Abstract
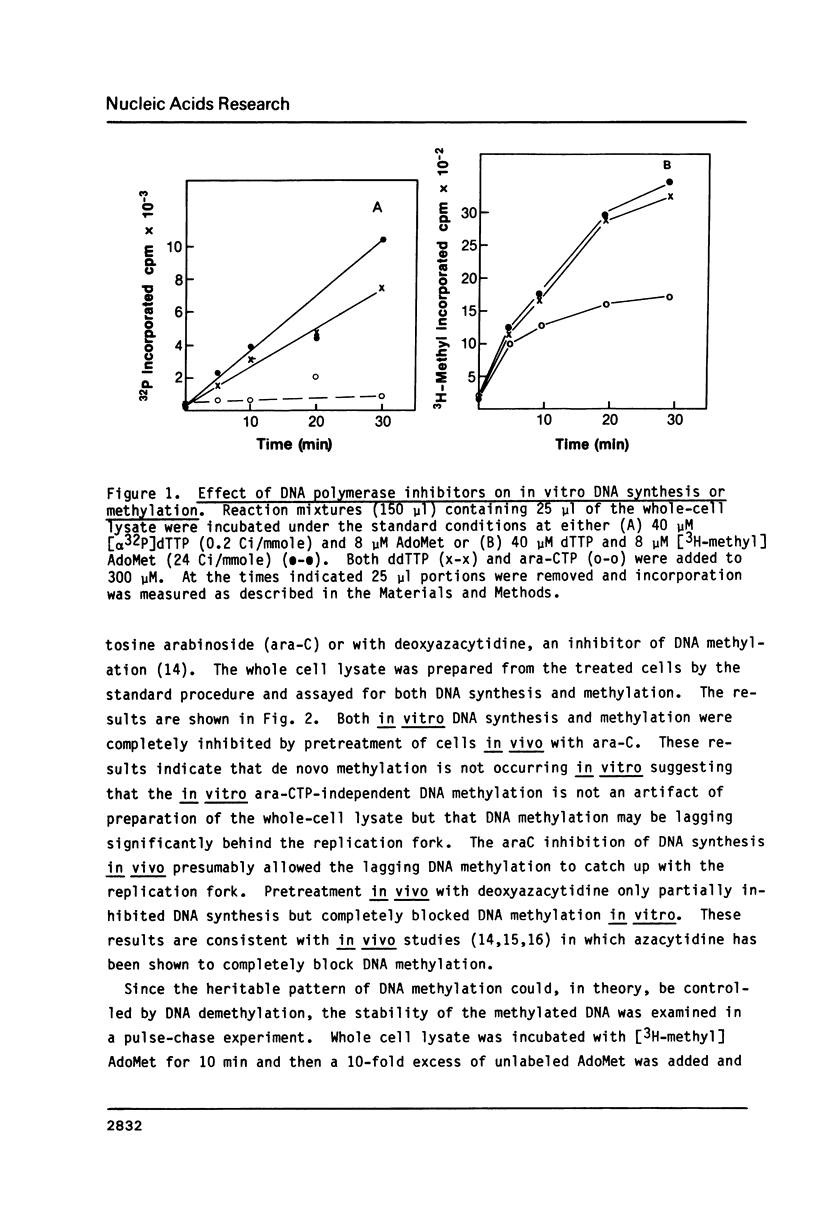
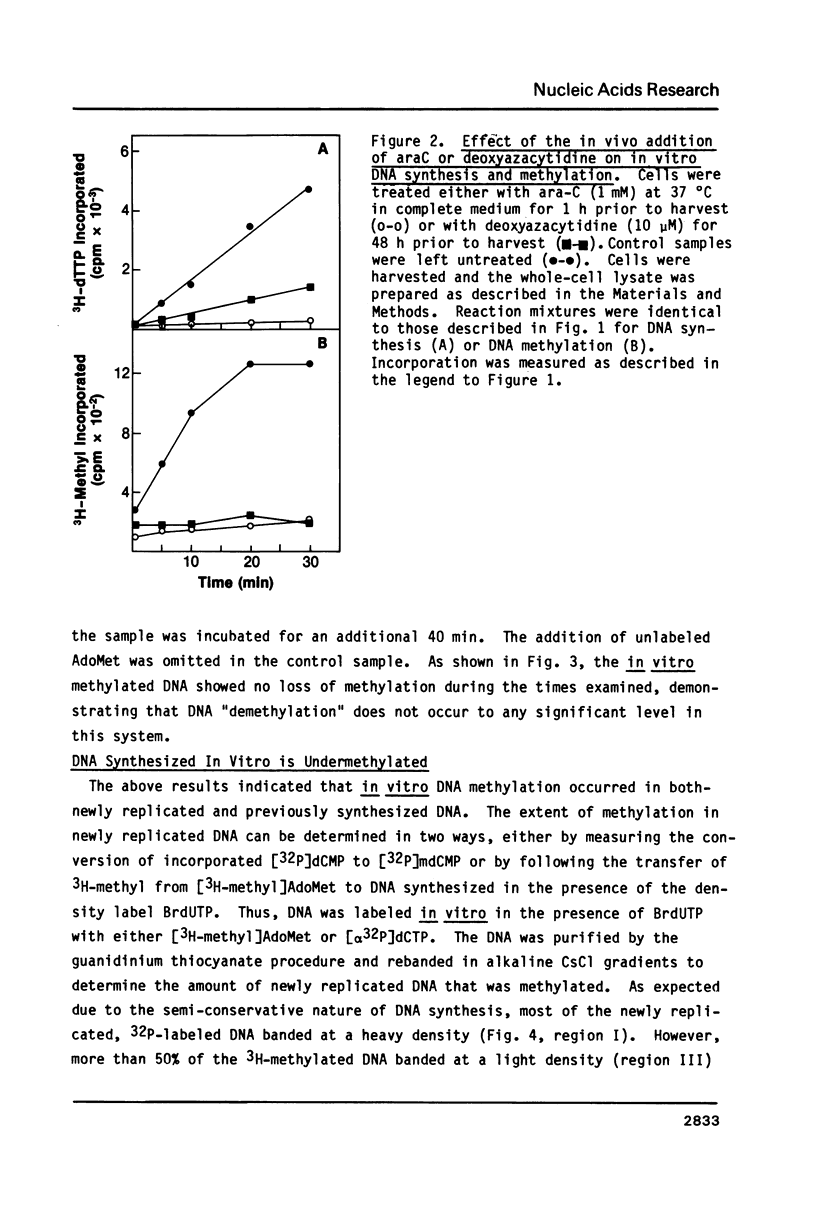
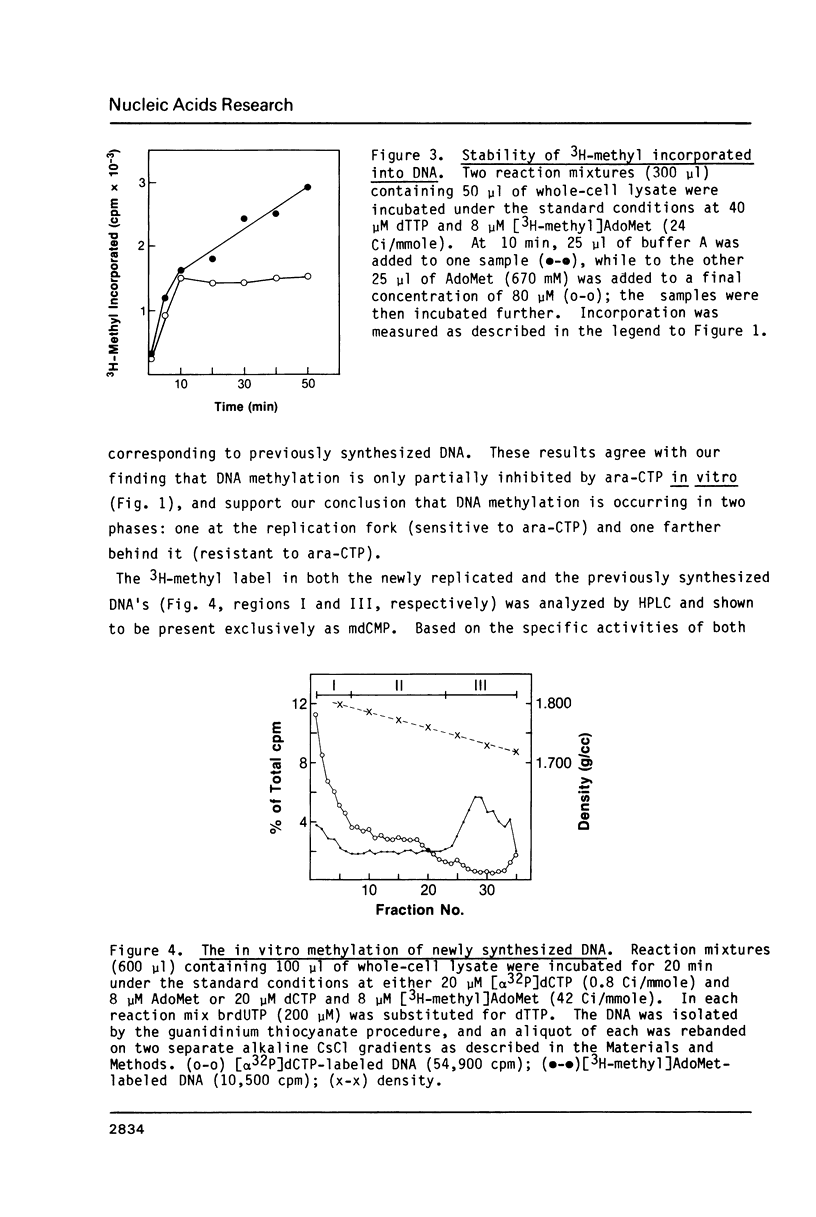
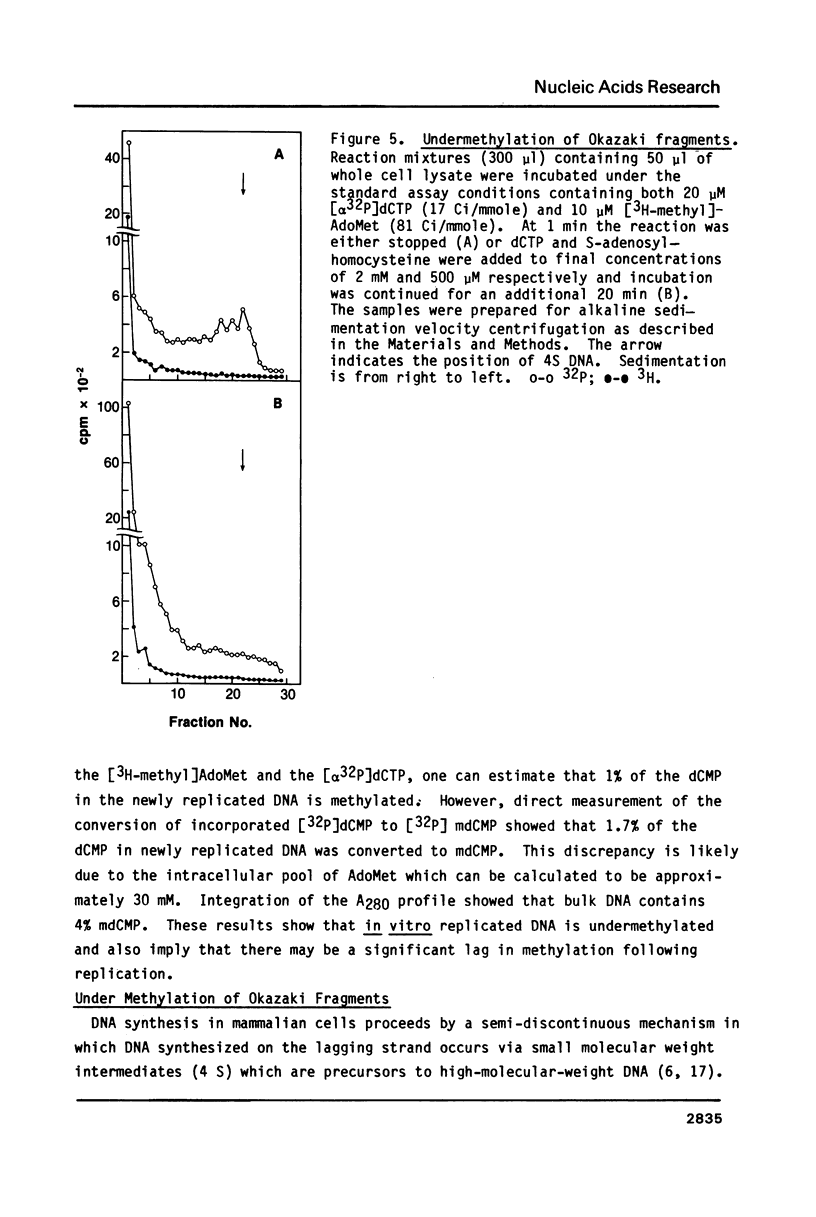
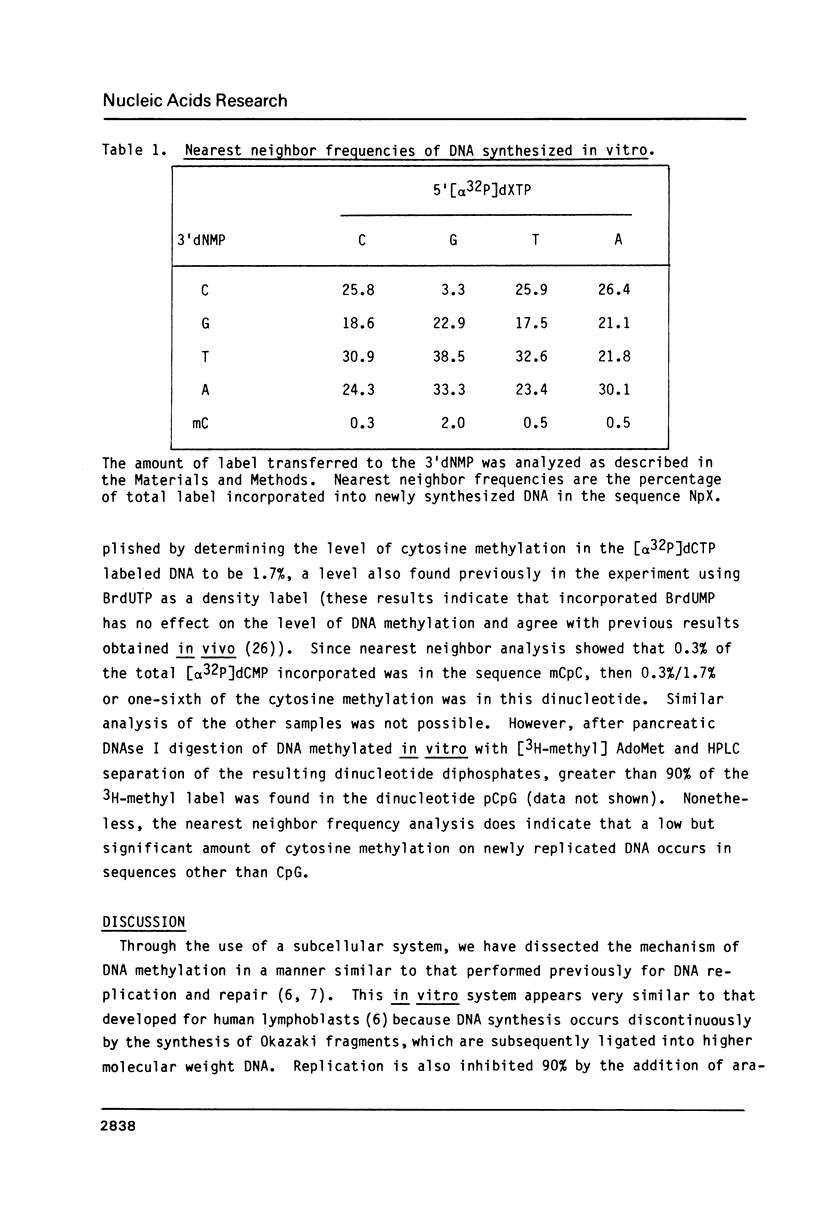
An in vitro DNA synthesizing system from mouse fibroblasts has been used to study DNA methylation. DNA methylation occurs in two phases, one at the replication fork and the other farther behind it. Although 4% of the dCMP residues in mouse cell DNA are mdCMP, only 1.7% of the total [alpha 32P]dCMP in newly replicated DNA is methylated in vitro. No methylation of Okazaki fragments was detected. Nearest neighbor analysis of the newly replicated DNA revealed that, although 40% of the CpG dinucleotides were methylated, significant amounts of cytosine methylation were also found in CpC, CpT, and CpA dinucleotides.
Full text
PDF





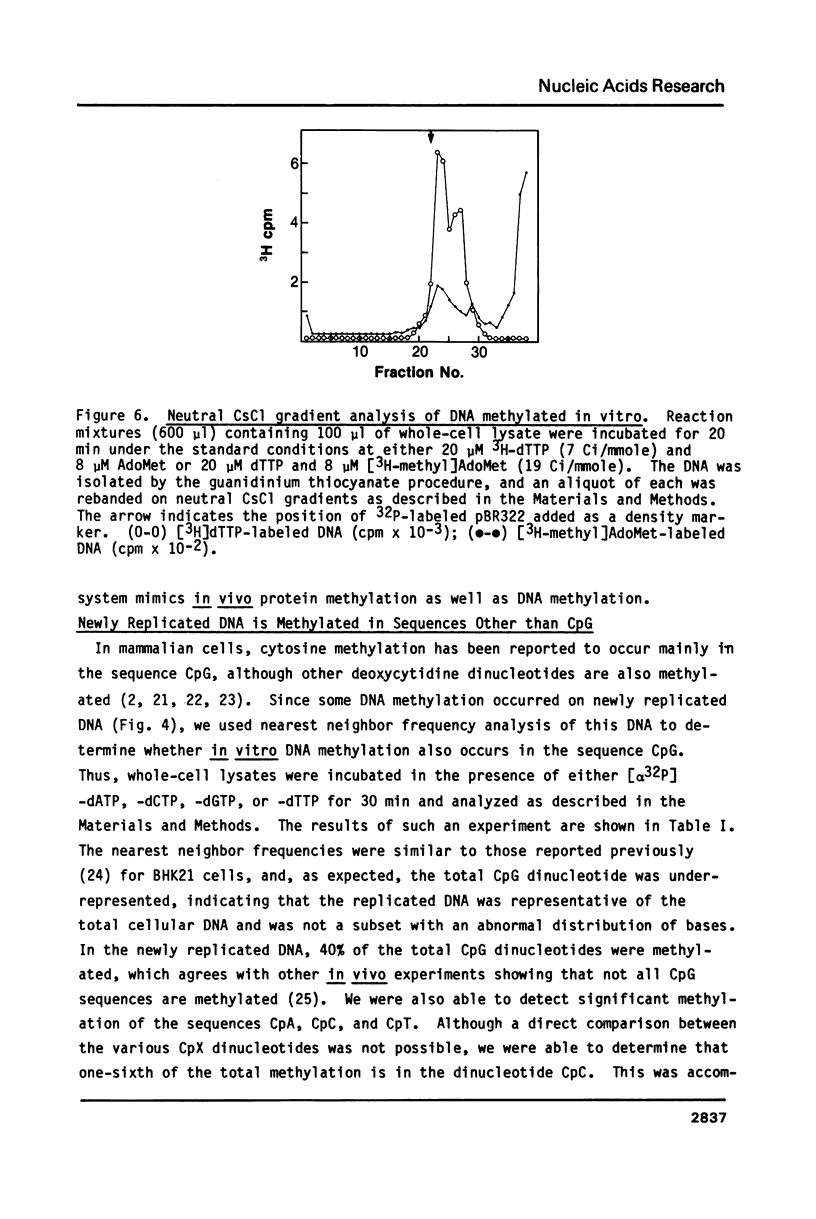




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Cami B., Dinh T. H., Igolen J., Kourilsky P. Processing of complex heteroduplexes in Escherichia coli and Cos-1 monkey cells. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5792–5796. doi: 10.1073/pnas.81.18.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. L., Hogarth C. DNA methylation in isolated nuclei: old and new DNAs are methylated. Biochim Biophys Acta. 1973 Dec 7;331(2):214–220. doi: 10.1016/0005-2787(73)90434-6. [DOI] [PubMed] [Google Scholar]
- Adams R. L. The relationship between synthesis and methylation of DNA in mouse fibroblasts. Biochim Biophys Acta. 1971 Dec 16;254(2):205–212. doi: 10.1016/0005-2787(71)90829-x. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman J. K. Separation of major and minor deoxyribonucleoside monophosphates by reverse-phase high-performance liquid chromatography: a simple method applicable to quantitation of methylated nucleotides in DNA. Anal Biochem. 1982 Jan 1;119(1):38–48. doi: 10.1016/0003-2697(82)90662-5. [DOI] [PubMed] [Google Scholar]
- Cox R., Prescott C., Irving C. C. The effect of S-adenosylhomocysteine on DNA methylation in isolated rat liver nuclei. Biochim Biophys Acta. 1977 Feb 16;474(4):493–499. doi: 10.1016/0005-2787(77)90070-3. [DOI] [PubMed] [Google Scholar]
- Creusot F., Acs G., Christman J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2'-deoxycytidine. J Biol Chem. 1982 Feb 25;257(4):2041–2048. [PubMed] [Google Scholar]
- Grafstrom R. H., Tseng B. Y., Goulian M. The incorporation of uracil into animal cell DNA in vitro. Cell. 1978 Sep;15(1):131–140. doi: 10.1016/0092-8674(78)90089-2. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Szyf M., Cedar H., Razin A. Methylation of replicating and post-replicated mouse L-cell DNA. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4919–4921. doi: 10.1073/pnas.80.16.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Hemimethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981 Jun 25;9(12):2933–2947. doi: 10.1093/nar/9.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryanov G. I., Kirnos M. D., Demidkina N. P., Alexandrushkina N. I., Vanyushin B. F. Methylation of DNA in L cells on replication. FEBS Lett. 1980 Apr 7;112(2):225–228. doi: 10.1016/0014-5793(80)80185-2. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. Adenine methylation of Okazaki fragments in Escherichia coli. J Bacteriol. 1976 Dec;128(3):853–854. doi: 10.1128/jb.128.3.853-854.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINSHEIMER R. L. The action of pancreatic deoxyribonuclease. II. Isomeric dinucleotides. J Biol Chem. 1955 Aug;215(2):579–583. [PubMed] [Google Scholar]
- Salomon R., Kaye A. M., Herzberg M. Mouse nuclear satellite DNA: 5-methylcytosine content, pyrimidine isoplith distribution and electron microscopic appearance. J Mol Biol. 1969 Aug 14;43(3):581–592. doi: 10.1016/0022-2836(69)90360-x. [DOI] [PubMed] [Google Scholar]
- Salomon R., Kaye A. M. Methylation of mouse DNA in vivo: di- and tripyrimidine sequences containing 5-methylcytosine. Biochim Biophys Acta. 1970 Apr 15;204(2):340–351. [PubMed] [Google Scholar]
- Singer J., Stellwagen R. H., Roberts-Ems J., Riggs A. D. 5-Methylcytosine content of rat hepatoma DNA substituted with bromodeoxyuridine. J Biol Chem. 1977 Aug 10;252(15):5509–5513. [PubMed] [Google Scholar]
- Sneider T. W. The 5'-cytosine in CCGG1 is methylated in two eukaryotic DNAs and Msp I is sensitive to methylation at this site. Nucleic Acids Res. 1980 Sep 11;8(17):3829–3840. doi: 10.1093/nar/8.17.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. Methylation of mammalian deoxyribonucleic acid. 3. Terminal versus internal location of 5-methylcytosine in oligodeoxyribonucleotides from Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1972 May 10;247(9):2872–2875. [PubMed] [Google Scholar]
- Subak-Sharpe H., Bürk R. R., Crawford L. V., Morrison J. M., Hay J., Keir H. M. An approach to evolutionary relationships of mammalian DNA viruses through analysis of the pattern of nearest neighbor base sequences. Cold Spring Harb Symp Quant Biol. 1966;31:737–748. doi: 10.1101/sqb.1966.031.01.094. [DOI] [PubMed] [Google Scholar]
- Tseng B. Y., Ahlem C. N. A DNA primase from mouse cells. Purification and partial characterization. J Biol Chem. 1983 Aug 25;258(16):9845–9849. [PubMed] [Google Scholar]
- Tseng B. Y., Erickson J. M., Goulian M. Initiator RNA of nascent DNA from animal cells. J Mol Biol. 1979 Apr 25;129(4):531–545. doi: 10.1016/0022-2836(79)90467-4. [DOI] [PubMed] [Google Scholar]
- Tseng B. Y., Goulian M. DNA synthesis in human lymphocyts: intermediates in DNA synthesis, in vitro and in vivo. J Mol Biol. 1975 Dec 5;99(2):317–337. doi: 10.1016/s0022-2836(75)80149-5. [DOI] [PubMed] [Google Scholar]
- Tseng B. Y., Goulian M. Initiator RNA of discontinuous DNA synthesis in human lymphocytes. Cell. 1977 Oct;12(2):483–489. doi: 10.1016/0092-8674(77)90124-6. [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Adams J. K., Cooper I. A. Characteristics of enzymatic DNA methylation in cultured cells of human and hamster origin, and the effect of DNA replication inhibition. Biochim Biophys Acta. 1982 Jan 26;696(1):15–22. doi: 10.1016/0167-4781(82)90004-5. [DOI] [PubMed] [Google Scholar]



