Abstract
Chronic sleep deprivation perturbs the circadian clock and increases susceptibility to diseases such as diabetes, obesity, and cancer. Increased inflammation is one of the common underlying mechanisms of these diseases, thus raising a hypothesis that circadian-oscillator components may regulate immune response. Here we show that absence of the core clock component protein cryptochrome (CRY) leads to constitutive elevation of proinflammatory cytokines in a cell-autonomous manner. We observed a constitutive NF–κB and protein kinase A (PKA) signaling activation in Cry1−/−;Cry2−/− cells. We further demonstrate that increased phosphorylation of p65 at S276 residue in Cry1−/−;Cry2−/− cells is due to increased PKA signaling activity, likely induced by a significantly high basal level of cAMP, which we detected in these cells. In addition, we report that CRY1 binds to adenylyl cyclase and limits cAMP production. Based on these data, we propose that absence of CRY protein(s) might release its (their) inhibition on cAMP production, resulting in elevated cAMP and increased PKA activation, subsequently leading to NF–κB activation through phosphorylation of p65 at S276. These results offer a mechanistic framework for understanding the link between circadian rhythm disruption and increased susceptibility to chronic inflammatory diseases.
Keywords: biological clock, immune system
In response to the rotation of the earth, many of the mammalian physiological processes such as sleep–wake cycle and feeding pattern undergo an ∼24-h oscillation referred as the circadian clock. Circadian oscillation helps organisms anticipate and adapt to predictable daily changes in the environment. The hypothalamic suprachiasmatic nucleus (SCN) functions as a master circadian oscillator, which controls behaviors. At molecular level, the circadian oscillator is based on a cell-autonomous transcription–translation feedback loop, in which transcriptional activators circadian locomotor output cycles kaput (CLOCK)/brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like (BMAL)1 activate the expression of Cryptochrome (Cry) and Period (Per), in turn their protein products repress BMAL1 and CLOCK activity, thus producing Cry and Per expression rhythms (1, 2). Reverse orientation c-erb (REV-ERB)s and RAR-related orphan receptor (ROR)s also participate in the rhythmic transcriptional activity of the molecular oscillator (3). Molecular components of the circadian clock network are conserved in the SCN and also in peripheral cells. The oscillator uses direct and indirect mechanisms to impose rhythms and regulate several physiological processes, such as rest–activity cycle, endocrine system, and metabolism (4).
Sleep loss or chronic sleep deprivation disrupts the circadian rhythm. It has been reported that people exposed to constant circadian disruption, such as shift-workers, show increased incidence of chronic diseases such as diabetes, obesity, and also cancer (5–7). Chronic inflammation is one of the important pathogenic features of these diseases, which prompted us to hypothesize that circadian clock components may play a role in the regulation of inflammation. Moreover, recent evidence demonstrated that major players in immune function, such as spleen, lymph node, and macrophage and natural killer (NK) cells, contain a cell-autonomous circadian clock (8–10). Moreover, secretion of cytokines, TNF-α and IL-6 has been reported to display circadian oscillation in macrophages, where ∼8% of transcriptome is under circadian regulation (10). Clinical evidence and sleep-loss studies have identified physiological connections between the circadian clock and immune system (11, 12). A molecular understanding of the mechanism of clock modulation of immune function, however, remains unclear.
In this study, we found that lack of a functional clock system due to absence of the core clock component cryptochrome (CRY) leads to elevated proinflammatory cytokine activation mediated through the NF–κB signaling pathway. We also found that absence of CRY proteins leads to constitutive activation of PKA, which results in phosphorylation of p65 at S276, which ultimately lead to increased NF–κB activation and expression of proinflammatory cytokines. Our results provide a possible molecular link between the arrhythmic clock system and increased inflammatory response.
Results
Absence of Cryptochrome Proteins Constitutively Activates the Proinflammatory Cytokine Expression.
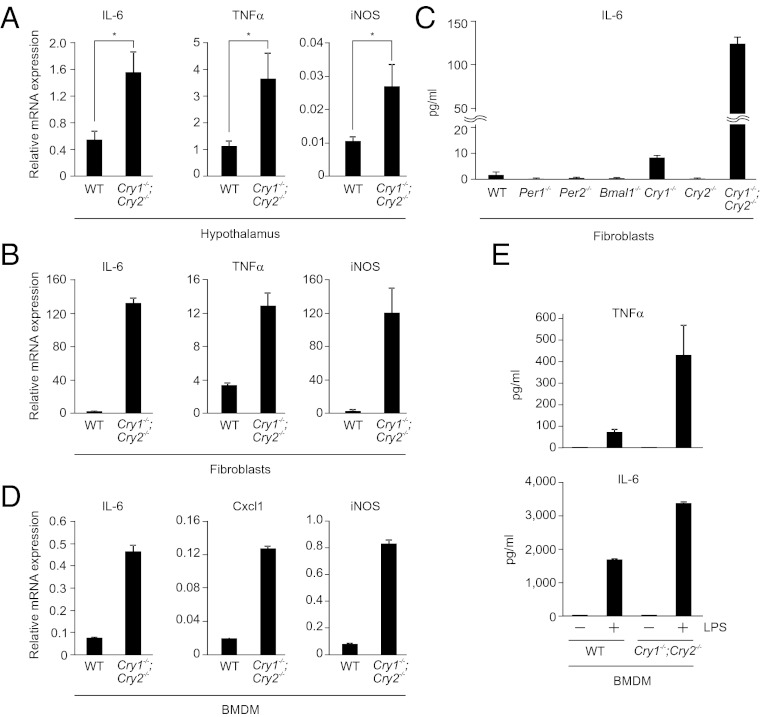
To gain insight into the role of circadian clock components on the immune function, we measured expression of inflammatory mediators in the hypothalamus of Cry1 and Cry2 double knockout (Cry1−/−;Cry2−/−) mice, which show arrhythmic behavior due to lack of a functional clock system (13, 14). Significant increase in expression of IL-6, TNF-α, and iNOS was observed in Cry1−/−;Cry2−/− mice relative to WT mice (Fig. 1A). We next tested whether the increased expression is cell-autonomous or systemic result of oscillator dysfunction. Fibroblasts possess a cell-autonomous and self-sustained circadian clock system (15, 16) that recapitulates key features of whole organism’s circadian function. Cry1−/−;Cry2−/− fibroblasts, which lack circadian oscillation (17), exhibited significantly increased expression of IL-6, TNF-α, and iNOS (Fig. 1B) and other genes involved in inflammation (Fig. S1A), thus exhibiting cell-autonomous roles of CRY proteins in regulation of key cytokines. Compared with Cry1−/− or Cry2−/− single knockout fibroblasts, cytokine expression showed a synergistic increase in double knockout Cry1−/−;Cry2−/− fibroblasts (Fig. S1B). IL-6 protein was detectable only in Cry-deficient fibroblasts, but not in other clock mutants that lack either Per1 or Per2 or Bmal1 (Fig. 1C). These results demonstrate that lack of CRY constitutively activates the proinflammatory cytokines, such as IL-6, TNF-α, and other inflammatory molecules, indicating a potential role for CRY proteins in regulation of inflammatory cytokine expression. Because IL-6 is inducible with TNF-α, we next asked if the high expression of IL-6 observed in Cry1−/−;Cry2−/− cells can be further induced by TNF-α. Both WT and Cry1−/−;Cry2−/− cells showed ∼8- to 10-fold increase of IL-6 expression following TNF-α induction (Fig. S1C), suggesting that both cells are responsive for an inflammatory stimuli.
Fig. 1.
Lack of a functional circadian clock system by absence of the circadian oscillator component CRY constitutively upregulates expression of inflammatory cytokines. (A) Estimation of mRNA levels of indicated genes by quantitative real-time PCR (qRT–PCR) in the hypothalamus region of brain of WT and Cry1−/−;Cry2−/− mice are shown. Data are mean ± SEM (n = 4 mice per group); *P < 0.05. (B) Quantification of mRNA levels of indicated genes by qRT–PCR in WT and Cry1−/−;Cry2−/− fibroblasts are shown. Data are mean ± SD (n = 3). (C) WT, Per1−/−, Per2−/−, Bmal1−/−, Cry1−/−, Cry2−/−, and Cry1−/−;Cry2−/− fibroblasts were left untreated for 12 h and the supernatants were analyzed for IL-6 protein levels by ELISA. Data are mean ± SD (n = 3). (D) BMDM from WT and Cry1−/−;Cry2−/− mice were analyzed for mRNA levels of indicated genes by qRT–PCR. Data are mean ± SD (n = 3). (E) WT and Cry1−/−;Cry2−/− BMDM cells were either untreated or treated with LPS (1 μg/mL for TNF-α and 10 ng/mL for IL-6 analysis) for 12 h and the supernatants were estimated for TNF-α or IL-6 protein levels by ELISA are shown. Data are mean ± SD (n = 4). Relative mRNA expression levels after normalization to actin are shown in A, B, and D.
Innate Immune System Gets Hypersensitive in the Absence of CRY.
Macrophages play a critical role in secretion of most inflammatory cytokines upon immune response. It has recently been shown that the expression of cytokines in macrophages is circadian clock–gated (10). Therefore, we next analyzed bone marrow–derived macrophages (BMDM) from WT and Cry1−/−;Cry2−/− mice for expression of immune mediators. BMDM from Cry1−/−;Cry2−/− mice showed a marked increase in expression of inflammatory cytokines such as IL-6, Cxcl1, and iNOS, compared with WT cells (Fig. 1D). Analysis of proinflammatory cytokines TNF-α and IL-6 by ELISA in the cell supernatants did not show any significant difference between WT and Cry1−/−;Cry2−/− BMDM cells (Fig. 1E), which prompted us to check whether Cry1−/−;Cry2−/− BMDM cells are hypersensitive to LPS stimulation. WT and Cry1−/−;Cry2−/− BMDM cells were treated with LPS and supernatants were collected after 12 h. ELISA analysis showed a marked increase in TNFα and IL-6 protein secretions in Cry1−/−;Cry2−/− BMDM cells compared with WT (Fig. 1E), which suggests that Cry-deficient macrophages are hypersensitive to immune response.
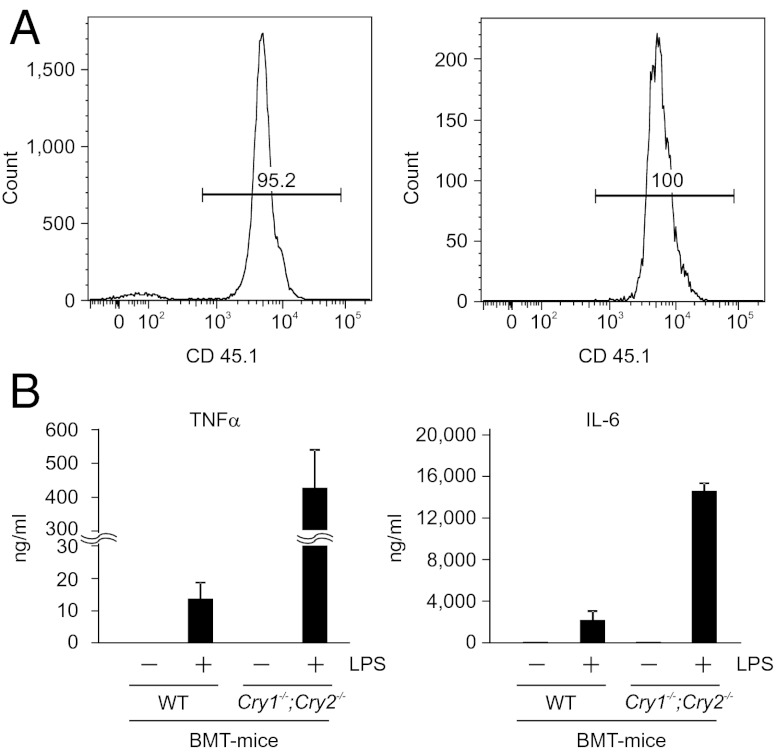
To determine the contribution of the hematopoietic cells from WT and Cry1−/−;Cry2−/− mice in vivo in systemic cytokine production during inflammatory response, we performed bone marrow transplantation (BMT) experiments. Bone marrow from either WT or Cry1−/−;Cry2−/− mice were transplanted and reconstituted into lethally irradiated recipient mice (B6.129S4-Il2rgtm1Wjl/J; Jackson Lab), which lack T, B, and NK cells and the macrophages in blood and spleen. Transplanted mice were allowed to repopulate for 8 wk and then the blood lymphocytes of these mice were analyzed by FACS for repopulation efficiency. FACS analysis showed that nearly 95% of lymphocytes in the transplanted mice were derived from donor mice, and in particular almost 100% of macrophages were derived from donor cells in transplanted mice (Fig. 2A). Furthermore, donor-derived T cells, B cells, monocytes, and macrophages were efficiently repopulated in both WT and Cry1−/−;Cry2−/− bone marrow–transplanted mice (Fig. S2 A–D). Eight weeks after transplantation, mice transplanted with either WT (BMT-WT) or Cry1−/−;Cry2−/− (BMT-KO) bone marrow were injected with PBS or LPS and serum samples of TNF-α and IL-6 protein were measured 2 h later. LPS-injected BMT-KO mice showed a significantly high level of both TNF-α and IL-6 (Fig. 2B) in their serum relative to LPS-injected BMT-WT mice. These results clearly indicate that absence of CRY proteins potentiates the immune system and significantly elevates secretion of proinflammatory cytokines in a physiological system upon LPS challenge.
Fig. 2.
Absence of cryptochrome potentiate immune system of the mice to secrete increased levels of inflammatory cytokines upon LPS challenge. (A) Reconstitution analysis by FACS of blood lymphocytes derived from recipient mice, B6.129S4-Il2rgtm1Wjl/J (CD45.2 positive), 8 wk after injection with bone marrow of donor mice (CD45.1 positive) of one representative animal. Left shows typical donor contribution to total lymphocytes. Right shows the percentage of CD45.1 positive cells (donor) gated on CD11b positive cells. (B) WT or Cry1−/−;Cry2−/− BMT were injected with either PBS or LPS (5 mg/kg) and the 2-h serum samples collected from these mice were analyzed for TNF-α and IL-6 protein levels by ELISA. Data are mean ± SEM (n = 3 mice per group for LPS injection and n = 1 for PBS injection).
NF–κB Signaling Pathway Is Constantly Activated in Cry1−/−;Cry2−/− Cells.
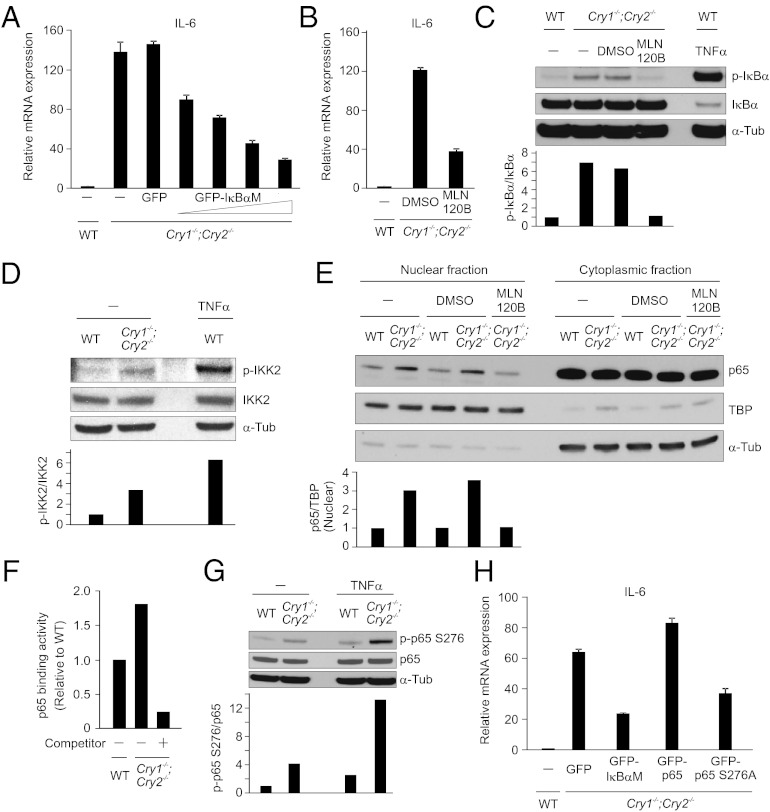
Next, we sought to identify the signaling pathway that is responsible for constitutive cytokine activation in Cry1−/−;Cry2−/− cells. Although CRY proteins are known to inhibit the transcription from CLOCK/BMAL1 target genes, IL-6 promoter is not known to be a CLOCK/BMAL1 target (18). Because NF–κB signaling is one of the major regulators and integrators of the inflammatory response in mammals (19, 20), we tested whether blocking of NF–κB signaling can suppress the constitutive IL-6 activation in Cry1−/−;Cry2−/− cells. In canonical NF–κB signaling, inducers such as TNF-α or LPS trigger inhibitor of nuclear factor-κB (IκB) kinase 2 (IKK2) activation, leading to phosphorylation, ubiquitination, and proteasomal degradation of IκBα, allowing the transport of NF–κB into the nucleus to activate its target genes (21). Cry1−/−;Cry2−/− cells were infected with either lentiviral GFP or lentiviral-GFP-tagged IκBα superrepressor (IκBαM), which is a nondegradable form of IκBα. Expression of a dominant negative form of IκBα, IκBαM led to significant reduction of IL-6 expression in Cry1−/−;Cry2−/− cells (Fig. 3A), which suggests that constitutive activation of the NF–κB signaling pathway likely underlies constitutive IL-6 activation in Cry1−/−;Cry2−/− cells. We next wondered whether IKK2 is also involved in constitutive NF–κB activation in Cry1−/−;Cry2−/− cells, because in most NF–κB signaling, IKK2 is necessary and sufficient for phosphorylation of IκBα and its degradation (21). Inhibition of IKK2 function by a chemical inhibitor MLN120B (10 μM) (22) showed a marked reduction of constitutive IL-6 expression in Cry1−/−;Cry2−/− (Fig. 3B), further suggesting that the constitutive activation of NF–κB pathway in Cry1−/−;Cry2−/− cells most likely signals, at least partly, through the canonical IKK2 kinase complex.
Fig. 3.
Blocking the canonical NF–κB pathway suppresses constitutive activation of IL-6 in Cry1−/−;Cry2−/− fibroblasts. (A) Estimation of IL-6 mRNA by qRT–PCR of Cry1−/−;Cry2−/− fibroblasts infected with lentivirus expressing GFP- or GFP-tagged superrepressor mutant of IκBα (GFP-IκBαM) with increasing dose are shown. (B) IL-6 mRNA expression was quantified by qRT–PCR of Cry1−/−;Cry2−/− fibroblasts treated with either DMSO or MLN120B (10 μM) for 2 h. (C) Total cell lysates of WT and Cry1−/−;Cry2−/− fibroblasts either untreated or treated with DMSO or with the IKK2 inhibitor, MLN 120B (10 μM, 1 h) were analyzed by Western blot for phospho-IκBα, IκBα, and tubulin. (D) Total cell lysates of WT and Cry1−/−;Cry2−/− fibroblasts analyzed by Western blot for phospho-IKK2, IKK2, and tubulin are shown. (E) Nuclear and cytoplasmic fractions of untreated or DMSO-treated WT and untreated or DMSO- or MLN120B-treated Cry1−/−;Cry2−/− fibroblasts analyzed by Western blot for p65, TATA-binding protein (TBP) and tubulin are shown. Detection of TBP and tubulin are used as markers of nuclear and cytoplasmic fractions, respectively. (F) Nuclear extracts (5 μg) from WT and Cry1−/−;Cry2−/− fibroblasts analyzed for p65 binding activity by ELISA are shown. Competition assay was performed with wild-type NF–κB consensus oligonucleotides. Values shown are relative to WT, value of WT p65 binding activity is set to 1, which is a representative of three independent experiments. (G) Western blot show the level phospho-p65 S276 in total cell lysates of WT and Cry1−/−;Cry2−/− fibroblasts either untreated or treated with TNF-α (10 ng/mL, 30 min). (H) Estimation of IL-6 mRNA levels by qRT–PCR of Cry1−/−;Cry2−/− fibroblasts expressing either GFP or superrepressor form of IκBα (GFP-IκBαM) or GFP-p65 or mutant form of p65 (GFP-p65 S276A) are shown. Relative mRNA expression levels after normalization to actin are shown in A, B, and H, and data are mean ± SD (n = 3).
The rate-limiting step in NF–κB signaling is phosphorylation of IκBα and its subsequent degradation (21). We assessed the phosphorylation status of IκBα in Cry1−/−;Cry2−/− cells. Compared with WT cells, we observed a markedly increased phospho-IκBα in Cry1−/−;Cry2−/− cells, which is substantially reduced by inhibition of IKK2 with MLN120B (Fig. 3C). This result is further supported by the high level of phosphorylated and active IKK2 in Cry1−/−;Cry2−/− cells compared with WT cells (Fig. 3D). Because increased IκBα degradation will lead to increased entry of p65 into the nucleus (21), we compared the nuclear p65 levels in WT and Cry1−/−;Cry2−/− cells. Western blot analysis after nucleocytoplasmic subcellular fractionation of WT and Cry1−/−;Cry2−/− cells showed a substantially elevated level of nuclear p65 in Cry1−/−;Cry2−/− cells, which is significantly reduced by inhibition of IKK2 with MLN120B (Fig. 3E).
Upon entering into the nucleus, transcription factor p65 binds to NF–κB binding sites to activate its target genes (21). We therefore analyzed the p65 binding activity in the nuclear lysates assessed by affinity to oligonucleotides containing a consensus NF–κB binding sequence. This assay showed a good functional efficiency and specificity for p65 binding in fibroblasts (Fig. S3). We observed a marked increase in p65-binding activity in Cry1−/−;Cry2−/− fibroblasts compared with WT cells, and this increased activity is dramatically reduced upon addition of competitor oligonucleotides (Fig. 3F). It has been reported that p65 is phosphorylated at S276 residue for its subsequent acetylation and increase of its transcriptional activity (23). We found a significant elevation of phospho-p65 at S276 in Cry1−/−;Cry2−/− cells compared with WT cells (Fig. 3G). Moreover, expression of a mutant form of p65 (p65 S276A) significantly suppressed IL-6 activation in Cry1−/−;Cry2−/− cells (Fig. 3H). Together, these results suggest that the increased phosphorylation of both IκBα and p65 at S276 may play an important role in the constitutive activation of IL-6 transcription in Cry1−/−;Cry2−/− cells.
PKA Signaling Activity Is Constitutive in Cry1−/−;Cry2−/− Cells.
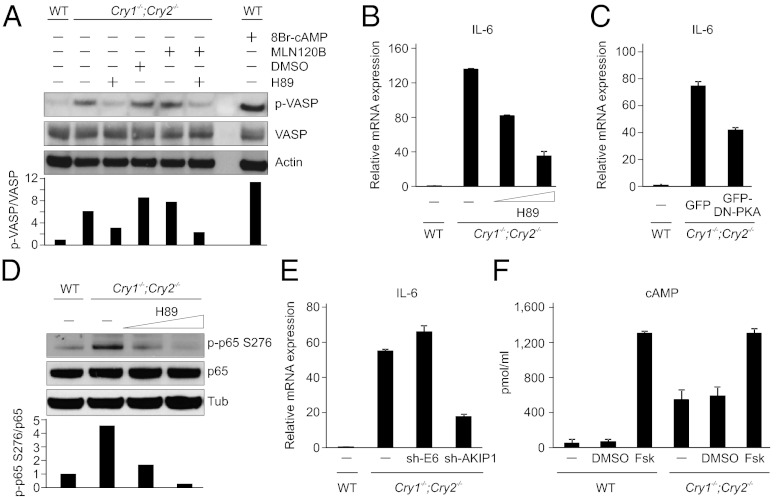
Protein kinase A (PKA) is involved in the phosphorylation of p65 at S276 (24). Owing to the significantly elevated phosphorylation level of p65 at S276 (Fig. 3G), we speculated that PKA signaling activity may also be high in Cry1−/−;Cry2−/− cells. We assessed the phosphorylation status of a known substrate of PKA, vasodialator-stimulated phosphoprotein (VASP) (25) and detected its increased phosphorylation in Cry1−/−;Cry2−/− cells (Fig. 4A). We also observed a near complete suppression of VASP phosphorylation in Cry1−/−;Cry2−/− cells by a PKA inhibitor H89 (20 μM), but not with the IKK2 inhibitor MLN120B (Fig. 4A), which suggests that PKA signaling is constitutively activated in Cry1−/−;Cry2−/− cells.
Fig. 4.
Increased cAMP and PKA signaling activity and also the PKA-mediated phosphorylation of p65 at S276 contributes to elevated NF–κB target gene activation in Cry1−/−;Cry2−/−. (A) WT fibroblasts were either untreated or treated with 8Br–cAMP (100 μM) for 30 min and Cry1−/−;Cry2−/− fibroblasts were either untreated or treated with H89 (20 μM), DMSO, or MLN120B (10 μM) for 1 h, and total cell lysates were analyzed by Western blot for phospho-VASP, VASP and actin. (B) Quantification of IL-6 mRNA expression by qRT–PCR of Cry1−/−;Cry2−/− fibroblasts treated with H89 (10 and 20 μM for 1 h). (C) Estimation of IL-6 mRNA expression by qRT–PCR of Cry1−/−;Cry2−/− fibroblasts expressed with a dominant–negative (GFP–DN–PKA) form of PKA. (D) Total cell lysates of untreated WT and untreated or H89 (20 and 30 μM, 1 h) treated Cry1−/−;Cry2−/− fibroblasts were analyzed by Western blot for phospho-p65 S276, p65 and tubulin. (E) RNA extracted from Cry1−/−;Cry2−/− fibroblasts infected with lentivirus expressing sh-RNA against a nonspecific target gene E6 as a control or against AKIP1 were analyzed for IL-6 mRNA expression by qRT–PCR. (F) Cellular concentration of cAMP measured by ELISA in the lysates of WT and Cry1−/−;Cry2−/− fibroblasts either untreated or treated with DMSO or forskolin (10 μM, 30 min) are shown. Data are mean ± SD (n = 4). Relative mRNA expression levels after normalization to actin are shown in B, C, and E, and data are mean ± SD (n = 3).
We next investigated whether inhibition of PKA signaling can suppress IL-6 activation detected in Cry1−/−;Cry2−/− cells. IL-6 mRNA activation was remarkably reduced when Cry1−/−;Cry2−/− cells were treated with increasing concentrations of the PKA inhibitor H89 (Fig. 4B). Consistent with this result with inhibitor, a marked reduction of IL-6 activation in Cry1−/−;Cry2−/− cells was also observed by expression of a mutant form of PKA (GFP-DN PKA) (Fig. 4C), which acts as a dominant negative protein by blocking the dissociation of regulatory subunit of PKA from its catalytic subunit (26). Inhibition of PKA signaling also resulted in a marked reduction in phospho-p65 at S276 in Cry1−/−;Cry2−/− cells (Fig. 4D).
A nuclear protein, A-kinase-interacting protein 1 (AKIP1), binds both to PKA (27) and p65 (28). This interaction has been suggested to enhance PKA-mediated transcriptional activity of p65 by promoting its nuclear retention and phosphorylation at S276, which potentially increases recruitment of transcriptional coactivators such as p300 to p65 (28). Given the constitutive activation of NF–κB and PKA signaling in Cry1−/−;Cry2−/− cells (Figs. 3 and 4 A–D), we tested whether knockdown of AKIP1 can suppress the constitutive activation of IL-6 in Cry1−/−;Cry2−/− cells. RNAi-mediated depletion of AKIP1 reduced IL-6 activation in Cry1−/−;Cry2−/− cells (Fig. 4E and Fig. S4). Thus, our data supports the notion that NF–κB activation through PKA contributes to the constitutive activation of IL-6 in Cry1−/−;Cry2−/− cells.
Rise in intracellular levels of cAMP leads to PKA activation (29). We compared cAMP concentration in WT and Cry1−/−;Cry2−/− cells before and following induction with forskolin, which triggers cAMP production by activating adenylyl cyclase (30). WT and Cry1−/−;Cry2−/− cells were first incubated with 3-Isobutyl-1-methylxanthine (IBMX) (500 μM), an inhibitor of phosphodiesterase, for 30 min, followed by addition of forskolin (10 μM) for 30 min, and cAMP levels were quantified. As anticipated, cAMP levels are elevated in WT and Cry1−/−;Cry2−/− cells upon forskolin treatment (Fig. 4F). However, surprisingly, we observed substantially high levels of cAMP even in uninduced state (control and DMSO treated) of Cry1−/−;Cry2−/− cells compared with WT cells, suggesting that the increase in PKA signaling activation in Cry1−/−;Cry2−/− cells is most likely due to high cAMP concentration at the basal level.
CRY1 Expression Limits cAMP Production Likely by Binding to and Inhibiting the Function of Adenylyl Cyclase.
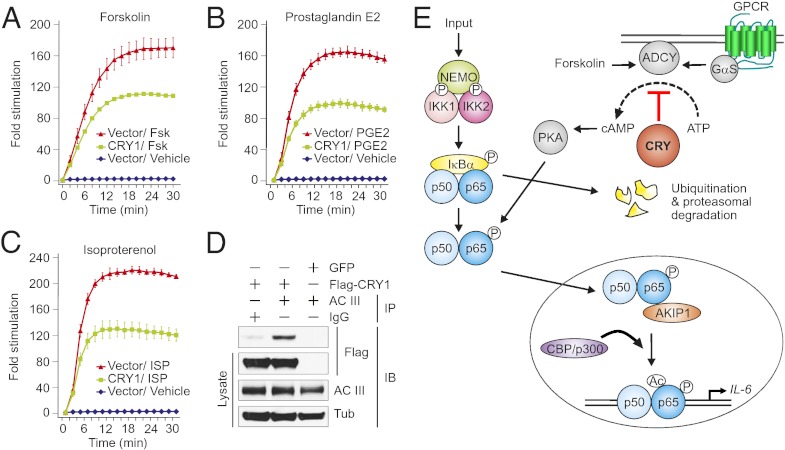
On the basis of above results, we hypothesized that CRY regulates the intracellular cAMP levels to regulate NF–κB pathway. We addressed whether overexpression of CRY1 protein can suppress the cAMP production induced by either forskolin, prostaglandin E2 (PGE2), or isoproterenol (each 10 μM). Stimulation with forskolin directly activates adenylyl cyclase (30); whereas, induction with PGE2 and isoproterenol activates their cognate Gαs–coupled receptors, which ultimately activate adenylyl cyclase leading to transient increase in intracellular cAMP (31, 32). Dynamic changes in the intracellular cAMP levels were measured by a cAMP-sensitive luciferase reporter system (GloSensor cAMP assay; Promega) expressed in 293T cells. Overexpression of CRY1 resulted in a significant reduction in cAMP production induced by forskolin, PGE2, and also by isoproterenol (Fig. 5 A–C). Because we observed a substantial suppression in cAMP production induced by all three stimuli tested, we hypothesize that CRY1 may acts on adenylyl cyclase to block its function, thereby suppressing cAMP production. To test this proposal, lysates of 293T cells overexpressing Flag-tagged CRY1 or GFP were analyzed by immunoprecipitation (IP). In 293T cell lysates, expressing Flag-CRY1 when adenylyl cyclase was immunoprecipitated with an antibody against adenylyl cyclase III (AC III), we detected Flag-CRY1 by immune blot (Fig. 5D). These results indicate that CRY1 bind to adenylyl cyclase to block its function and suppress the cAMP production.
Fig. 5.
CRY1 inhibits the forskolin-, PGE2-, and isoproterenol-induced generation of intracellular cAMP, likely by binding to and inhibiting the function of adenylyl cyclase. (A–C) Luciferase assay performed to measure the kinetics of cAMP production after stimulation in the presence of 500 μM of IBMX in 293T cells are shown. Fold stimulation of cAMP production are shown after stimulation with either forskolin, PGE2, or isoproterenol (each 10 μM) in the absence (control vector) or presence of CRY1 expression. Data are mean ± SD (n = 3). (D) CRY1 interacts with adenylyl cyclase. All of the indicated immunoprecipitate and lysate samples analyzed by Western blot for Flag tag (CRY1), adenylyl cyclase III and tubulin are shown. (E) The proposed model showing the likely function of CRY in binding to and suppressing the action of adenylyl cyclase in cAMP production and the pathways activated in the absence of CRY proteins.
Discussion
In summary, results presented here indicate that both PKA and IKK2–IκBα-dependent phosphorylation of NF–κB contributes to increased activation of inflammatory cytokines, for example IL-6, in Cry1−/−;Cry2−/− fibroblasts. Here, we propose that circadian clock modulates NF–κB activity via CRY-mediated control of PKA activity. Circadian clock–controlled expression of CRY proteins regulate the production of cAMP, which in turn contributes to the observed circadian rhythms in cytosolic cAMP, which is a necessary component to core clock function in SCN neurons and in peripheral cell types (33). We presume that temporally gated oscillations of cAMP levels (33) and resultant PKA activity temporally modulates the basal activation state of the NF–κB pathway and may explain the well-described circadian rhythms in immune responses in mammals (34). Our data support the hypothesis that when CRY proteins are present they bind to adenylyl cyclase (Fig. 5D) and downregulate cAMP production (Fig. 5 A–C). In the absence of CRY, inhibition on adenylyl cyclase is relieved, which subsequently leads to increased cAMP production (Fig. 4F) and PKA activation (Fig. 4A) that cause increased phosphorylation of p65 at S276 (Fig. 4D), ultimately resulting in activation of its target genes such as IL-6 (Fig. 5E).
We observed increased activation of TNF-α in Cry1−/−;Cry2−/− fibroblasts and also in the hypothalamus of Cry1−/−;Cry2−/− mice. Hashiramoto et al. has also previously shown high expression of TNF-α in Cry1−/−;Cry2−/− fibroblasts (35). We also observed increased phospho-IKK2 in Cry1−/−;Cry2−/− fibroblasts (Fig. 3D). We put forward that the most likely initiator of phosphorylation and activation of IKK2 signaling complex in Cry1−/−;Cry2−/− cells is the increased activation of TNF-α (a well-known activator of IKK2 complex), which has been observed in these cells. This activated IKK2 complex subsequently leads to phosphorylation (Fig. 3C) and degradation of IκBα, which in turn cause increased p65 accumulation into the nucleus (Fig. 3E) and high level of DNA binding activity (Fig. 3F), ultimately resulting in increased expression of NF–κB target genes such as IL-6. Phosphorylation and degradation of IκBα is a critical step required to release p65 and PKA from IκBα, which allows PKA to phosphorylate p65 (36) and enter into nucleus, which is why overexpression of superrepressor mutant form of IκBα (IκBαM) dramatically suppressed up to 80% of IL-6 expression in Cry1−/−;Cry2−/− fibroblasts (Fig. 3A).
Recent reports have demonstrated the role of CRY in regulation of metabolism and immune function. CRY proteins are known to regulate glucose homeostasis by at least two different mechanisms. They bind to and repress glucocorticoid receptors (37), which regulate transcription of gluconeogenic genes. CRY proteins also bind to and inhibit Gαs function, thereby attenuating GPCR-activation-dependent increase in intracellular cAMP (38). However, we detected CRY1 in complex with adenylyl cyclase (Fig. 5D), and overexpression of CRY1 reduced cAMP production in response to PGE2, isoproterenol, and even the direct adenylyl cyclase activator, forskolin (30) (Fig. 5A). These results clearly suggest an additional cell-autonomous role of CRY in modulating cellular response to different stimuli, which contributes to our understanding of altered immune response under circadian disruption. It has previously been shown that p53 mutant mice lacking functional CRY proteins are more susceptible to TNF-α-initiated apoptosis through NF–κB activation (39). Monje et al. had reported that depression like behavior in mice induced by constant darkness is associated with increased levels of IL-6 in plasma and hippocampus mediated through NF–κB signaling pathway (40). Interestingly, NF–κB has been shown to play a role in metabolic adaptation both in normal and also in cancer cells by regulation of mitochondrial oxidative phosphorylation (41), and the current finding will suggest an additional CRY–NF–κB-mediated pathway in metabolic adaptation of cancer cells.
In this study, we analyzed the inflammatory response of mammalian cells only lacking a functional circadian clock system, without any additional manipulations. Our results strongly indicate that an arrhythmic clock system, induced by the absence of CRY proteins, alone is sufficient to increase the stress levels of cells leading to constant expression of inflammatory cytokines and causing a low-grade chronic inflammatory status called metaflammation (42) and Cry-deficient cells could be a good model system to study this phenomenon. Compelling evidence has shown that low-grade constant inflammation could be the underlying cause for chronic diseases such as diabetes, obesity, and also cancer (43–45). Moreover, people exposed to frequent circadian clock disturbances, such as night-shift workers, are increasingly susceptible to these diseases. Therefore, further understanding connection between the circadian clock, metabolism, and inflammation will help to identify therapeutic drug targets to cure diseases like metabolic syndrome and cancer.
Materials and Methods
All animal experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Salk Institute. Male C57BL/6J and Cry1−/−;Cry2−/− mice were used (12 wk old). TRIzol (Invitrogen) reagent was used to extract RNA from animal tissues by following the manufacturer’s protocol. Immortalized fibroblasts and 293T cells were cultured in Dulbecco-modified Eagle medium (Invitrogen) supplemented with 10% (vol/vol) FBS (Atlanta Biologicals) and 1% (vol/vol) of antibiotic–antimycotic (Invitrogen). The production of lentiviruses was performed according to the protocol described elsewhere (46). Additional details are in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (NIH), Ipsen/Biomeasure, Sanofi-aventis, and the H. N. and Frances C. Berger Foundation. The project described was supported by Grant R37AI048034 from the National Institute of Allergy and Infectious Diseases. R.N. was supported in part by a grant from the Swiss National Science Foundation (Bern, Switzerland). S.P. was supported by NIH Grant DK091618; M.H. was supported by Japan Society for Promotion of Science Fellowship. S.K.N. was supported by NIH Grant P30 CA014195-38. I.M.V. is an American Cancer Society Professor of Molecular Biology, and holds the Irwin and Joan Jacobs Chair in Exemplary Life Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209965109/-/DCSupplemental.
References
- 1.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- 3.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saini C, Suter DM, Liani A, Gos P, Schibler U. The mammalian circadian timing system: Synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol. 2011;76:39–47. doi: 10.1101/sqb.2011.76.010918. [DOI] [PubMed] [Google Scholar]
- 5.Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10:199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajaratnam SM, Arendt J. Health in a 24-h society. Lancet. 2001;358:999–1005. doi: 10.1016/S0140-6736(01)06108-6. [DOI] [PubMed] [Google Scholar]
- 7.Willyard C. Hungry for sleep. Nat Med. 2008;14:477–480. doi: 10.1038/nm0508-477. [DOI] [PubMed] [Google Scholar]
- 8.Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174:7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30:621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- 10.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass J, Turek FW. Sleepless in America: A pathway to obesity and the metabolic syndrome? Arch Intern Med. 2005;165:15–16. doi: 10.1001/archinte.165.1.15. [DOI] [PubMed] [Google Scholar]
- 12.Bollinger T, Bollinger A, Oster H, Solbach W. Sleep, immunity, and circadian clocks: A mechanistic model. Gerontology. 2010;56:574–580. doi: 10.1159/000281827. [DOI] [PubMed] [Google Scholar]
- 13.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 14.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 18.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 20.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 21.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Hideshima T, et al. MLN120B, a novel IkappaB kinase beta inhibitor, blocks multiple myeloma cell growth in vitro and in vivo. Clin Cancer Res. 2006;12:5887–5894. doi: 10.1158/1078-0432.CCR-05-2501. [DOI] [PubMed] [Google Scholar]
- 23.Chen LF, et al. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 25.Butt E, et al. cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J Biol Chem. 1994;269:14509–14517. [PubMed] [Google Scholar]
- 26.Clegg CH, Correll LA, Cadd GG, McKnight GS. Inhibition of intracellular cAMP-dependent protein kinase using mutant genes of the regulatory type I subunit. J Biol Chem. 1987;262:13111–13119. [PubMed] [Google Scholar]
- 27.Sastri M, Barraclough DM, Carmichael PT, Taylor SS. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci USA. 2005;102:349–354. doi: 10.1073/pnas.0408608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao N, Asamitsu K, Hibi Y, Ueno T, Okamoto T. AKIP1 enhances NF-kappaB-dependent gene expression by promoting the nuclear retention and phosphorylation of p65. J Biol Chem. 2008;283:7834–7843. doi: 10.1074/jbc.M710285200. [DOI] [PubMed] [Google Scholar]
- 29.Taylor SS, et al. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Seamon KB, Padgett W, Daly JW. Forskolin: Unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc Natl Acad Sci USA. 1981;78:3363–3367. doi: 10.1073/pnas.78.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furuyashiki T, Narumiya S. Stress responses: The contribution of prostaglandin E(2) and its receptors. Nat Rev Endocrinol. 2011;7:163–175. doi: 10.1038/nrendo.2010.194. [DOI] [PubMed] [Google Scholar]
- 32.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrushesky WJ, Langevin T, Kim YJ, Wood PA. Circadian dynamics of tumor necrosis factor alpha (cachectin) lethality. J Exp Med. 1994;180:1059–1065. doi: 10.1084/jem.180.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashiramoto A, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol. 2010;184:1560–1565. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 36.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 37.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Sancar A. Regulation of apoptosis by the circadian clock through NF-kappaB signaling. Proc Natl Acad Sci USA. 2011;108:12036–12041. doi: 10.1073/pnas.1108125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monje FJ, et al. Constant darkness induces IL-6-dependent depression-like behavior through the NF-kappaB signaling pathway. J Neurosci. 2011;31:9075–9083. doi: 10.1523/JNEUROSCI.1537-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauro C, et al. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13:1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 43.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 44.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 45.Sun B, Karin M. Obesity, inflammation, and liver cancer. J Hepatol. 2012;56:704–713. doi: 10.1016/j.jhep.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.