Abstract
Collagenases of the matrix metalloproteinase (MMP) family play major roles in morphogenesis, tissue repair, and human diseases, but how they recognize and cleave the collagen triple helix is not fully understood. Here, we report temperature-dependent binding of a catalytically inactive MMP-1 mutant (E200A) to collagen through the cooperative action of its catalytic and hemopexin domains. Contact between the two molecules was mapped by screening the Collagen Toolkit peptide library and by hydrogen/deuterium exchange. The crystal structure of MMP-1(E200A) bound to a triple-helical collagen peptide revealed extensive interactions of the 115-Å–long triple helix with both MMP-1 domains. An exosite in the hemopexin domain, which binds the leucine 10 residues C-terminal to the scissile bond, is critical for collagenolysis and represents a unique target for inhibitor development. The scissile bond is not correctly positioned for hydrolysis in the crystallized complex. A productive binding mode is readily modeled, without altering the MMP-1 structure or the exosite interactions, by axial rotation of the collagen homotrimer. Interdomain flexing of the enzyme and a localized excursion of the collagen chain closest to the active site, facilitated by thermal loosening of the substrate, may lead to the first transition state of collagenolysis.
Keywords: extracellular matrix, X-ray crystallography, protease
The interstitial collagens I, II, and III are the major structural proteins in connective tissues such as skin, bone, cartilage, tendon, and blood vessels (1). They consist of three α chains with repeating Gly-X-Y triplets (X and Y are often proline and hydroxyproline, respectively) that intertwine each other to form a triple helix of ∼300 nm in length (2). Interstitial collagens are resistant to most proteolytic enzymes, but vertebrate collagenases cleave them at a single site approximately three-quarters of the way from the N terminus of the triple helix, thus initiating collagenolysis (3). Owing to this unique activity, collagenases play important roles in embryo development, morphogenesis, tissue remodeling, wound healing, and human diseases, such as arthritis, cancer, and atherosclerosis (4, 5).
Matrix metalloproteinase 1 (MMP-1) is a typical vertebrate collagenase (3). It consists of an N-terminal catalytic (Cat) domain containing an active-site zinc ion and a C-terminal hemopexin (Hpx) domain comprised of a four-bladed β-propeller, which are connected by a linker region (6, 7). Although the Cat domain can cleave a number of noncollagenous proteins including heat-denatured collagen (gelatin), its activity on native triple-helical collagen is negligible. The combination of the Cat and Hpx domains is required for MMP-1 to be able to degrade native collagen, and the same is true for all other collagenolytic MMPs, namely MMP-2, MMP-8, MMP-13, and MMP-14 (3). How collagenases interact with collagen and how the Hpx domain endows these enzymes with collagenolytic activity is not clearly understood. Another enigma of collagenolysis became apparent when the crystal structures of MMP-1Cat (8–10), MMP-8Cat (11, 12), and full-length pig MMP-1 (6) were solved: The catalytic cleft of these enzymes is too narrow to accommodate collagen in its native triple-helical conformation. A number of hypotheses have been proposed to explain how collagenases may destabilize and cleave triple-helical collagen (13–16), and we have experimentally demonstrated that MMP-1 unwinds triple-helical collagen locally before peptide bond hydrolysis (17, 18). A recent study mapped the interaction sites in MMP-1 and in triple-helical collagen by using NMR and proposed a model of collagen binding and unwinding (19). However, because the interactions between MMP-1 and collagen were not observed directly, a number of assumptions had to be made to derive a mechanism of collagenolysis.
In this study, we have combined biochemical experiments with crystallographic structure determination to show how MMP-1 recognizes its unique cleavage site in interstitial collagens. Our results suggest that collagenolysis relies on multiple exosite interactions, which not only serve to position the scissile bond near the active site, but also assist in the local unfolding of the collagen triple helix that is required for cleavage to occur.
Results
Cat and Hpx Domains of MMP-1 Cooperate in Collagen Binding.
We first examined a catalytically inactive MMP-1(E200A) mutant (residue numbering based on the proMMP-1 sequence) and its individual domains for binding to native collagen I. Full-length MMP-1(E200A) showed saturable collagen binding with an apparent KD of 0.4 μM, whereas the catalytic domain Cat(E200A) showed no detectable binding and the Hpx domain bound only weakly (Fig. 1A). The Cat(E200A) domain did not bind to collagen I even when it was added together with increasing concentrations of the Hpx domain (SI Appendix, Fig. S1A), suggesting that the Cat and Hpx domains bind collagen cooperatively only when they are linked together.
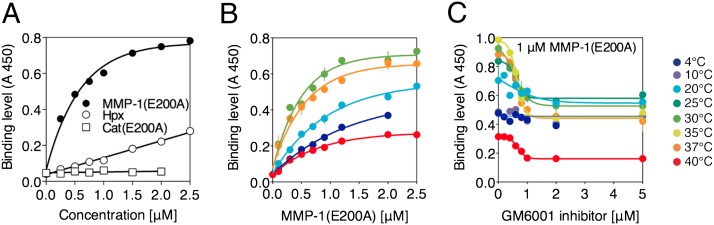
Fig. 1.
Binding of MMP-1(E200A) to immobilized collagen I. (A) Binding of full-length MMP-1(E200A) and its individual Cat(E200A) and Hpx domains at 20 °C. (B) Binding of MMP-1(E200A) at different temperatures. (C) Binding of 1 μM MMP-1(E200A) in the presence of the active-site inhibitor GM6001 at different temperatures.
The binding of MMP-1(E200A) to collagen increased with temperature until collagen denatured above 37 °C (Fig. 1B), indicating that MMP-1 prefers a looser triple helix but not denatured collagen. The temperature-dependent enhancement of MMP-1(E200A) binding to collagen was substantially reduced by the active-site inhibitor GM6001 (Fig. 1C). This effect was not observed below 10 °C, where little collagen cleavage occurs (17). No significant temperature-dependent increase in collagen binding was observed with either the Cat(E200A) or Hpx domain alone (SI Appendix, Fig. S1 B and C). These results suggest that the active site cleft of MMP-1 participates in collagen binding at body temperature by stabilizing and/or inducing a looser collagen conformation.
MMP-1–Binding Motif in Interstitial Collagens.
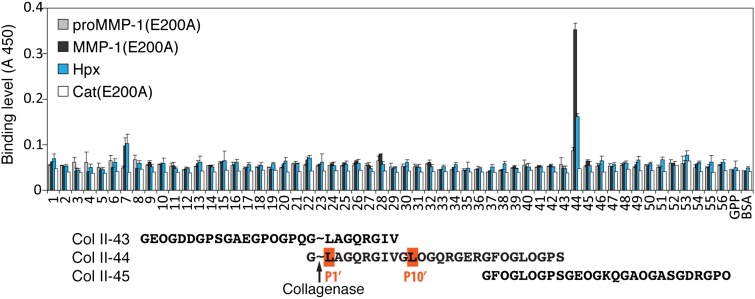
To identify the collagen residues interacting with MMP-1, we screened the Collagen Toolkit library of triple-helical peptides encompassing the entire collagen II sequence (20) for binding to MMP-1(E200A). Only one peptide, Col II-44, starting at the collagenase cleavage site G∼LAGQR… (∼ indicates the scissile bond) was recognized by the enzyme (Fig. 2). Col II-43, which also contains the cleavage site but with fewer C-terminal residues, did not bind MMP-1(E200A). Further experiments with truncated and alanine-substituted peptides identified the leucines at the P1′ and P10′ positions (nomenclature after Schechter and Berger; ref. 21) of collagen as important for MMP-1(E200A) binding (SI Appendix, Fig. S2A). A sequence alignment showed that the leucines at these two positions are conserved in all α chains of collagens I, II, and III from different species (SI Appendix, Fig. S2B). The consensus sequence for collagenase binding is G∼(L/I)(A/L)-GXY-GXY-GL(O/A) (P1′ and P10′ positions are bold; O, hydroxyproline). This motif is unique and not found elsewhere in collagens I, II, and III.
Fig. 2.
Screening of a triple-helical peptide library of collagen II with proMMP-1(E200A), MMP-1(E200A), Hpx domain, and Cat(E200A) domain. Error bars show SDs from three repeats. Critical Leu and Ile residues at P1′ and P10′ subsites are highlighted in the collagen II portion of the peptide sequences surrounding the collagenase cleavage site. ∼, bond cleaved by collagenases.
Mapping of the Collagen-Binding Regions of MMP-1 in Solution.
We then used hydrogen/deuterium exchange mass spectrometry (H/DXMS) to map the regions in MMP-1 involved in collagen binding. Five MMP-1 regions showed delayed deuterium incorporation in the presence of collagen I (Fig. 3A and SI Appendix, Fig. S3 and Table S1): Residues 145–161 (site 1) and 205–215 (site 3) in the Cat domain and residues 266–277 (site 4), 283–297 (site 5), and 330–346 (site 6) in the Hpx domain. Sites 4 and 5 were reported to reduce deuterium exchange upon binding to a triple-helical peptide (22). Residues 179–193 (site 2) showed increased deuterium incorporation in the presence of collagen, suggesting that this region, which includes the loop between the fifth β-strand and the second α-helix of the Cat domain, is more exposed to the solvent in the collagen-bound state. This loop contains a collagenase-characteristic cis-peptide bond (23, 24), and it was identified as important for the collagenolytic activity of MMP-1 (23). Our data suggest that this loop participates in collagenolysis dynamically. The combined H/DXMS results indicate the general orientation of the collagen triple helix binding to MMP-1 (Fig. 3, dashed line).
Fig. 3.
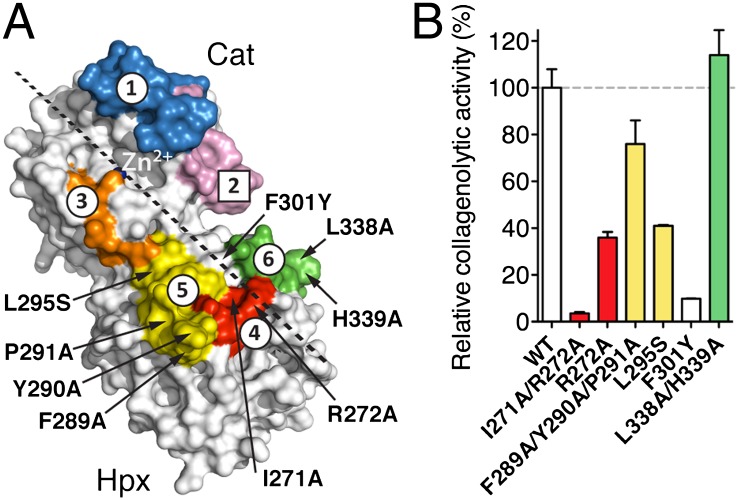
Collagen I footprint on MMP-1(E200A) determined by H/DXMS and mutagenesis. (A) Sites 1 and 3–6 are protected from deuterium incorporation, and site 2 shows enhanced deuterium incorporation upon collagen binding. The sites are mapped onto the crystal structure of MMP-1(E200A) (7). Residues potentially involved in collagenolysis that were mutated are indicated. Dashed line, predicted collagen binding direction. (B) Relative initial velocities of collagen I cleavage by MMP-1 and its variants normalized to WT.
S10′ Exosite in the Hpx Domain.
Considering the H/DXMS results and the importance for MMP-1(E200A) binding of the two leucines at the P1′ and P10′ positions of collagen, we searched for hydrophobic sites in MMP-1 that could accept these residues. For MMP-1 to cleave the Gly(P1)-Leu(P1′) bond, the P1′ Leu must be located close to the S1′ pocket of the Cat domain. Using this constraint, we docked a modeled triple-helical peptide to MMP-1 and identified candidate residues in the Hpx domain that might be involved in binding the P10′ Leu (Fig. 3A). They were mutated and the mutant proteins tested for their collagenolytic activity (Fig. 3B). The most dramatic reduction of collagenase activity was observed with the F301Y single mutant and the I271A/R272A double mutant, which exhibited 10% and 4% of wild-type activity, respectively. Together, Phe301, Ile271, and Arg272 in the Hpx domain participate in forming the hydrophobic S10′ binding pocket, or exosite. Ile271 and Arg272 were identified in the H/DXMS experiment as part of site 4. Phe301 is located between H/DXMS sites 5 and 6. It was not detected by H/DXMS analysis most likely because its backbone amide is buried in the Hpx domain.
Crystal Structure of an MMP-1(E200A)–Collagen Peptide Complex.
Based on the results of the collagen peptide binding studies, we designed a triple-helical collagen peptide for cocrystallization with MMP-1(E200A). The peptide spans positions P7-P17′ of the collagen II cleavage site and is capped by two and three nonnative Gly-Pro-Hyp triplets at the N and C termini, respectively, to enhance triple helicity (Fig. 4A). The melting temperature of this peptide was 31 °C (SI Appendix, Fig. S4). The peptide formed a stable complex with MMP-1(E200A) in solution (SI Appendix, Fig. S5). Crystals of the complex were grown at room temperature and used to determine the structure at 3.0-Å resolution (SI Appendix, Table S2).
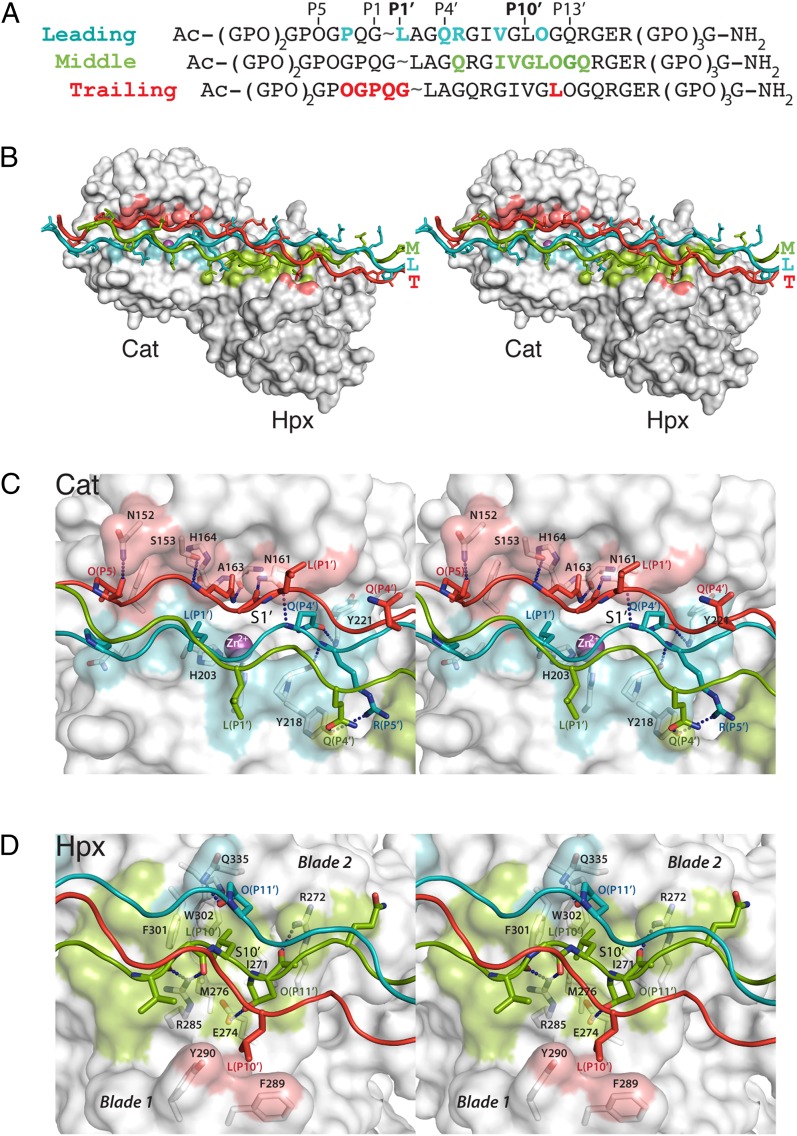
Fig. 4.
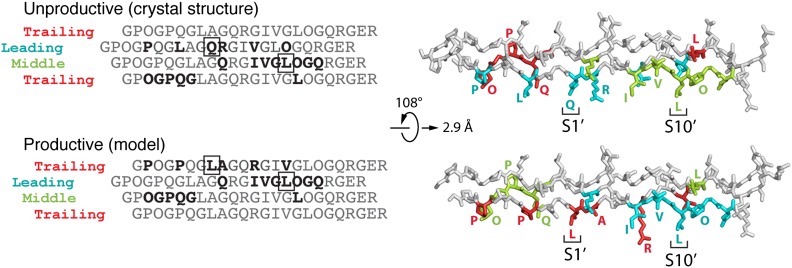
Crystal structure of the MMP-1(E200A)–triple helical collagen peptide complex. (A) Sequence of the collagen peptide used for cocrystallization with MMP-1(E200A). Residues within 4 Å distance of the enzyme in the complex are colored. The subsite designation is indicated for the leading chain. (B) Stereoview of the MMP-1(E200A)–collagen peptide complex. The collagen chains are colored cyan (L), green (M), and red (T), and the enzyme is shown as a gray surface with areas within 4 Å distance of the L, M, and T chains colored correspondingly. Magenta sphere, active-site zinc ion. (C) Stereoview of the interactions between collagen chains (colored as in B) and the active site cleft of MMP-1(E200A). Dashed lines indicate hydrogen bonds. (D) Stereoview of the interactions of the collagen chains with the Hpx domain. Selected residues making enzyme-substrate contacts are labeled with respective colors and shown in stick representation (N, dark blue; O, red).
The overall structure of human MMP-1(E200A) in complex with collagen is similar to those reported for full-length pig (6) and human (7) MMP-1 without collagen. The collagen peptide in the complex with MMP-1(E200A) is an uninterrupted triple helix ∼115 Å in length and bent by ∼10° near the Cat-Hpx junction (Fig. 4B and SI Appendix, Fig. S6). The three chains of the collagen triple helix are arranged with the characteristic one residue stagger, resulting in a leading (L), middle (M), and trailing (T) chain (25). All three chains contribute to MMP-1(E200A) binding, creating an extensive interaction surface spanning nearly 60 Å in length. Complex formation buries 480 Å2 of solvent-accessible surface of the L chain, 440 Å2 of the M chain, and 370 Å2 of the T chain.
Interactions Between MMP-1(E200A) and the Collagen Peptide.
The majority of the interactions between the collagen peptide and the Cat domain are of a polar nature (Fig. 4C and SI Appendix, Fig. S7). Of the three collagen chains, the L chain is closest to the active site of MMP-1, but the S1′ pocket remains empty. The residue closest to the S1′ pocket is Gln(P4′L) (residues are identified by their position relative to the scissile bond and a subscript designating the collagen chain). The side chain of Gln(P4′L) forms a hydrogen bond with the backbone amide of Tyr221 at the entrance of the S1′ pocket, and the backbone amide of Gln(P4′L) forms a hydrogen bond with the side chain of Asn161. Leu(P1′L) is 9 Å away from the S1′ pocket and stabilized by a hydrophobic contact with the zinc ligand His203. The Arg(P5′L) side chain and backbone amide form hydrogen bonds with the side chain of Tyr218 and the carbonyl oxygen of Pro219, respectively. The M chain makes only a single contact with the Cat domain, between the side chain of Gln(P4′M) and the side chain of Tyr218. The T chain, however, makes extensive interactions with the upper part of the substrate binding cleft, including five hydrogen bonds (Fig. 4C and SI Appendix, Fig. S7). Some of these hydrogen bonds are identical to those formed with a cysteine-switch sequence (26–29) in proMMPs and with a predicted peptide substrate (30).
The interactions between the collagen peptide and the Hpx domain are a combination of polar and apolar contacts, and are mediated mainly by the M chain (Fig. 4D and SI Appendix, Fig. S7). Consistent with our mutagenesis results, the side chain of Leu(P10′M) is bound in a hydrophobic S10′ exosite pocket delineated by Ile271, Met276, Phe301, Trp302, and the alkyl portion of Arg272 (Fig. 4D). The S10′ pocket is surrounded by polar residues (Arg272, Glu274, Arg285, and Gln335) that make a total of five hydrogen bonds with the collagen peptide (SI Appendix, Fig. S7). The interactions at the S10′ exosite are critical for collagen recognition and collagenolysis by MMP-1 (Fig. 3B). In addition, Ile(P7′M) forms hydrophobic interactions with Val300 and Phe301. The M and T chains make van der Waals contacts with Phe289 and Tyr290, but these interactions evidently are not critical, because the triple mutant MMP-1(F289A/Y290A/P291A) retains 76% of wild-type collagenase activity (Fig. 3B).
Implications for Collagenolysis.
The way in which the collagen peptide is bound to the active-site cleft of MMP-1(E200A) is more compatible with cleavage of the Gly(P3′L)-Gln(P4′L) bond instead of the canonical Gly(P1)-Leu(P1′) bond (31). To test whether MMP-1 can cleave also before Gln(P4′), we performed N-terminal sequencing of the products generated by active MMP-1 at 25 °C and 37 °C. We found that only the Gly(P1)-Leu(P1′) bond was cleaved in the peptide, as in native collagen II, and concluded that the mode of collagen binding in the crystallized complex is unproductive. A productive mode could be achieved by a substantial movement of the Cat domain relative to the Hpx domain, but this necessitates the breaking of the Cat-Hpx interface (19). We favor an alternative mechanism that leaves the interface intact. The helical symmetry of homotrimeric collagen dictates that axial rotation of the collagen homotrimer (in other words, binding of MMP-1 to a different set of collagen chains) changes the register of the chain passing through the active site cleft, but leaves all other interactions unchanged (Fig. 5 and SI Appendix, Fig. S8). If Leu(P10′L) instead of Leu(P10′M) is made to occupy the hydrophobic S10′ exosite, the T chain becomes the chain that interacts most closely with the active site, and now Leu(P1′T) is situated near the S1′ pocket. Notably, this productive binding mode does not alter any of the extensive interactions with the upper part of the substrate binding cleft (now made by the M chain) or with the S10′ exosite in the Hpx domain (now made by the L chain) (Fig. 5 and SI Appendix, Fig. S8). The energy difference between the observed unproductive and putative productive complex is likely to be small, and the two forms may coexist in solution. The unproductive complex may be favored by the E200A mutant of MMP-1 or it may have crystallized preferentially in our experiment.
Fig. 5.
Modes of collagen binding to MMP-1. Shown is relationship between the unproductive and productive collagen binding modes (see text). In the sequence alignment on the left, the trailing chain is shown twice to emphasize the circular nature of the chain arrangement. The P1′ and P10′ residues are boxed, and residues interacting with MMP-1 are in bold. The structural models on the right show that the unproductive and productive binding modes are related by a simple rotation/translation of the collagen triple helix. Note that only the interactions with one collagen chain are changed (L in the unproductive mode, T in the productive mode); the interactions with the other two chains are the same in both modes.
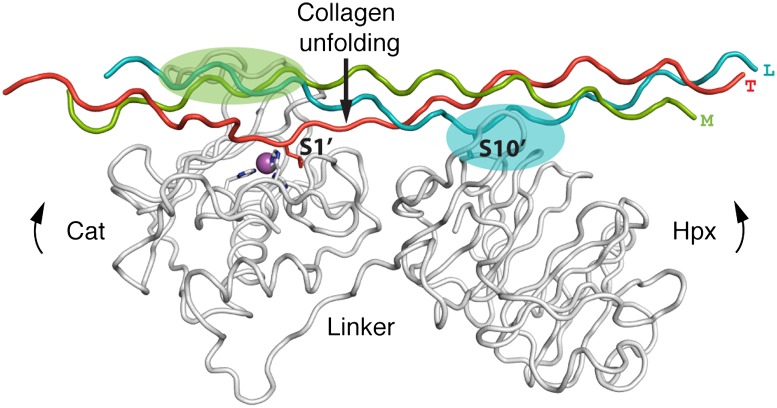
Starting from the modeled productive complex (SI Appendix, Fig. S8), it proved straightforward to achieve the first transition state of collagenolysis, in which one collagen chain is inserted into the active site (Fig. 6). By stretching the P1′-P6′ region of the T chain and reorienting three residues preceding the scissile bond, it was possible to insert Leu(P1′T) into the S1′ pocket in a manner that fully agrees with the observed binding mode of single-chain peptidomimetic MMP inhibitors (8, 32). The Cα atom of Leu(P1′T) moves by less than 4 Å during this localized excursion from the regular triple-helical structure, and only four collagen interchain hydrogen bonds are broken in the transition state. We propose that the energy penalty for this local unfolding event is compensated by the favorable interactions that the enzyme forms with the looped-out collagen chain.
Fig. 6.
Model of the first transition state of collagenolysis based on the productive complex. The upper rim of the catalytic site cleft anchors the M chain (green), and the Hpx domain anchors the L chain (cyan). These interactions position the T chain above the active site. Interdomain flexing of the enzyme bends the collagen triple helix and facilitates the insertion of Leu(P1′) of the T chain into the S1′ pocket.
Discussion
Our study has revealed an unprecedented proteinase–substrate interaction. The most striking feature of the MMP-1–collagen complex is the extensive interaction of all three collagen chains with the two domains of MMP-1, which explains the strict requirement of the Hpx domain for efficient collagenolysis (3). The individual Cat or Hpx domains of MMP-1 showed very little collagen binding in our experiments, but when linked together, they acted cooperatively in a temperature-dependent manner. The affinity for collagen I increased up to 37 °C, a temperature that would not cause collagen to denature within the time frame of the experiment (∼2 h) (33). Above this temperature the binding sharply decreased. This temperature dependence, which is also unusually pronounced in the rate of collagen cleavage by MMP-1 (34), can be explained by the relative thermoflexibility of the triple helix at the collagenase cleavage site (35–38) and the contribution of interdomain flexibility of MMP-1. The cooperative binding is therefore energetically most favored close to body temperature, when the triple helix is locally looser but the overall fold of the substrate is still maintained. A complete loss of triple helicity makes collagen bind less tightly to MMP-1, as evidenced from a Km value of ∼1 μM for collagen I compared with 4–7 μM for gelatin (39). The small active-site inhibitor GM6001 abolished the temperature-dependent increase of collagen binding, indicating that the catalytic site of MMP-1 is not only involved in the final hydrolysis step, but also in the unfolding machinery, which uses the active site to extract one collagen chain to form the first transition state of collagenolysis (Fig. 6).
Apart from providing a favorable environment for the loose triple helix at the cleavage site, what other mechanism(s) might contribute to collagenolysis? Comparison of the crystal structure of proMMP-1 (29) with the structure of the activated form of MMP-1(E200A) (6, 7) suggested that the Cat-Hpx linker region confers interdomain flexibility. Such flexing was further demonstrated for MMP-1 by NMR and small angle X-ray scattering studies (40, 41) and seems to be a common property of MMPs (42, 43). Our crystal structure of MMP-1 bound to collagen has a more closed conformation than free MMP-1 (SI Appendix, Fig. S9). This change in interdomain angle is accompanied by the bending of the collagen peptide at a point halfway between the Cat and Hpx domains. Thus, domain motions in MMP-1 may facilitate structural changes in the triple helix. Our structural results show how the two domains of MMP-1 anchor two of the collagen chains (M and L in the modeled productive complex) and position the third (T) chain near the active site. Stretching of the T chain by interdomain flexing of the enzyme may assist in local collagen unfolding. Because the P1′ residue of the trailing chain is poised above the S1′ pocket, transient excursions of the scissile bond can be efficiently captured by the enzyme, leading to the stabilization of the first transition state of collagenolysis (Fig. 6). Although thermal relaxation of the cleavage site is important for MMP-1 binding, this alone is not sufficient for efficient collagenolysis (17, 37). We propose that mammalian collagenases do not serve merely as passive acceptors of spontaneously occurring unfolded collagen states (44, 45), but they promote a local perturbation of the triple helix that facilitates collagenolysis. The recently reported NMR study supports this notion by demonstrating a weakening of the collagen interchain contacts in the complex with MMP-1 (19).
Our mechanism of collagenolysis (Fig. 6) differs from that derived from NMR experiments (19) in that it does not require a separation of the Cat and Hpx domains. Bertini et al. (19), who also used the E200A mutant of MMP-1, found that the Cat domain interacted mainly with the L chain and the Hpx domain mainly with the L and M chains. These results agree with our crystal structure of an unproductive complex. In their modeling, Bertini et al. (19) imposed a productive interaction of the L chain with the S1′ pocket, which forced the breaking of the Cat-Hpx interface. Our interpretation requires only small conformational changes in the enzyme but postulates the coexistence of productive and unproductive binding modes.
The crystal structure of a bacterial collagenase comprising of a N-terminal activator domain, a glycine-rich linker, and a peptidase domain has been reported (46). Like MMPs, this bacterial collagenase requires an ancillary activator domain (unrelated to the Hpx domain of MMPs) to cleave triple-helical collagen. The authors proposed that the enzyme clamps collagen between the activator and peptidase domains of the saddle-shaped structure. Our MMP-collagen structure shows that MMP-1 binds collagen in an extended manner, rather than surrounding it. These differences notwithstanding, interdomain plasticity seems to be a common feature of enzymes that can cleave the intractable collagen triple helix.
Most MMPs are believed to have specific substrate recognition sites distant from the catalytic site, which are referred to as exosites. Apart from the Hpx domains of collagenases, examples include the fibronectin type II repeats in MMP-2 and MMP-9, which contribute to the binding and degradation of elastin, gelatin, and type IV collagen (47). Recent NMR and mutagenesis studies of the Cat domain of MMP-12 characterized several exosites for elastin (48, 49). Similar studies were carried out with the isolated Hpx domain of MMP-1 and a triple-helical collagen peptide (40) and suggested a role for Phe282 (Phe301 in the numbering used in ref. 40) in binding of collagen. However, the exact modes of interaction between the enzyme and the target substrate have not been elucidated in these studies. Our crystal structure has revealed in detail the interactions that occur between full-length MMP-1 and a triple-helical peptide long enough to define all relevant contacts. Phe282 is buried in the Cat-Hpx interface in our structure and not involved in collagen binding. Defining the exosites in proteolytic enzymes has important implications for the development of specific inhibitors. The hydrophobic S10′ exosite in MMP-1 is a particularly attractive target because it is essential for collagenolysis. Recent studies of Robichaud et al. (50) showed the importance of Leu at the P10′ subsite of collagen III for the cleavage by MMP-1. Noncollagenous extracellular matrix and nonmatrix substrates are cleaved by the Cat domain of MMP-1 (16). Therefore, typical inhibitors targeting the S1′ pocket block all MMP-1 activities, whereas inhibitors directed against the S10′ exosite are predicted to selectively inhibit collagenolysis. This exosite of MMP-1 is largely conserved in other MMPs, but to our knowledge it is specific for collagenolytic activity. We propose that molecules specifically binding the S10′ exosite be therapeutically useful in diseases associated with accelerated collagen breakdown such as arthritis, cancer, aneurysm, and atherosclerosis.
Materials and Methods
Proteins and Peptides, Binding Assays, H/DXMS Analysis, and Enzyme Activity Assay.
Recombinant proMMP-1(E200A), its Cat(E200A) and Hpx domains, and MMP-1 variants were produced in Escherichia coli. Collagen I was purified from Guinea pig skin and treated with pepsin. The library of triple-helical peptides of human collagen II (Toolkit II) and all other collagen peptides were synthesized by Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. Collagen and peptide binding assays were carried out in ELISA format by using biotinylated proteins and detected by streptavidin-horseradish peroxidase system. Local hydrogen/deuterium exchange kinetics of MMP-1(E200A) free and bound to collagen I were analyzed by digestion with pepsin and mass spectrometry and quantified by using HX-Express software. The collagenolytic activity of MMP-1 variants was determined by SDS/PAGE and densitometric analyses of collagen I cleavage products. Detailed methods are described in SI Appendix.
Crystallization, Data Collection, and Structure Determination.
MMP-1(E200A) and the collagen peptide were mixed in a 1:1.5 molar ratio, and the complex was purified by size exclusion chromatography. Crystals of the complex were obtained at pH 8.5 and diffracted to 3.0 Å resolution. The structure was solved by molecular replacement and refined to a free R-factor of 0.273. Detailed methods are described in SI Appendix. Data processing and refinement statistics are listed in SI Appendix, Table S2. The coordinates of the MMP-1(E200A)–collagen complex have been deposited in the Protein Data Bank (ID code 4AUO).
Supplementary Material
Acknowledgments
We thank Noriko Ito for construction of MMP-1 mutants, Ida B. Thøgersen for peptide sequencing, the staff at Diamond beamline I24 for help with X-ray data collection, and Peter Brick for many helpful discussions. Protein visualization and modeling was done in PyMOL (www.pymol.org). The work was supported by Wellcome Trust Grants 068724, 075473, 094470, and 083942; the Medical Research Council, Arthritis Research UK; and Cancer Research UK. E.H. is a Wellcome Trust Senior Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4AUO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204991109/-/DCSupplemental.
References
- 1.Kadler KE, Baldock C, Bella J, Boot-Handford RP. Collagens at a glance. J Cell Sci. 2007;120:1955–1958. doi: 10.1242/jcs.03453. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 3.Nagase H, Visse R. Triple helicase activity and the structural basis of collagenolysis. In: Parks WC, Mecham RP, editors. Extracellular Matrix Degradation, Biology of Extracellular Matrix. Heidelberg: Springer; 2011. pp. 95–122. [Google Scholar]
- 4.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: A tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 5.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, et al. Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed β-propeller. Structure. 1995;3:541–549. doi: 10.1016/s0969-2126(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 7.Iyer S, Visse R, Nagase H, Acharya KR. Crystal structure of an active form of human MMP-1. J Mol Biol. 2006;362:78–88. doi: 10.1016/j.jmb.2006.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borkakoti N, et al. Structure of the catalytic domain of human fibroblast collagenase complexed with an inhibitor. Nat Struct Biol. 1994;1:106–110. doi: 10.1038/nsb0294-106. [DOI] [PubMed] [Google Scholar]
- 9.Lovejoy B, et al. Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor. Science. 1994;263:375–377. doi: 10.1126/science.8278810. [DOI] [PubMed] [Google Scholar]
- 10.Spurlino JC, et al. 1.56 Å structure of mature truncated human fibroblast collagenase. Proteins. 1994;19:98–109. doi: 10.1002/prot.340190203. [DOI] [PubMed] [Google Scholar]
- 11.Bode W, et al. The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J. 1994;13:1263–1269. doi: 10.1002/j.1460-2075.1994.tb06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stams T, et al. Structure of human neutrophil collagenase reveals large S1′ specificity pocket. Nat Struct Biol. 1994;1:119–123. doi: 10.1038/nsb0294-119. [DOI] [PubMed] [Google Scholar]
- 13.Bode W. A helping hand for collagenases: The haemopexin-like domain. Structure. 1995;3:527–530. doi: 10.1016/s0969-2126(01)00185-x. [DOI] [PubMed] [Google Scholar]
- 14.De Souza SJ, Pereira HM, Jacchieri S, Brentani RR. Collagen/collagenase interaction: Does the enzyme mimic the conformation of its own substrate? FASEB J. 1996;10:927–930. doi: 10.1096/fasebj.10.8.8666171. [DOI] [PubMed] [Google Scholar]
- 15.Ottl J, et al. Recognition and catabolism of synthetic heterotrimeric collagen peptides by matrix metalloproteinases. Chem Biol. 2000;7:119–132. doi: 10.1016/s1074-5521(00)00077-6. [DOI] [PubMed] [Google Scholar]
- 16.Overall CM. Molecular determinants of metalloproteinase substrate specificity: Matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 17.Chung L, et al. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han S, et al. Molecular mechanism of type I collagen homotrimer resistance to mammalian collagenases. J Biol Chem. 2010;285:22276–22281. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertini I, et al. Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis. J Am Chem Soc. 2012;134:2100–2110. doi: 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farndale RW, et al. Cell-collagen interactions: The use of peptide Toolkits to investigate collagen-receptor interactions. Biochem Soc Trans. 2008;36:241–250. doi: 10.1042/BST0360241. [DOI] [PubMed] [Google Scholar]
- 21.Schechter I, Berger A. On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem Biophys Res Commun. 1968;32:898–902. doi: 10.1016/0006-291x(68)90326-4. [DOI] [PubMed] [Google Scholar]
- 22.Lauer-Fields JL, et al. Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J Biol Chem. 2009;284:24017–24024. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung L, et al. Identification of the (183)RWTNNFREY(191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J Biol Chem. 2000;275:29610–29617. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- 24.Brandstetter H, et al. The 1.8-Å crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition. J Biol Chem. 2001;276:17405–17412. doi: 10.1074/jbc.M007475200. [DOI] [PubMed] [Google Scholar]
- 25.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 26.Becker JW, et al. Stromelysin-1: Three-dimensional structure of the inhibited catalytic domain and of the C-truncated proenzyme. Protein Sci. 1995;4:1966–1976. doi: 10.1002/pro.5560041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgunova E, et al. Structure of human pro-matrix metalloproteinase-2: Activation mechanism revealed. Science. 1999;284:1667–1670. doi: 10.1126/science.284.5420.1667. [DOI] [PubMed] [Google Scholar]
- 28.Elkins PA, et al. Structure of the C-terminally truncated human ProMMP9, A gelatin-binding matrix metalloproteinase. Acta Crystallogr D Biol Crystallogr. 2002;58:1182–1192. doi: 10.1107/s0907444902007849. [DOI] [PubMed] [Google Scholar]
- 29.Jozic D, et al. X-ray structure of human proMMP-1: New insights into procollagenase activation and collagen binding. J Biol Chem. 2005;280:9578–9585. doi: 10.1074/jbc.M411084200. [DOI] [PubMed] [Google Scholar]
- 30.Grams F, et al. X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design. Eur J Biochem. 1995;228:830–841. doi: 10.1111/j.1432-1033.1995.tb20329.x. [DOI] [PubMed] [Google Scholar]
- 31.Billinghurst RC, et al. Enhanced cleavage of type II collagen by collagenases in osteoarthritic articular cartilage. J Clin Invest. 1997;99:1534–1545. doi: 10.1172/JCI119316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovejoy B, et al. Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors. Nat Struct Biol. 1999;6:217–221. doi: 10.1038/6657. [DOI] [PubMed] [Google Scholar]
- 33.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welgus HG, Jeffrey JJ, Eisen AZ. Human skin fibroblast collagenase. Assessment of activation energy and deuterium isotope effect with collagenous substrates. J Biol Chem. 1981;256:9516–9521. [PubMed] [Google Scholar]
- 35.Brown RA, Hukins DW, Weiss JB, Twose TM. Do mammalian collagenases and DNA restriction endonucleases share a similar mechanism for cleavage site recognition? Biochem Biophys Res Commun. 1977;74:1102–1108. doi: 10.1016/0006-291x(77)91632-1. [DOI] [PubMed] [Google Scholar]
- 36.Fields GB. A model for interstitial collagen catabolism by mammalian collagenases. J Theor Biol. 1991;153:585–602. doi: 10.1016/s0022-5193(05)80157-2. [DOI] [PubMed] [Google Scholar]
- 37.Fiori S, Saccà B, Moroder L. Structural properties of a collagenous heterotrimer that mimics the collagenase cleavage site of collagen type I. J Mol Biol. 2002;319:1235–1242. doi: 10.1016/S0022-2836(02)00365-0. [DOI] [PubMed] [Google Scholar]
- 38.Stultz CM. Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. J Mol Biol. 2002;319:997–1003. doi: 10.1016/S0022-2836(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 39.Welgus HG, Jeffrey JJ, Stricklin GP, Eisen AZ. The gelatinolytic activity of human skin fibroblast collagenase. J Biol Chem. 1982;257:11534–11539. [PubMed] [Google Scholar]
- 40.Bertini I, et al. Interdomain flexibility in full-length matrix metalloproteinase-1 (MMP-1) J Biol Chem. 2009;284:12821–12828. doi: 10.1074/jbc.M809627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold LH, et al. The interface between catalytic and hemopexin domains in matrix metalloproteinase-1 conceals a collagen binding exosite. J Biol Chem. 2011;286:45073–45082. doi: 10.1074/jbc.M111.285213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenblum G, et al. Insights into the structure and domain flexibility of full-length pro-matrix metalloproteinase-9/gelatinase B. Structure. 2007;15:1227–1236. doi: 10.1016/j.str.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 43.Bertini I, et al. Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12. J Am Chem Soc. 2008;130:7011–7021. doi: 10.1021/ja710491y. [DOI] [PubMed] [Google Scholar]
- 44.Nerenberg PS, Salsas-Escat R, Stultz CM. Do collagenases unwind triple-helical collagen before peptide bond hydrolysis? Reinterpreting experimental observations with mathematical models. Proteins. 2008;70:1154–1161. doi: 10.1002/prot.21687. [DOI] [PubMed] [Google Scholar]
- 45.Salsas-Escat R, Nerenberg PS, Stultz CM. Cleavage site specificity and conformational selection in type I collagen degradation. Biochemistry. 2010;49:4147–4158. doi: 10.1021/bi9021473. [DOI] [PubMed] [Google Scholar]
- 46.Eckhard U, Schönauer E, Nüss D, Brandstetter H. Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat Struct Mol Biol. 2011;18:1109–1114. doi: 10.1038/nsmb.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: Structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 48.Palmier MO, et al. NMR and bioinformatics discovery of exosites that tune metalloelastase specificity for solubilized elastin and collagen triple helices. J Biol Chem. 2010;285:30918–30930. doi: 10.1074/jbc.M110.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulcher YG, Van Doren SR. Remote exosites of the catalytic domain of matrix metalloproteinase-12 enhance elastin degradation. Biochemistry. 2011;50:9488–9499. doi: 10.1021/bi2009807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robichaud TK, Steffensen B, Fields GB. Exosite interactions impact matrix metalloproteinase collagen specificities. J Biol Chem. 2011;286:37535–37542. doi: 10.1074/jbc.M111.273391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.