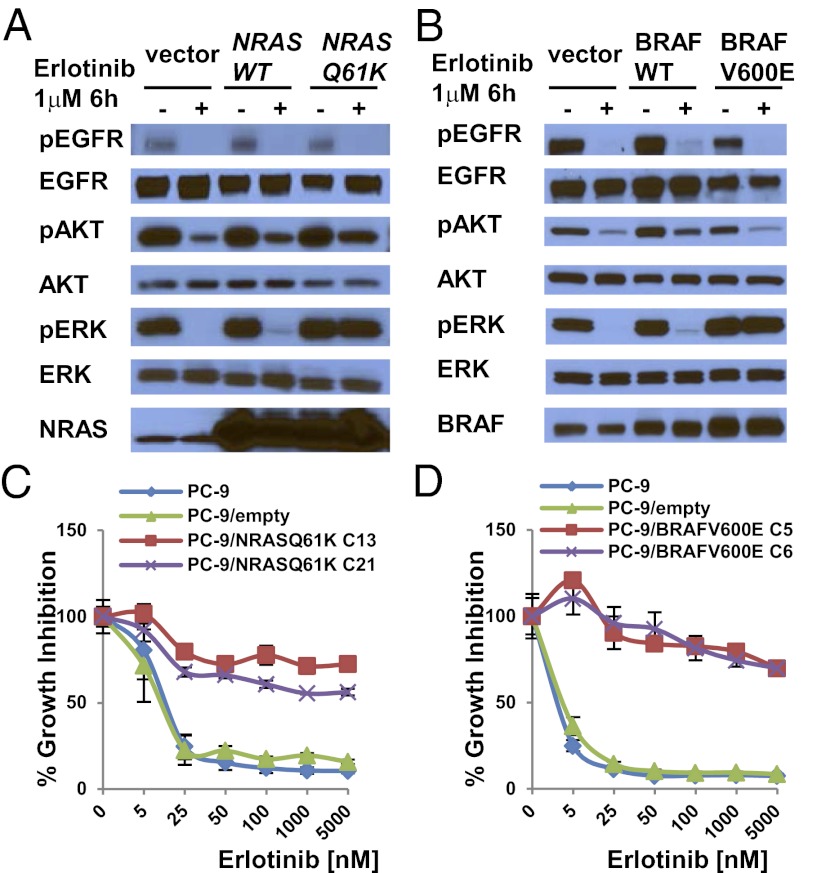
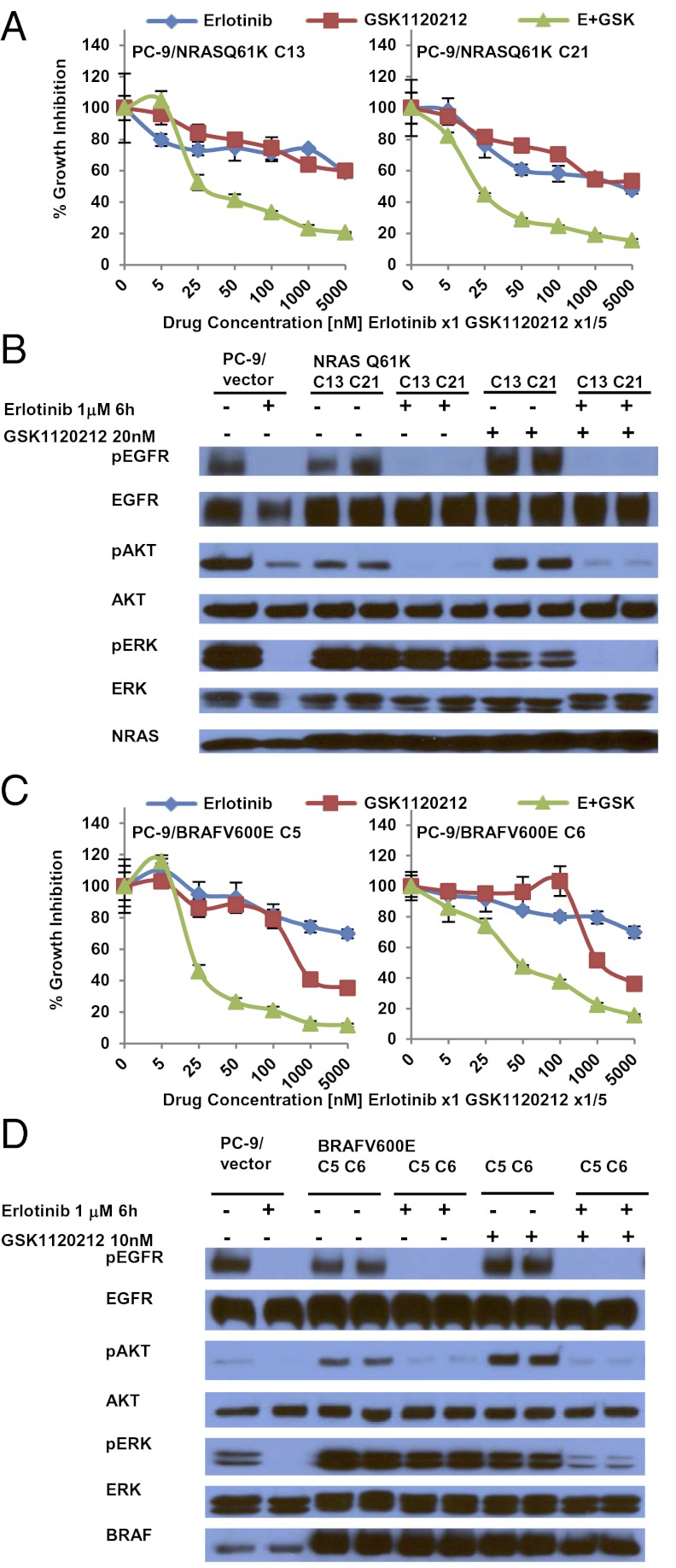
To confirm that mutations in genes in the RAS signaling pathway could confer acquired resistance to EGFR-TKIs, we introduced cDNAs encoding NRAS Q61K or BRAF G469A in PC-9 cells and BRAF V600E in both PC-9 and PC-9R cells and treated cell transfectants with various inhibitors. We did not express mutant forms of KRAS or MEK1, because we did not find correlative human data. Ectopic expression of these mutants but not their wild-type counterparts led to constitutive pERK activation, even in the presence of erlotinib or afatinib (Fig. 3 A and B and Fig. S6 A and B). Consistent with these data, EGFR-mutant cells with stable expression of either mutant were resistant to growth inhibition by erlotinib (Fig. 3 C and D), and phospho-ERK activation was maintained in the presence of drug (Fig. S6 C and D). In the stable transfectants, addition of MEK inhibitors to erlotinib led to greater growth inhibition (Fig. 4 A and C) and enhanced reduction of phospho-ERK levels compared with either drug alone (Fig. 4 B and D). In the mutated BRAF transfectants, addition of the BRAF inhibitor vemurafenib to erlotinib induced greater growth inhibition than either drug alone (Fig. S7).
Fig. 3.
Ectopic expression of NRAS Q61K or BRAF V600E in EGFR mutant cells mediates resistance to EGFR TKIs. (A and B) PC-9 cells were transiently transfected with expression plasmids encoding NRAS wild type or NRAS Q61K (A) or BRAF wild type or BRAF V600E (B) and were cultured in the absence or presence of erlotinib for 6 h. Corresponding cell lysates were subjected to immunoblotting with the indicated antibodies. Lysates from cells harboring NRAS wild type or NRAS Q61K displayed higher levels of total NRAS than seen in control-transfected cells, but only cells transfected with NRAS Q61K displayed enhanced phospho-ERK expression in the presence of erlotinib. Similarly, lysates from cells harboring BRAF wild type or BRAF V600E displayed higher levels of total BRAF than seen in control-transfected cells, but only cells transfected with BRAF V600E displayed enhanced phospho-ERK expression in the presence of erlotinib. (C and D). PC-9 cells were stably transfected with control plasmids or expression plasmids encoding NRAS Q61K (C) or BRAF V600E (D). Ectopic expression of NRAS Q61K (C) or BRAF V600E (D) mediated resistance of PC-9 cells to erlotinib. C13, clone 13; C21, clone 21; C5, clone 5; C6, clone 6. Data shown are mean ± SD of three independent experiments performed in hextuplicate.
Fig. 4.
MEK inhibition restores the sensitivity of PC-9/NRAS Q61K or PC-9/BRAF V600E stable clones to erlotinib. C13, clone 13; C21, clone 21; C5, clone 5; C6, clone 6. (A and C) The combination of erlotinib and GSK1120212 leads to greater inhibition of cell growth in PC-9 cells stably expressing NRAS Q61K (PC-9/NRAS Q61K cells) (A) or BRAF V600E (PC-9/BRAF V600E cells) (C) than seen either drug alone. Data shown are mean ± SD of three independent experiments performed in hextuplicate. (B and D) In stable transfectants the combination of erlotinib plus GSK1120212 leads to a greater reduction in phospho-ERK levels than either drug alone. PC-9/NRAS Q61K (B) or PC-9/BRAF V600E (D) cells were cultured in the absence or presence of erlotinib or/and GSK1120212 for 6 h; corresponding cell lysates were subjected to immunoblotting with the indicated antibodies.
Discussion
Elucidating how patients with EGFR-mutant lung tumors develop acquired resistance to EGFR TKIs has been an active area of investigation. More than half of patients develop a second-site EGFR mutation, i.e., the T790M amino acid change, which results in altered binding of drug to the ATP-binding pocket (6–9). Rarer mechanisms (1–10%) include MET amplification, PIK3CA mutation, and changes in tumor morphology (i.e., transformation to small cell lung cancer or development of features of EMT) (9–13). Up to 40% of cases are still unexplained.
Knowledge of these mechanisms coupled with prospective testing in the relevant clinical samples has led to the creation of rational strategies to overcome acquired resistance. For example, using mouse lung tumor models of drug-sensitive and -resistant EGFR-mutant alleles, we previously showed that dual inhibition of EGFR with the second-generation EGFR TKI afatinib plus the anti-EGFR antibody cetuximab could eradicate T790M-driven tumors (40). A trial based on these data has now shown a 40% partial response rate in patients with acquired resistance (41).
Here, we performed in vitro modeling to determine additional mechanisms of resistance. Surprisingly, we found that one human EGFR-mutant tumor cell line acquired a new mutation in NRAS. Because these models have recapitulated findings in humans (notably T790M mutations and MET amplification), and because a variety of RAS signaling pathway genes have been associated with acquired resistance to other targeted therapies in other solid tumors, we systematically screened for recurrent mutations in KRAS/NRAS/BRAF/MEK1 in nearly 200 tumor samples from patients with acquired resistance to EGFR TKIs. Although no KRAS, NRAS, or MEK1 mutations were detected, we did find one case with concurrent EGFR exon19 deletion and EGFR T790M and BRAF V600E mutations and another case with EGFR exon19 deletion and the BRAF G469A mutation (2/195, 1.0%). Although our cell-line models indicated that the NRAS Q61K and EGFR L858R mutations were in the same cell, we could not determine if the BRAF and EGFR mutations were in the same or different tumor cells in the patient samples. In case they were present in the same cells, we studied further the biological and therapeutic consequences of acquired NRAS and BRAF mutations in EGFR-mutant lung tumor cells and showed that these tumor cells were resistant to erlotinib alone but were sensitive to combination treatment with EGFR and MEK inhibition. Transfectants with mutant BRAF were sensitive to the combination of EGFR and BRAF inhibition as well. Collectively, these data suggest that RAS pathway gene mutations are rare but do occur, with BRAF gene mutations found in 1% of these patients.
In previous smaller studies, with only 6, 37, and 14 patients, respectively, we and others showed that RAS mutations are not found in lung cancers from patients with acquired resistance to EGFR TKIs (7, 12, 29). The sample sizes were too small to be definitively conclusive. Only 37 tumors have been examined for BRAF mutations (7, 12, 29), and MEK1 status has not been assessed. By contrast, PIK3CA mutations have been found with other drivers in lung cancers, including in tumors from patients with acquired resistance (12, 19, 20). Why RAS signaling pathway gene mutations are infrequent in lung cancers as opposed to other types of solid tumors treated with targeted therapies is unclear. One possibility is that introduction of mutant RAS or BRAF into EGFR mutant lung cells is toxic in most instances, whereas other cell types are more permissive. Another possibility is that other, unexamined alterations in the pathway, such as amplification (e.g., involving KRAS) or loss of downstream regulatory genes (e.g., NF1) either by genetic or epigenetic mechanisms, occur specifically in lung cancer but not in other cancers.
In the 11-18R cell-line model of acquired resistance, we found from biochemical and pharmacologic studies that the NRAS Q61K mutation acts in the ERK pathway downstream of mutant EGFR. Consistent with these data, a recent report showed that chronic exposure of cells harboring EGFR T790M to an EGFR T790M-selective irreversible pyrimidine EGFR kinase inhibitor, WZ4002, led to the development of ERK2 amplification. Combined treatment of resistant cells with WZ4002 and a MEK inhibitor or ERK2 knockdown suppressed cell growth (42). These results demonstrate that activation of the RAS signaling pathway can mediate resistance; such activation may become more frequent in patients as better ways to inhibit EGFR T790M are developed. Thus, it will be interesting to determine if patients who develop acquired resistance to afatinib/cetuximab or EGFR T790M-specific inhibitors harbor activation of the RAS signaling pathway via either gene mutation or amplification.
In summary, preclinical modeling coupled with emerging data about acquired resistance to targeted therapies in solid tumors led us to reexamine, in the largest collection of tumor samples to date, the contribution of mutations in RAS signaling pathway genes as mediators of acquired resistance to EGFR TKIs in lung cancer. Such samples occasionally harbored BRAF mutations but lacked recurrent mutations in KRAS, NRAS, or MEK1. Although the percent of cases with BRAF mutation is small (1%), the positive findings coupled with the negative results provide deeper insight into mechanisms of acquired resistance to EGFR TKIs in lung cancer, inform ongoing clinical trials designed to overcome resistance, and narrow the list of genes that should be routinely screened for in samples from patients with acquired resistance. As more data emerge on mechanisms of resistance to targeted therapies in various cancers, the findings reported here further demonstrate that, even though colorectal cancers, melanomas, GISTs, and lung cancers share common signaling cascades, each disease must be examined independently to determine disease-specific mediators of acquired resistance.
Materials and Methods
Cell Culture.
EGFR-mutant PC-9 (EGFR exon19del E746–A750), HCC827 (EGFR exon19del E746–A750), HCC4006 (EGFR exon19del L747–E749), HCC4011 (EGFR L858R), 11-18 (EGFR L858R), and H1299 (NRAS Q61K) cells were cultured in RPMI 1640 medium (Mediatech) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals) and penicillin-streptomycin solution (final concentration 100 U/mL penicillin, 100 μg/mL streptomycin) (Mediatech). Cells were grown in a humidified incubator with 5% CO2 at 37 °C. To create EGFR TKI-resistant lines, parental cells were cultured with increasing concentrations of TKIs starting with the IC30. Doses were increased in a stepwise pattern when normal cell proliferation patterns resumed. Fresh drug was added every 72–96 h. Resistant cells that grew in 5 μM erlotinib were derived after 3–6 mo of culturing with drug. 11-18R cells were maintained initially as polyclonal populations under constant TKI selection. DNA identity testing on both the parental and resistant cells confirmed that the cells were derived from the same origin. Clonal resistant cells were isolated by limiting dilution.
Growth Inhibition Assay.
For cell growth-inhibition experiments, cells were seeded in 96-well plates at a density of 3,000 cells per well and on the following day were exposed to drugs alone or in combination. Cell Titer Blue Reagent (Promega) was added 72 h after drug addition, and fluorescence was measured on a Spectramax spectrophotometer (Molecular Devices) according to the manufacturer's instructions. All experimental points were set up in hextuplicate replicates and were performed at least three independent times. Erlotinib and afatinib were synthesized by the Memorial Sloan-Kettering Cancer Center (MSKCC) Organic Synthesis Core. AZD6244, GSK1120212, vemurafenib, SGX-523, and GDC-0941 were purchased from Selleck Chemicals.
Antibodies and Immunoblotting.
The following antibodies were obtained from Cell Signaling Technology: phospho-EGFR, EGFR, MET, phospho-ERK, ERK, phospho-AKT, AKT, HRP-conjugated anti-mouse, and HRP-conjugated anti-rabbit. NRAS and BRAF antibody were purchased from Santa Cruz. For immunoblotting, cells were harvested, washed in PBS, and lysed in 50 mmol/L Tris⋅HCl (pH 8.0), 150 mmol/L sodium chloride, 5 mmol/L magnesium chloride, 1% Triton X-100. 0.5% sodium deoxycholate, 0.1% SDS, 40 mmol/L sodium fluoride, 1 mmol/L sodium orthovanadate, and complete protease inhibitors (Roche Diagnostics). Lysates were subjected to SDS/PAGE followed by blotting with the indicated antibodies and detection by Western Lightning ECL reagent (Perkin-Elmer).
RAS Activation Assay.
11-18 and 11-18R cells were serum starved overnight and supplemented with 1 μM of erlotinib for 6 h. RAS activity was measured using the Ras-binding domain of Raf-1 to pull down active Ras according to the manufacturer’s protocol (Cell BioLabs). Following separation by SDS PAGE, proteins were transferred to membranes which were probed with an anti-NRAS antibody.
Patient Samples and Data.
Tumor specimens were obtained with patients’ consent under Institutional Review Board (IRB)-approved protocols in each institution. Samples were frozen and stored at −80 °C in institutional tumor banks or in formalin-fixed, paraffin-embedded blocks. Mutational profiling results were performed within Clinical Laboratory Improvement Amendments-certified laboratories on multiple samples as part of routine standard of care and/or on IRB-approved protocols.
Sequencing of NRAS and Systematic Mutation Screening.
Genomic DNA was extracted from patient samples (>70% tumor cells) and cell lines using standard procedures. NRAS exons 2 and 3 were amplified from genomic DNA and were sequenced directly. NRAS mutations, along with other mutations, also were screened using a SNapShot-based (32, 38) or mass spectrometry-based (Sequenom) assays (39).
siRNA Experiments.
EGFR, NRAS, and negative control oligos (Dharmacon) were used at a concentration of 10 nM and transfected with Lipofectamine RNAimax according to the manufacturer's protocol (Invitrogen).
Expression Constructs and Transfections.
A cDNA for NRAS was purchased from Origene and subcloned into a Flag-N-CMV6 entry vector (Origene). A cDNA for BRAF was kindly provided by David Solit (MSKCC) and was subcloned into a pcDNA3.1 vector (Invitrogen). The NRAS Q61K and BRAF V600E and BRAF G469A mutations were introduced into the cDNAs using site-directed mutagenesis (Agilent) with mutant-specific primers according to the manufacturer's instructions. The cDNAs were fully resequenced to ensure that no additional mutations were introduced. Plasmid transfections into PC-9 or PC-9R cells were performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Selection of cells was started 48 h later in 96-well plates with appropriate antibiotics.
Xenograft Studies.
Nude mice (nu/nu; Harlan Laboratories) were used for in vivo studies and were cared for in accordance with guidelines approved by the MSKCC Institutional Animal Care and Use Committee and Research Animal Resource Center. Eight-week-old female mice were injected s.c. with 15 million 11-18R cells together with Matrigel. Once tumors reached an average volume of 150 mm3, mice were randomized and dosed via oral gavage with either erlotinib (MSKCC Organic Synthesis Core), GSK1120212 (Selleck Chemicals), or the combination at the indicated doses. A uniform volume for administration (200 μL) was used for each group. Mice were observed daily throughout the treatment period for signs of morbidity/mortality. Tumors were measured twice weekly using calipers, and volume was calculated using the formula length × width2 × 0.52. Body weight also was assessed twice weekly. The experiment was terminated after 4 wk of treatment.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Cancer Institute (NCI) Grants R01-CA121210, P01-CA129243, and U54-CA143798. W.P. received additional support from Vanderbilt’s Specialized Program of Research Excellence in Lung Cancer Grant CA90949 and from Vanderbilt-Ingram Cancer Center Core Grant P30-CA68485. L.V.S. received additional support from NCI Grant 1R21-156000 and from Uniting Against Lung Cancer: New England and the Marjorie E. Korff Fund.
Footnotes
Conflict of interest statement: L.V.S. received consulting fees from Clovis Oncology, GlaxoSmithKline, and Celgene Corporation. J.C.-H.Y. received consulting fees from Boehringer Ingelheim. V.A.M. is an employee at Foundation Medicine and has equity value in the company. M.G.K. has received consulting fees from Boehringer Ingelheim and research funding for other projects from Pfizer and Boehringer. J.A.E. has received consulting fees and has stock option ownership in Agios Pharmaceuticals and has received research funding for other projects from Novartis, GlaxoSmithKline, and AstraZeneca. D.D.-S. received consulting fees from Bio-Reference Laboratories. W.P. has received consulting fees from MolecularMD, AstraZeneca, Bristol-Myers Squibb, Symphony Evolution, and Clovis Oncology and research funding for other projects from Enzon, Xcovery, AstraZeneca, and Symphogen. W.P. and V.A.M. are part of a patent regarding EGFR T790M mutation testing that was licensed by Memorial Sloan-Kettering Cancer Center to MolecularMD.
This article is a PNAS Direct Submission.
See Author Summary on page 12282 (volume 109, number 31).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203530109/-/DCSupplemental.
References
- 1.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, et al. North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 7.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balak MN, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 9.Arcila ME, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 11.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakowski MF, Ladanyi M, Kris MG. Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 14.Sequist LV, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22:2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paik PK, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29:2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchetti A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 18.Marks JL, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto H, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaft JE, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012;11:485–491. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks JL, et al. Mutational analysis of EGFR and related signaling pathway genes in lung adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4. PLoS ONE. 2007;2:e426. doi: 10.1371/journal.pone.0000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kris MG, et al. The NCI’s Lung Cancer Mutation Consortium (LCMC) Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma (abstract CRA7506) J Clin Oncol 2011. 29(Suppl) Available at http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview=abst_detail_view&confID=102&abstractID=81670. [Google Scholar]
- 23.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Rocck W, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–756. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 25.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti EGFR therapy in colorectal cancer. Nature. 2012 doi: 10.1038/nature11156. 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agaram NP, et al. Novel V600E BRAF mutations in imatinib-naive and imatinib-resistant gastrointestinal stromal tumors. Genes Chromosomes Cancer. 2008;47:853–859. doi: 10.1002/gcc.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosaka T, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 30.Politi K, Fan PD, Shen R, Zakowski M, Varmus H. Erlotinib resistance in mouse models of epidermal growth factor receptor-induced lung adenocarcinoma. Dis Model Mech. 2010;3:111–119. doi: 10.1242/dmm.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chmielecki J, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Z, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino A, et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 34.Ichihara E, et al. Effects of vandetanib on lung adenocarcinoma cells harboring epidermal growth factor receptor T790M mutation in vivo. Cancer Res. 2009;69:5091–5098. doi: 10.1158/0008-5472.CAN-08-4204. [DOI] [PubMed] [Google Scholar]
- 35.Suda K, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res. 2010;16:5489–5498. doi: 10.1158/1078-0432.CCR-10-1371. [DOI] [PubMed] [Google Scholar]
- 36.Suda K, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol. 2011;6:1152–1161. doi: 10.1097/JTO.0b013e318216ee52. [DOI] [PubMed] [Google Scholar]
- 37.Li D, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dias-Santagata D, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 40.Regales L, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janjigian YY, et al. 2011. Activity and tolerability of afatinib (BIBW 2992) and cetuximab in NSCLC patients with acquired resistance to erlotinib or gefitinib (abstract 7525^), J Clin Oncol 29 (Suppl). Available at http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview=abst_detail_view&confID=102&abstractID=78057.
- 42.Ercan D, et al. 2011. Amplification of ERK2 mediates resistance to the novel irreversible EGFR inhibitor WZ4002 (abstr 4736^). American Association for Cancer Research annual meeting. Available at http://www.abstractsonline.com/Plan/ViewAbstract.aspx?sKey=5dc0a975-14e5-47f5-80d3-4dbd39f02a11&cKey=ffa124f6-5a91-4a2a-9b2f-97f0f288b5d2&mKey=%7b507D311A-B6EC-436A-BD67-6D14ED39622C%7d.