Abstract
The stability of human embryonic stem cells (hESCs) is of critical importance for both experimental and clinical applications. We find that as an initial response to altered culture conditions, hESCs change their transcription profile for hundreds of genes and their DNA methylation profiles for several genes outside the core pluripotency network. After adaption to conditions of feeder-free defined and/or xeno-free culture systems, expression and DNA methylation profiles are quite stable for additional passaging. However, upon reversion to the original feeder-based culture conditions, numerous transcription changes are not reversible. Similarly, although the majority of DNA methylation changes are reversible, highlighting the plasticity of DNA methylation, a few are persistent. Collectively, this indicates these cells harbor a memory of culture history. For culture-induced DNA methylation changes, we also note an intriguing correlation: hypomethylation of regions 500–2440 bp upstream of promoters correlates with decreased expression, opposite to that commonly seen at promoter-proximal regions. Lastly, changes in regulation of G-coupled protein receptor pathways provide a partial explanation for many of the unique transcriptional changes observed during hESC adaptation and reverse adaptation.
Keywords: demethylation, methylcytosine, G-protein-signaling modulator 3, murine embryonic fibroblast, hypermethylation
Transcriptional changes are expected as cells adapt to different culture conditions, even if there are no overt morphological changes and no evidence of differentiation. However, in human embryonic stem cells (hESCs) it is unclear whether transcriptional changes are accompanied, or preceded, by changes in DNA methylation. Further, if there are adaptive changes in DNA methylation, are they reversible when the cells are returned to the original culture environment? Stable alterations in gene expression can come from feedback loops involving diffusible factors, but one attractive model is that DNA methylation often aids the persistent memory of environmental cues, thus helping a cell remember its developmental history. There is considerable evidence supporting this idea (1, 2), but it has been known for some time that DNA methylation is dynamic, with occasional methylation loss and then repair by de novo mechanisms (3). Recently it has become clear that, at least at some sites, 5-methylcytosine can be actively removed (4). Therefore, epigenetic states are persistent, but also changeable.
In the context of pluripotent hESCs, especially when grown in larger scale for clinical trials, epigenetic stability is crucial. With this in mind we have studied HES-2 hESCs, measuring both transcription and DNA methylation genomewide as cells are passaged and transferred to different growth conditions. Working under the accepted notion that clinical utility requires hESCs to be grown in fully defined and preferably xeno-free culture conditions (5–7), we selected two of the most widely used and commercially available defined systems for our cell cultures. Both Matrigel/mTesR1 and CELLstart/STEMPRO are adhesive matrix and medium combinations used as alternatives to feeder-based hESC cultures. Matrigel is derived from mouse sarcoma-cell extracts and is highly enriched in extracellular matrix (ECM) components and a variety of growth factors (5). CELLstart is a humanized substrate considered to be the first xeno-free option for hESC cultures (5). Both STEMPRO and mTesR1 media are complex formulations that use FGF- and TGF-β-signaling pathways to maintain pluripotency. We report here on the stability and adaptability of epigenetic changes under these culture conditions. Subsequent to DNA methylation changes seen upon initial adaptation, we find that both transcription levels and DNA methylation patterns to be remarkably stable. Although relatively few, regions of differential methylation are nevertheless significant and in some instances irreversible. The large dataset generated by this study confirms the usual negative correlation between DNA methylation and transcription levels at promoters near transcription start sites and the positive correlation in gene bodies, but also identifies hypomethylation in promoter-distal regions to be associated with decreased gene expression.
Results
Experimental Approach.
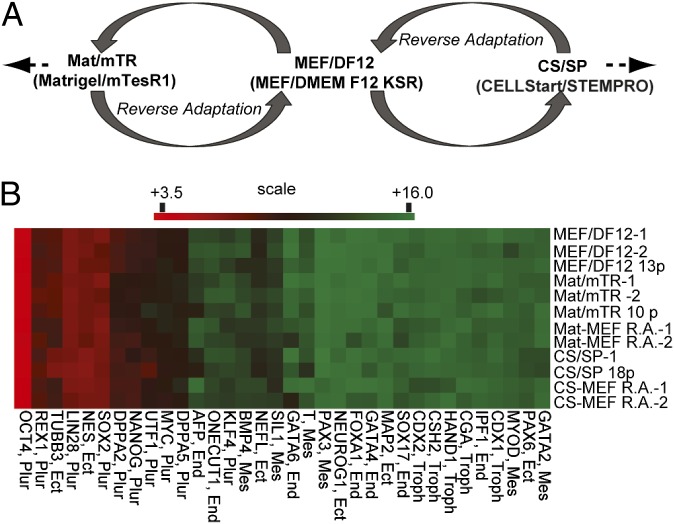
The purpose of this work was to study the specificity, stability, and reversibility of any culture-induced DNA methylation changes in hESCs. Our strategy is outlined in Fig. 1A and complete sample details are provided in Table S1. HES-2 hESCs were first grown on murine embryonic fibroblast (MEF)/DMEM F12 with knockout serum replacement (MEF/DF12), then adapted to Matrigel/mTesR1 (Mat/mTR), and, after several passages, reverse adapted. That is, hESCs were returned to original MEF/DF12 conditions. This experimental approach was then repeated using CELLstart/STEMPRO (CS/SP). It is important to note that hESCs under all conditions exhibited normal karyotype (Table S1), high pluripotency marker expression, and minimal expression of differentiation markers (Fig. 1B, Fig. S1 A and B, and Table S1). Microarray expression results for key pluripotency markers were validated by real-time qRT–PCR and protein immunofluorescence (Fig. S1 A and B). To minimize any influence from low-level spontaneous differentiation and/or any feeder cell contamination, flow-cytometric isolation of cells positive for Tra-1–60 was carried out for all samples. Transcription levels were measured by use of NimbleGen 12 × 135 k expression arrays covering 45,033 transcripts. DNA methylation was assessed by use of methylated island recovery assay (MIRA) (8) followed by hybridization on tiling microarrays that extended 2440 bp upstream and 610 bp downstream of all annotated promoters and CpG islands. Technical and biological replicates were all highly correlated (Table S2). No methylation differences were identified between samples at pluripotency genes (Table S3).
Fig. 1.
Overview of hESC culture adaptation strategy and validation of pluripotency marks. (A) Overview of culture strategy/sample collection. HES-2 hESCs grown on MEFs were adapted to Mat/mTR- or CS/SP-culture conditions and then reverse adapted to MEF/DF12. Additional samples were collected after more passaging and from separate hESC recoveries. (B) Heat map illustrating relative gene expression of pluripotent and lineage-specific markers by microarray analysis. Highly expressed genes are in red; minimally expressed genes are shaded green. Plur = pluripotency; Mes = mesoderm; End = endoderm; Ect = ectoderm; and Troph = trophoblast. Scale is log2 expression value.
hESCs Fine-Tune Transcriptional Profiles to Reflect Culture Environment.
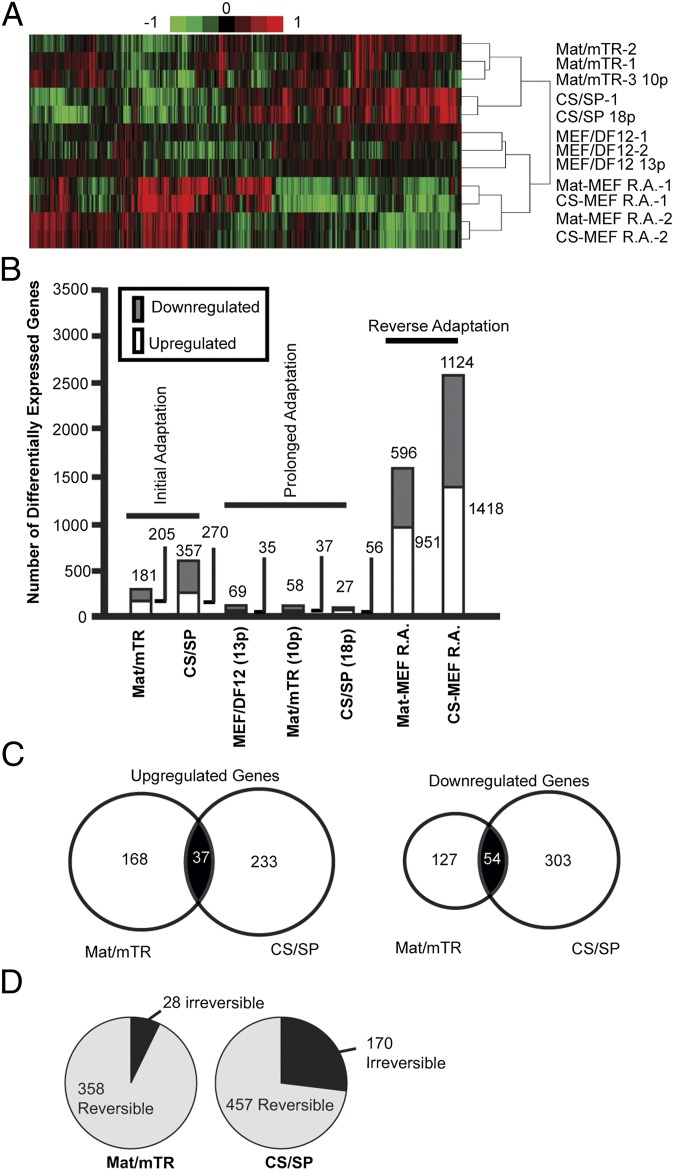
Initial results of genomewide transcriptional profiling showed linear correlation values exceeding 0.9 (R2) for all technical and biological replicates, and between pairwise sample comparisons (Table S2). Unsupervised hierarchical clustering grouped transcriptional profiles according to their culture environment regardless of passage number or separate recovery (Fig. 2A). In total, 386 and 627 transcripts were differentially expressed in Mat/mTR and CS/SP adapted cells, respectively, compared with start MEF/DF12 cultures (Fig. 2B). These results are well in line with previously published work on these culture systems (9). Details of differentially expressed transcripts are provided in Dataset S1. The majority of transcriptional changes were culture specific; however, 37 up-regulated and 54 down-regulated transcripts were shared between Mat/mTR and CS/SP cultures (Fig. 2C). Gene ontology results imply a substrate-specific reconfiguration of cell–cell and cell–ECM interactions (Fig. S1 C and D and Dataset S1). We note up-regulation of secreted factors and ECM components, including collagen subtypes, fibronectin, laminin, and a variety of cadherins that likely facilitate cellular adhesion unique to Matrigel and CELLstart matrices. These results are coherent with the general morphology of hESCs grown on Matrigel and CELLstart, in which the absence of feeders is associated with flatter colony morphology, and in the case of CELLstart, looser cell–cell contacts as evidenced by additional cell spreading (Fig. S1A) (5). With respect to medium formulation, commensurate expression responses are noted in Table S4. Large expression variation in factors controlling TGF-β and Wnt signaling is seen, consistent with both pathways being targeted by mTesR1 and STEMPRO media (5). Down-regulation of metallothionein factors, of which many were shared between Mat/mTR and CS/SP, may reflect the lack of trace metal additive in mTesR1 and STEMPRO media (Fig. S1 C and D, Table S4, and Dataset S1). Additional adaptive expression changes are outlined in Table S4.
Fig. 2.
Gene expression changes during hESC culture adaptation. (A) Hierarchical clustering of expression data. Low intensity genes were removed if failing to meet the criteria of log2 intensity value of 8 in at least 10% of samples. Filtered genes were subjected to clustering with Pearson’s correlation and complete linkage using Cluster v3.0. (B) Numbers of differentially expressed genes during hESC culture adaptation (>twofold change, P < 0.01, moderated t test). Initial adaptation: Mat/mTR- or CS/SP-adapted culture vs. start MEF/DF12 culture; prolonged adaptation: final passage vs. initial adaptation or original start culture for MEF/DF12 samples; reverse adaptation: reverse-adapted cells vs. Mat/mTR or CS/SP intermediate culture conditions. (C) Venn diagrams showing overlap of differentially expressed genes between Mat/mTR- and CS/SP-adapted cultures. (D) Reversibility of expression changes. Differential gene expression identified during Mat/mTR or CS/SP acclimation was considered irreversible if still significantly up- or down-regulated in final MEF/DF12 reverse-adapted samples compared with Mat/mTR or CS/SP intermediates cultures.
Reverse Adaptation Demonstrates Lack of Transcriptional Reversibility.
Clearly, cells undergo specific transcriptional changes to facilitate adaptation to novel culture conditions. However, reversion of culture environments from Mat/mTR or CS/SP back to MEF/DF12 did not result in the original hESC MEF/DF12 transcriptional profile and these samples appear quite distinct (Fig. 2A). First, as compared to initial or prolonged adaptation, the number of differentially expressed genes was several times higher after reverse adaptation (Fig. 2B). Second, numerous transcripts differentially expressed during adaptation to Mat/mTR or CS/SP remained so despite 7 and 10 passages from Mat/mTR and CS/SP into MEF/DF12 conditions, respectively (Fig. 2D). Details of irreversible transcripts by culture are provided in Dataset S1C and SI Results, but in general reflect irreversibility of many transcripts involved in cell adhesion. For the several hundred additional transcriptional changes that uniquely accompany reverse adaptation, regardless of Mat/mTR or CS/SP intermediate cultures, gene ontology results highlight cellular stress responses, including p53 signaling, and involve genes in cellular homeostasis, cell adhesion, and metabolism (see Fig. S3 and Dataset S1D).
5-Methylcytosine Changes During Adaptation and Passaging.
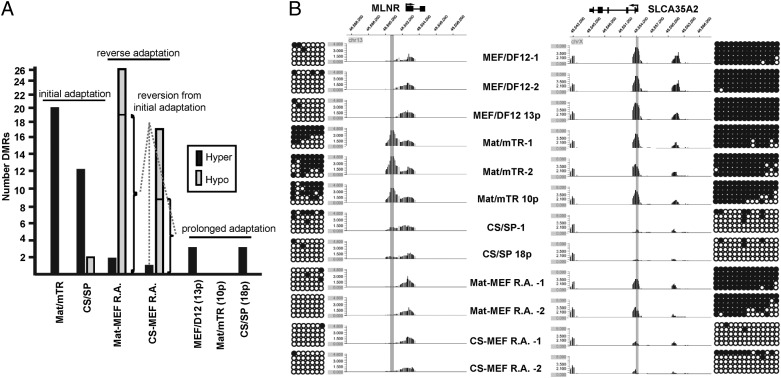
Initial comparative results indicated DNA methylation patterns across samples to be remarkably similar, with Pearson’s correlation values between any two samples exceeding 0.96 (P < 0.0001). A chromosomewide (chr 1) view is shown in Fig. S2A. Consistent with existing literature (8, 10), we observed gene promoters as predominately unmethylated; whereas, DNA methylation was more prevalent at intragenic and intergenic CpG islands (Table S3A). Pluripotency markers lacked promoter DNA methylation and displayed no methylation differences between samples. (Table S3B and Fig. S2B). Regions of differential DNA methylation (DMRs) were subsequently identified between samples (Fig. 3A; see Materials and Methods for statistical approach). As an example, Mat/mTR-specific and reversible hypermethylation at the motilin receptor (MLNR) promoter is shown in Fig. 3B. Also, the promoter of the solute carrier family 35 (UDP-galactose transporter), member A2 (SLC35A2) illustrates a CS/SP-specific and rare irreversible hypomethylation event (Fig. 3B). MIRA-chip results for numerous DMRs were validated by bisulfite sequencing and a direct correlation between methylation peak score and percent methylation was observed (Fig. 3B and Fig. S2 B and C). Details of identified DMRs are provided in Table S5 and primers used for methylation validation in Table S6.
Fig. 3.
DNA methylation in hESCs during culture adaptation. (A) Summary of DMRs across samples. Mat/mTR and CS/SP initial DMRs were identified through comparison with original MEF/DF12-cultured cells. Prolonged DMRs were identified in higher passages versus initial adaptation to Mat/mTR or CS/SP, or in the case of MEF/DF12, p82 versus start culture (p69). Sample descriptions provide passage number postadaptation. Reversible DMRs are noted. Hyper = hypermethylation; Hypo = hypomethylation. (B) Examples of and validation of culture-specific DMRs. A hypermethylated Mat/mTR-specific DMR is shown on the left at the MLNR gene promoter. The peak was identified in three separate Mat/mTR samples and is reversible in accordance with culture conditions. A hypomethylated CS/SP DMR at the SLCA35A2 promoter region is provided on the right. The DMR is an example of a rare irreversible methylation change. MIRA-chip results were validated by bisulfite sequencing of sequences centric to identified DMRs (highlighted in gray). Circles represent consecutive CpG dinucleotides. Dark circles: methylated CpG sites. Open circles: unmethylated CpG sites.
Despite the hundreds of transcriptional changes that accompanied initial adaptation from feeder-based cultures to Mat/mTR and CS/SP, only 20 and 14 DMRs were identified, respectively. DMRs under both culture adaptations were overwhelming sites of hypermethylation (Fig. 3A). Indeed, only two CS/SP DMRs were hypomethylated during culture acclimation. Although the sites of differential methylation represent only a tiny fraction of all regions tested, they appear to be highly specific and stable over additional culture time. In fact, only three subsequent DMRs (all hypermethylation) were observed in separate recovery and prolonged cultures of cells in CS/SP (18 passages) and MEF/DF12 (13 passages). Such accumulation of DNA methylation over passaging has been previously observed (11). No additional DMRs were noted in Mat/mTR (10 passages) (Fig. 3A). In all cases, DMRs identified originally were sustained through subsequent passages. When taken together, initial and prolonged adaptive responses to shifting culture environment involve predominately site-specific and stable DNA hypermethylation.
Reverse Adaptation and the Reversibility of Epigenetic Changes.
Given the unique expression profiles that accompany reverse adaptation and the potential consequences of altered DNA methylation landscapes, we sought to determine whether DMRs formed in the first round adaptation were reversible. How plastic is DNA methylation in hESCs? We found that nearly all DMRs are reversible. The same changes were seen in two independent reverse-adaptation experiments for both Mat/mTR and CS/SP reverse adaptation. Specifically, 19/20 Mat/mTR and 11/14 CS/SP DMRs were reversible (Fig. 3A, Fig. S2E, and Table S5). Cumulatively, this is ∼90% reversion of culture-induced DNA methylation changes. Furthermore, whereas initial adaptation is marked by DNA site-specific hypermethylation, reverse adaptation is marked by nonrandom DNA hypomethylation. It was intriguing to note that there were several additional sites of DNA hypomethylation, of which most were clustered on chromosome X p11.4 and shared between Mat-MEF R.A. and CS-MEF R.A. samples (Fig. S2 F and G). These hypomethylation sites uniquely accompany reverse adaptation, perhaps as part of a stress response to shifting culture environments (Fig. S1E and Dataset S1D). This region has been shown to be prone to escape from X-chromosome inactivation (XCI) (12, 13). HES-2 is known to have undergone XCI, but may lose some markers of XCI after significant passaging (P > 90) (14, 15). Consistent with this, XIST expression was detectable at similar levels across all samples (Fig S2H). Our data indicate that most X-linked promoters and CpG islands, including the Xist promoter, are partially methylated and do not change with culture conditions, as expected for XCI. Therefore, it is unlikely that global changes in XCI have occurred, but rather through reverse adaptation X p11.4 may be uniquely escaping XCI.
It appears that hESCs adopt a precise DNA methylation profile in accordance to their culture environment and can undergo site-specific reprogramming to reflect these environments. The process appears however, to be imperfect. Indeed, a few sites of residual DNA methylation may persist as a culture-induced “memory” of cellular manipulation. Specifically, there was one shared irreversible DMR after reverse adaptation from Mat/mTR or CS/SP and two irreversible DMRs uniquely from CS/SP conditions (Fig. 3 A and B and Table S5). These DMRs were restricted to promoters (Fig. S2E) and the genes they were associated with may be of particular consequence. The shared Mat/mTR and CS/SP irreversible methylation gain was observed at the PHF17 (JADE-1) gene promoter, which is a candidate tumor suppressor in renal tumorigenesis through a Wnt-signaling connection (16). Further, irreversible methylation was noted at monocyte-differentiation-associated miRNAs mir-503 and mir-424 in CS/SP cultures (17).
Closer examination of individual DMRs leads to several interesting observations (Table S5B). First, as with the case of differential gene expression, we noted differential methylation at several adhesion genes (e.g., MXRA5, PCDHB18, CDH22, and, PLEKHA2). Although Mat/mTR and CS/SP represent distinct culture systems, they have several common ingredients (Table S4) and both mandate hESC growth without the adhesion, nutrition, and signaling from feeder cells (5). It is perhaps not surprising, then, that several DMRs identified during adaptation, from and back to MEF/DF12, were shared between Mat/mTR- and CS/SP-cultured hESCs (Fig. S2 E–G). Despite normal pluripotent expression patterns and the selection of only Tra-1–60 (+) cells (Fig. 1B, Fig. S1 A and B, and Table S1), we identified culture-associated DMRs at a few development genes in addition to the miRNAs mentioned previously. Examples include: HOXB13, NOTCH4, and ALXI; these observations are considered in the discussion within the context of stem cell lineage propensity. Last, we identified multiple culture-induced DMRs at genes involved in G-protein-coupled signaling, including MLNR, RAB14, RAB5C, ARRB2, and GPSM3. These genes likely play key roles in regulating cellular sensitivity to ligand-induced G-protein signaling cascades (18–20), and therefore might contribute to many of the expression changes observed during culture adaptation.
Differential DNA Methylation of Promoter-Proximal Region Is Correlated with Reduced Gene Expression, but Upstream Promoter-Distal Regions Show an Opposite Correlation.
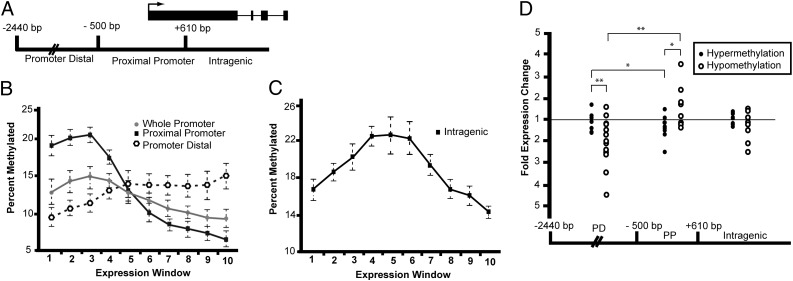
DNA methylation at promoters is generally considered a gene-silencing epigenetic modification (21, 22). Consistent with expectation, we observed that high levels of gene promoter methylation are associated with lower levels of transcription genomewide (Fig. 4B and Fig. S3 A–C). Also consistent with previous work (10), we find that intragenic DNA methylation increases with expression until moderate expression levels are reached, followed by a decrease in methylation for highly expressed genes (Fig. 4C). We did not observe a clear tendency for culture-change-induced intragenic DMRs to influence positively or negatively gene expression (Fig. 4D and Table S5).
Fig. 4.
Correlations of DNA methylation with gene expression. (A) Schematic map illustrating defined boundaries for classifying DNA methylation as related to a generic gene structure. Intergenic DMRs fall in CpG islands annotated anywhere outside of gene-associated regions as defined within the schematic. Arrow indicates TSS. (B) The percent genes with methylated promoter-proximal, promoter-distal, and whole promoters is shown for each of 10 expression windows; 1 = no expression, 10 = highly expressed. Data points are mean ± SD (error bars) of all samples for each expression window. (C) The percent of total genes with intragenic DNA methylation is shown for each of 10 expression windows. (D) Collective expression analysis of genes with associated DMRs. Any gene-associated DMRs identified across samples were pooled into a single graph examining DMR associations with gene-expression changes based on DMR location and the type of methylation change. Fold gene-expression change is displayed for each corresponding DNA methylation change. *P value < 0.05; **P value < 0.005 (Student's t test).
The extended array tiling coverage allowed us to separately analyze the promoter-proximal and the promoter-distal regions. The expected correlation between promoter methylation and transcriptional repression is most prominently observed for the promoter-proximal region [−500 to +610 bp of transcription start sites (TSS)] and to a lesser extent across the whole promoter (Fig. 4 A and B) (22). In contrast, in upstream promoter-distal regions (−501 to −2,440 bp), DNA methylation trended slightly toward increased gene expression (Fig. 4B and Fig. S3 A–C). Given these observations, we next examined all identified culture-change-induced DMRs for corresponding gene expression. As expected, promoter-proximal hypermethylation was predominately associated with a decrease in gene expression; whereas, hypomethylation was associated with increased gene expression (Fig. 4D and Table S5). In the promoter-distal region, although DMR hypermethylation was not clearly associated with unidirectional gene-expression changes, the loss of DNA methylation is well correlated with expression down-regulation, in direct contrast to the transcriptional behavior of genes harboring culture-induced promoter-proximal hypomethylation. Thus, we note a correlation in which loss of DNA methylation in promoter-distal regions is associated with gene silencing.
Culture-Induced DMR Associated with G-Protein Signaling Is Linked with the Lack of Transcriptional Reversibility.
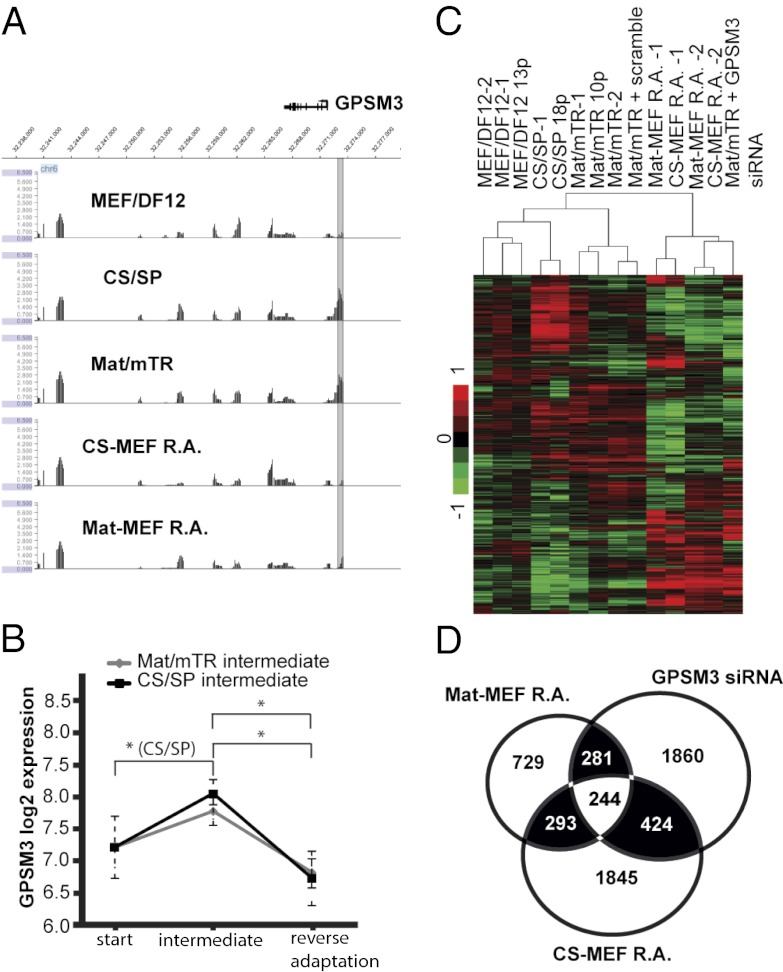
Given the number of G-protein-associated genes with DMRs (Table S5), and the role for G-protein-signaling modulator 3 (GPSM3) in regulating G-protein-coupled receptor signaling pathways (19, 20), we hypothesized that GPSM3 knockdown by siRNA may induce a transcriptional response similar to that in Mat-MEF R.A. and CS-MEF R.A. samples (Fig. 2 A and B). Previously, we had noted both Mat/mTR and CS/SP hESC adaptation induces reversible GPSM3 promoter-distal DNA methylation with concurrent changes in gene expression (Table S5 and Fig. 5 A and B). By siRNA treatment we obtained ∼50% GPSM3 knockdown in hESCs grown on Mat/mTR, effectively mimicking the approximately twofold reduction in GPSM3 expression upon reverse adaptation (Fig. S3D and Fig. 5B). Unsupervised hierarchical clustering demonstrated GPSM3 knockdown samples to be most similar to reverse-adapted cells, and scramble control transfection to minimally affect genomewide transcription (Fig. 5C). Of the 2,809 differentially expressed genes ∼34% are shared with either CS-MEF R.A. or Mat-MEF R.A. samples (Fig. 5D). Given the wide scope of differential gene expression unique to our reverse-adapted samples, we cannot conclude that promoter-distal hypomethylation of GPMS3 is the primary cause for GPSM3 down-regulation in these cells; the effect may be secondary to expression changes in related signaling pathways and possibly through differential DMRs outside of our array coverage. However, it appears culture-associated changes in GPSM3 expression, which may involve GPMS3 differential methylation, are in part responsible for the transcriptional character observed in reverse-adapted cells.
Fig. 5.
Knockdown of GPSM3 results in similar transcriptional deregulation seen in reverse-adapted cells. (A) GPSM3 is reversibly promoter-distal hypermethylated during adaptation from MEF/DF12. P value data are displayed at and surrounding each gene for initial and reverse-adapted samples. (B) GPSM3 expression through culture adaptation. Average expression values ± SD for all MEF/DF12, Mat/mTR, CS/SP, and reverse-adapted cells are displayed. (C) Hierarchical clustering of expression results. See Fig. 2 for description of clustering approach. GPSM3 knockdown samples cluster alongside reverse-adapted cells. (D) Many differentially expressed genes stemming from GPSM3 knockdown are differentially expressed in reverse-adapted cells. Venn diagram illustrates overlap of gene expression changes across three conditions.
Discussion
hESCs hold enormous promise for regenerative medicine, but it is well accepted that for human therapy, hESCs must first be grown in defined and, if possible, xeno-free conditions (6, 7). In this report we have investigated the epigenetic stability of hESCs as they maneuver a series of culture-environment shifts including two of the most widely used feeder-independent culture systems. Our results therefore have important implications for the clinical utility of pluripotent cells, as well as providing insight into the plasticity and function of DNA methylation in these cells. We have confirmed that, as expected, transcriptional changes take place in hESCs when culture conditions are changed. The role of DNA methylation in hESCs is not known, but it is recognized that mouse ESCs with no DNA methylation are viable (23). Further, repression of retrotransposons, normally viewed as an essential DNA methylation function, is primarily regulated through a methylation-independent mechanism in mouse ESCs (24). It is therefore possible that DNA methylation plays little or no role in hESCs until differentiation is triggered and serves primarily to set the stage for lineage switching. Do changes in DNA methylation accompany (or cause) changes of transcription? We find that at least some DNA methylation changes do accompany changes in transcription, with the usual inverse correlation of promoter-proximal DNA methylation with transcription level (Fig. 4B) (22). It is clear that epigenetic changes do occur, but how stable are they? We find that after initial adaptation to the new environmental conditions, very few changes in either transcription or DNA methylation take place with continued passaging (Figs. 2B and 3A). As long as culture conditions are carefully controlled, the epigenetic stability of hESCs is probably adequate for the requirements of clinical trial scaleup.
One of our unique findings was obtained by reverse adaptation to the original culture conditions. We find that, notwithstanding the general perspective that DNA methylation is persistent, most DNA methylation changes in our system are highly specific and plastic, reverting back to the original methylation state when given the appropriate environmental cue. Upon transfer from feeder layers to either CS/SP or Mat/mTR almost all changes were de novo methylation. Upon reverse adaptation to growth on MEF/DF12 essentially all changes were demethylation. It is currently unclear why a few DMRs were irreversible, but this finding is consistent with recent studies indicating that induced pluripotent stem cells harbor residual DNA methylation and other epigenetic “memories” of their tissues of origin (25). In our studies, although only a few DMRs did not revert, these irreversible epigenetic marks may provide a basis for which even the same hESC line can have unique growth and differentiation properties across different laboratories (5, 15). Extending this idea, it may be of particular significance that a few DMRs were also located at or near developmental genes. With the exception of mir-503 and mir-424, all DMRs at these genes were reversible. However, even with these changeable methylation marks, the influence they may have on induced differentiation and lineage propensity is unknown and the potential consequences on cell differentiation via altered DNA methylation at lineage switches is currently being investigated in our laboratory.
By cycling culture environments and tracking corresponding DNA methylation changes, we identified a correlation between DNA methylation changes in promoter-distal regions and gene expression. Noting that the classical inverse association of promoter DNA methylation and gene expression is more specifically constrained to regions closest to the transcriptions start site (Fig. 4B and Fig. S3 A–C) (22), we considered the possibility of a divergent role for DNA methylation outside of promoter-proximal regions. Genomewide, a positive trend toward promoter-distal DNA methylation and gene expression was noticeably less clear than the inverse correlation of promoter-proximal DNA methylation; however, in examining promoter-distal DMR–associated gene-expression changes, we see significant down-regulation of genes undergoing loss of DNA methylation (Fig. 4D). These observations are well in line with the ideas of Lister et al. (26), in providing evidence that the positive correlation between gene body methylation and gene expression often seen may more accurately be described as the loss of DNA methylation and corresponding gene expression brought about by differentiation. It is also known that in response to methyltransferase-inhibiting drugs, promoter hypomethylation causes both gene activation and gene silencing (27). When taken together, the role of DNA methylation in hESCs continues to be difficult to precisely identify.
Last, we see that multiple hESC culture adaptations may have unintended consequences. Although DNA methylation appears predominately reversible, we observed several hundred transcripts as irreversible or differentially expressed only in reverse-adapted cells. Reverse adapted cells have clearly adopted expression and methylation profiles unique from the original MEF/DF12 hESC cultures. For expression changes, gene ontology analysis indicates a stress response and potentially oncogenic pathways. Concerns for cell-transplant therapy are obvious. These altered transcriptional states are not necessarily due to DNA methylation changes, but it was intriguing that multiple DMRs were observed in a handful of important G-protein signal-regulating genes. Among them we selected GPSM3 for knockdown analysis and partially recovered a reverse-adapted hESC transcriptional profile despite conducting the knockdown on Mat/mTR (Fig. 5 C and D). Although the GPSM3 promoter-distal DMR and corresponding gene expression is reversible (Fig. 5 A and B), the complexities of assembling G-protein-coupled receptors (GPCRs) and effectors subsequent to culture associated amendments to transcription and translation could provide a basis by which a change in GPSM3 expression can have particularly potent effects on genomewide transcription (18–20). At this time, given the proprietary nature of some mTesR1 and STEMPRO media components and the presence of unknown and variably secreted factors from feeder cells, it is unclear which ligands may be directly acting on GPCRs involving GPSM3 regulation. What is clearer is that hESCs in unique environments have different adhesive, metabolic, and proliferative characteristics. Therefore, the inter- and intracellular signaling networks are expected to also be distinct, and when subjected to an expression change for a key signal regulator, whether epigenetically controlled or through knockdown, the downstream effect is likely to include wide-ranging transcriptional changes.
Materials and Methods
Cell Culture.
MEF/DMEM F12 KSR (MEF/DF12) cultures.
HES-2 (ES02) human embryonic stem cells were grown on mitotically inactive MEF feeders in accordance with WiCell standard operating procedures. Cells were maintained in medium as described by WiCell, but with no serum supplement.
Mat/mTR cultures and CS/SP cultures.
HES-2 cells were adapted from MEF/DF12 conditions to matrigel substrate (BD Biosciences) and mTesR1 medium (STEMCELL Technologies), or separately to CELLStart substrate and STEMPRO medium (Invitrogen). Cultures were maintained in accordance to manufacturers’ protocols. An overview of culture-environment adaptation is provided in Fig. 1A. Complete sample details can be found in Table S1.
MIRA Chip, Gene Expression, and Gene Ontology Analysis.
Methylated DNA was enriched using the MethylCollector Ultra Kit (Active Motif), which is based on MIRA (28). Amplified input and methyl-enriched DNA was cohybridized on NimbleGen CpG Island Plus RefSeq Promoter Arrays. For gene expression, total RNA was extracted for hybridization to NimbleGen 12 × 135 k human gene-expression arrays (Roche NimbleGen). Microarrays were processed at the University of California, Los Angeles (UCLA) DNA Microarray Core. Informatics approaches for identifying methylated regions, DMRs, and differential gene expression are described in SI Materials and Methods. Details on validating microarray results are also provided in the supplement and can be seen in Fig. 3B and Figs. S1 and S2 B and C. The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 using functional annotation was used for gene ontology analysis (29)
SI Materials and Methods include additional details on study design and materials.
Supplementary Material
Acknowledgments
We thank Xinmin Li and Jian Zhou for their excellent microarray processing at UCLA’s DNA Microarray Core. We also acknowledge the Analytical Cytometry and Bioinformatics Core for their assistance in cell sorting and statistical analysis, respectively.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209620109/-/DCSupplemental.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Riggs AD. DNA methylation and cell memory. Cell Biophys. 1989;15:1–13. doi: 10.1007/BF02991574. [DOI] [PubMed] [Google Scholar]
- 3.Riggs AD, Xiong Z. Methylation and epigenetic fidelity. Proc Natl Acad Sci USA. 2004;101:4–5. doi: 10.1073/pnas.0307781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell Res. 2011;21:1649–1651. doi: 10.1038/cr.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akopian V, et al. International Stem Cell Initiative Consortium Comparison of defined culture systems for feeder cell free propagation of human embryonic stem cells. In Vitro Cell Dev Biol Anim. 2010;46:247–258. doi: 10.1007/s11626-010-9297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallon BS, Park KY, Chen KG, Hamilton RS, McKay RD. Toward xeno-free culture of human embryonic stem cells. Int J Biochem Cell Biol. 2006;38:1063–1075. doi: 10.1016/j.biocel.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 8.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci USA. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon TM, et al. Human embryonic stem cells (hESCs) cultured under distinctive feeder-free culture conditions display global gene expression patterns similar to hESCs from feeder-dependent culture conditions. Stem Cell Rev. 2010;6:425–437. doi: 10.1007/s12015-010-9158-x. [DOI] [PubMed] [Google Scholar]
- 10.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allegrucci C, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- 12.Teichroeb JH, Betts DH, Vaziri H. Suppression of the imprinted gene NNAT and X-chromosome gene activation in isogenic human iPS cells. PLoS ONE. 2011;6:e23436. doi: 10.1371/journal.pone.0023436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito T, et al. Escape from X inactivation of two new genes associated with DXS6974E and DXS7020E. Genomics. 1997;43:183–190. doi: 10.1006/geno.1997.4797. [DOI] [PubMed] [Google Scholar]
- 14.Hall LL, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chitalia VC, et al. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol. 2008;10:1208–1216. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forrest AR, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 18.Dupré DJ, Robitaille M, Rebois RV, Hébert TE. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu Rev Pharmacol Toxicol. 2009;49:31–56. doi: 10.1146/annurev-pharmtox-061008-103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giguère PM, Laroche G, Oestreich EA, Siderovski DP. G-protein signaling modulator-3 regulates heterotrimeric G-protein dynamics through dual association with Gβ and Gαi protein subunits. J Biol Chem. 2012;287:4863–4874. doi: 10.1074/jbc.M111.311712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao P, Nguyen CH, Chidiac P. The proline-rich N-terminal domain of G18 exhibits a novel G protein regulatory function. J Biol Chem. 2010;285:9008–9017. doi: 10.1074/jbc.M109.057174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 22.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakaue M, et al. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- 24.Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pompeia C, et al. Microarray analysis of epigenetic silencing of gene expression in the KAS-6/1 multiple myeloma cell line. Cancer Res. 2004;64:3465–3473. doi: 10.1158/0008-5472.CAN-03-3970. [DOI] [PubMed] [Google Scholar]
- 28.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 29.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.