Abstract
Hepatocytes generated from human induced pluripotent stem cells (hiPSCs) are unprecedented resources for pharmaceuticals and cell therapy. However, the in vitro directed differentiation of human pluripotent stem cells into mature hepatocytes remains challenging. Little attention has so far been paid to variations among hiPSC lines in terms of their hepatic differentiation. In the current study, we developed an improved hepatic differentiation protocol and compared 28 hiPSC lines originated from various somatic cells and derived using retroviruses, Sendai viruses, or episomal plasmids. This comparison indicated that the origins, but not the derivation methods, may be a major determinant of variation in hepatic differentiation. The hiPSC clones derived from peripheral blood cells consistently showed good differentiation efficiency, whereas many hiPSC clones from adult dermal fibroblasts showed poor differentiation. However, when we compared hiPSCs from peripheral blood and dermal fibroblasts from the same individuals, we found that variations in hepatic differentiation were largely attributable to donor differences, rather than to the types of the original cells. These data underscore the importance of donor differences when comparing the differentiation propensities of hiPSC clones.
Keywords: endodermal differentiated hiPSCs, DNA methylation, regenerative medicine
Human induced pluripotent stem cells (hiPSCs) are generated from somatic cells through the ectopic expression of defined factors (1). HiPSCs are similar to human embryonic stem cells (hESCs) in terms of their infinite proliferation in vitro and pluripotency and the ability to differentiate into cells of all three germ layers (2). In addition, hiPSCs can be generated from individual donors with known characteristics. Therefore, hiPSCs are expected to be promising materials for cell therapy, drug development, and studies on pathogenesis.
Hepatocytes are promising target cells for medical applications of hiPSCs. Hepatocytes derived from genetically matched hiPSCs might make it possible to overcome interindividual differences in drug metabolism, which are a major cause of unpredictable side effects in the pharmacological field, and also enable cell transplantation therapy for metabolic or life-threatening liver diseases without the need for intensive immunosuppressive treatment.
Several multistep hepatic differentiation protocols using hESCs have been reported, mimicking the hepatogenesis process that occurs during development (3–6). In these protocols, endodermal cells are initially induced by a high concentration of activin A, followed by hepatoblast and hepatocyte differentiation. Similar multistep protocols have been applied to hiPSCs (7–9). The phenotypes of inherited metabolic liver diseases were partially recapitulated in hepatic cells differentiated from patient-derived hiPSCs (10). Recently, functional hepatocytes were efficiently generated from hES/iPSCs by adenovirus vector-mediated overexpression of liver-related transcription factors (11). However, the hepatic cells differentiated from hES/iPSCs only partially mimic the authentic hepatocytes in terms of their global gene expression patterns and functions.
To achieve robust hepatic differentiation from hES/iPSCs, the quality of the stem cells used is at least as important as the differentiation procedures. Variations among hESCs in their differentiation propensity toward certain lineages have been reported (12, 13). This diversity may be attributable to the genetic backgrounds of the donor cells, as well as to different culture conditions. Variations among iPSCs may be even greater, because their quality can also be affected by variations in the technology used for iPSC generation, such as the types of original cells, reprogramming factors used, and factor delivery methods (14, 15). In particular, iPSCs may retain epigenetic memories of the donor cells that skew their differentiation tendency toward the original cell type (16–18). It remains to be determined which of these factors play major roles in determining the propensity of hiPSCs for hepatic differentiation.
In the current study, we developed a modified protocol that efficiently and stably generated endodermal cells from hES/iPSCs. We then examined the propensity for hepatic differentiation of various hiPSC clones, including those from different tissue origins from the same donors, as well as hESC clones. Our data indicated that donor differences are an important determinant of the propensity for hepatic differentiation.
Results
Variable Hepatic Differentiation Among hiPS/ESC Clones.
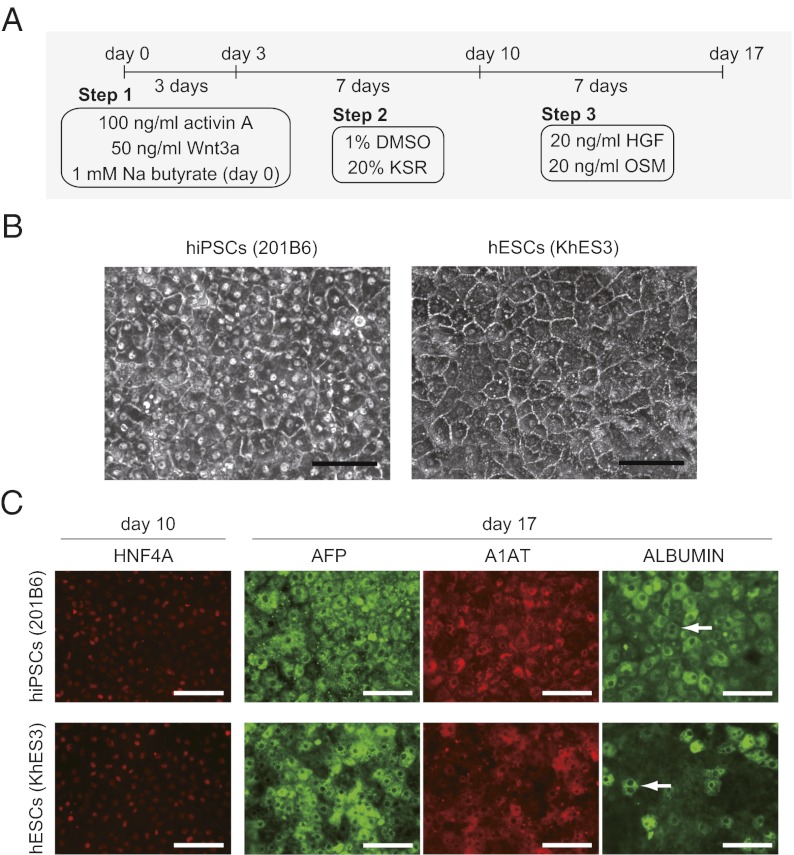
To generate hepatic differentiated cells from hiPS/ESCs, we used a previously reported differentiation protocol designed for hESCs (5), with minor modifications (Fig. 1A). We investigated five “sibling” hiPSC clones that had been generated from the same adult human dermal fibroblasts (aHDFs), as well as two hESC clones. Three hiPSC clones (201B2, -B6, and -B7) were generated by retroviral transduction of four reprogramming factors (OCT3/4, SOX2, KLF4, and c-MYC) (1), whereas two clones (253G1 and -G4) were generated using three factors (devoid of c-MYC) (19). Two hESC lines (KhES1 and -3) were previously established in Japan (20).
Fig. 1.
Hepatic differentiation of representative hiPS/ESC clones. (A) Schematic representation of the directed hepatic differentiation protocol used in this study. (B) Morphology of hepatic differentiated hiPS/ESCs on day 17. (Scale bar: 100 μm.) (C) Immunostaining analysis of hepatic differentiated hiPS/ESCs (day 10, HNF4A; day 17, AFP, A1AT, and ALBUMIN). (Scale bar: 100 μm.) Arrows indicate mature hepatic binuclear cells.
After a 17-d differentiation period, definite areas of hepatocyte-like cell morphology, characterized by large cuboidal cells with prominent compact nuclei, were observed in three clones (201B2, 201B6, and KhES3) (Fig. 1B). They also expressed albumin (Fig. 2A) and liver-related genes, including HNF4A, A1AT, AFP, ALBUMIN, TDO2, and ASGR1 (Fig. 2B). In contrast, the remaining clones (201B7, 253G1, 253G4, and KhES1) showed only a few hepatocyte-like cell clusters. They expressed lower levels of albumin and liver-related genes (Fig. 2 A and B). The marker of undifferentiated cells, OCT3/4, was down-regulated in all clones (Fig. 2B).
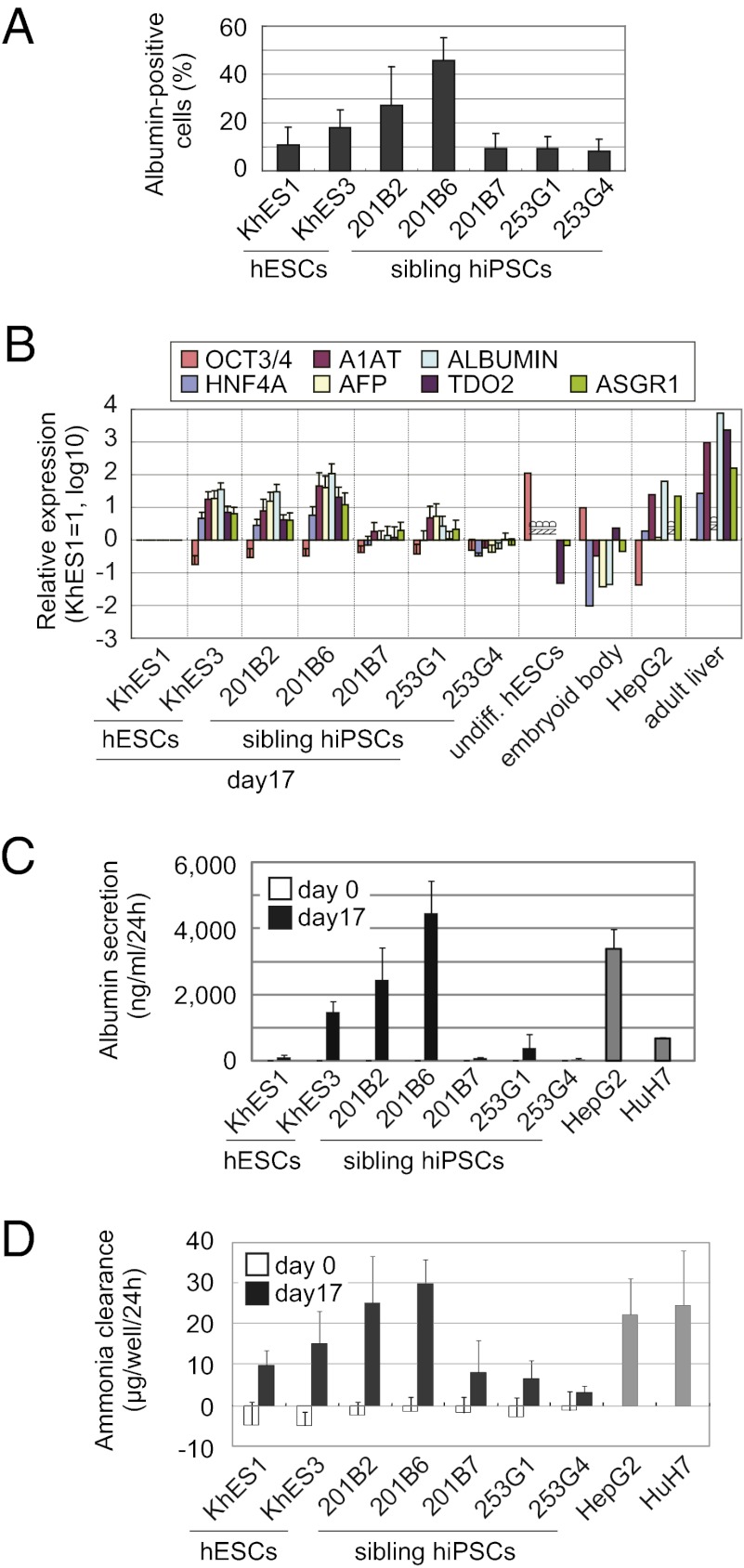
Fig. 2.
Diverse hepatic differentiation among hiPS/ESC clones. (A) Percentage of albumin-positive cells detected by a flow cytometric analysis after 17 d of hepatic differentiation. (B) Real-time PCR analysis of liver-related and undifferentiated gene expression in the hepatic differentiated hiPS/ESCs on day 17. The graph represents the fold expression of genes relative to KhES1 on day 17. ND, not determined. (C) Albumin secretion potential of the undifferentiated (day 0) and hepatic differentiated hiPS/ESCs on day 17 as determined by an ELISA. (D) Comparison of the ammonia clearance activity of the undifferentiated (day 0) and hepatic differentiated hiPS/ESCs on day 17. Error bars indicate the SD (n = 3).
We further characterized the hepatic cells derived from hiPSCs (201B6) and hESCs (KhES3). They expressed various CYP450 mRNAs, such as CYP1A1, CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP7A1. They also expressed mature hepatocyte markers, including ABCC2 (MRP2), ABCB11 (MDR/TAP), and UGT1A1 (Fig. S1A). Immunofluorescent staining also confirmed that these cells expressed liver-related markers, such as HNF4A on day 10 and AFP, A1AT, and ALBUMIN on day 17 (Fig. 1C). Some binuclear cells indicating a mature hepatic phenotype were also found in those clones. The cells were also positive for periodic acid-Shiff (PAS) staining (Fig. S1B). The albumin secretion level in the culture media, ammonia clearance capacity, and CYP3A4 activity were comparable to those obtained from HepG2 and HuH7, human hepatoma cell lines (Fig. 2 C and D and Fig. S1C). Taken together, these findings confirm that hiPSCs and hESCs can differentiate into the hepatic lineage, but that the differentiation propensity varies significantly among clones.
Modified Protocol Allowing for Effective Endodermal Differentiation.
We hypothesized that the variable hepatic differentiation may be attributable to our protocol, in which differentiation medium is added to undifferentiated hiPS/ESC clusters. Because we cannot control sizes of the cell clusters, the cell-to-cell interactions and cellular responses to growth factors supplemented in the differentiation medium may be skewed, impairing the reproducibility among experiments. To overcome this problem, we modified the protocol so that we could start with hiPS/ESCs dissociated into single cells by Accutase (Fig. 3A). To support the survival of cells, a ROCK inhibitor (Y27632) was added to the culture medium (21). We also decreased the concentration of sodium butyrate (NaB) from 1 to 0.5 mM to enhance cell survival.
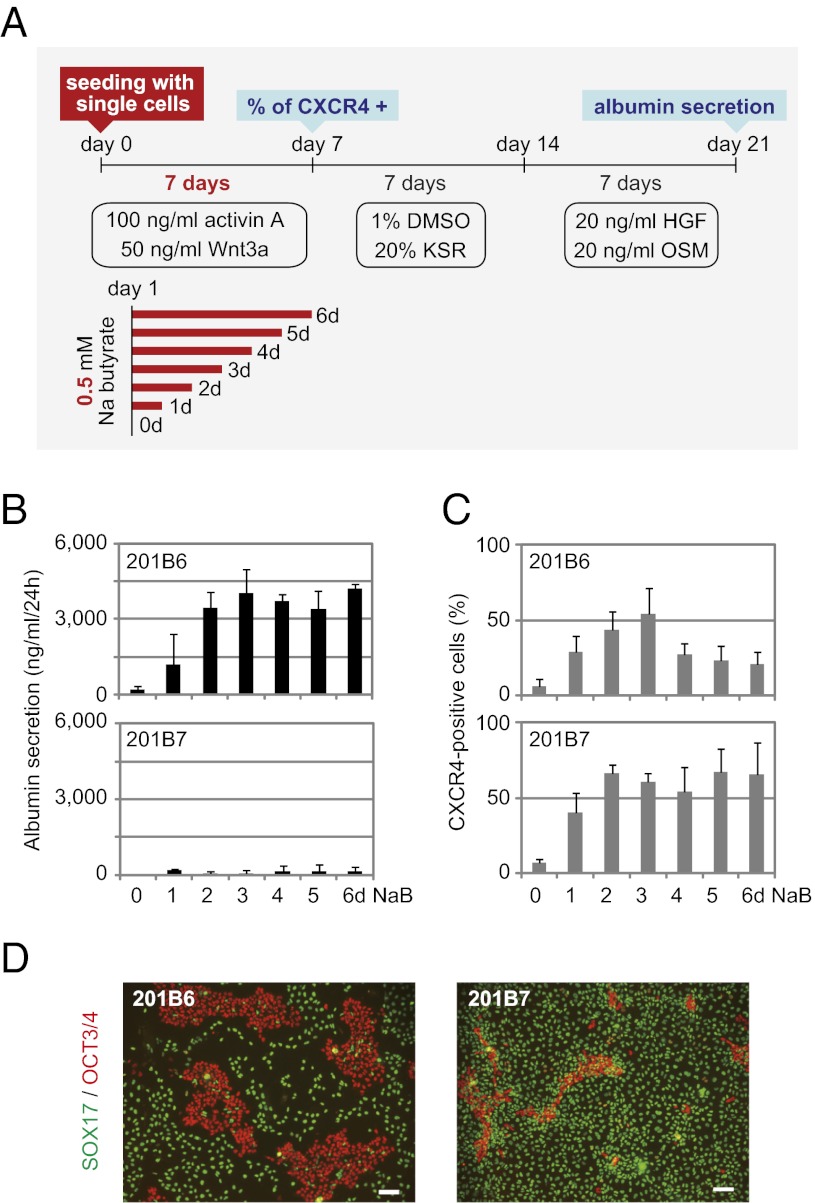
Fig. 3.
Close comparison of the propensity for hepatic differentiation between sibling hiPSC lines 201B6 and 201B7. (A) Schematic representation of the modified protocol used for hepatic differentiation. In this protocol, hiPS/ESCs were enzymatically digested into single cells and plated on Matrigel-coated dishes. For endodermal cell induction, the cells were cultivated with activin A and Wnt3a for 7 d. A total of 0.5 mM sodium butyrate (NaB) was supplemented from day 1 for various durations (0–6 d). (B) Albumin secretion level after 21 d of hepatic differentiation. The error bars indicate the SD (n = 3). (C) Percentage of CXCR4-positive cells after 7 d of endodermal differentiation as determined by flow cytometry. Error bars indicate the SD (n = 3). (D) Immunostaining of SOX17 and OCT3/4 after 7 d of endodermal differentiation. NaB was added for 3 d. (Scale bar: 100 μm.)
We applied the modified procedure to the sibling hiPS clones, 201B6 and 201B7, which represent clones with good and poor hepatic differentiation, respectively. With this modified protocol, more albumin-positive hepatic cells were observed in derivatives of clone 201B6 than in those of clone 201B7 at the end of the differentiation protocol (day 21) (Fig. S2). The albumin secretion level of clone 201B6 on day 21 was also much higher than that of clone 201B7 for any administration period of NaB (Fig. 3B), indicating that clone 201B6 favors differentiation into the hepatic lineage, whereas clone 201B7 is refractory to hepatic differentiation.
However, in contrast to the results regarding final hepatic differentiation, 201B7 hiPSCs differentiated into CXCR4-positive endodermal cells (3, 22) with >50% efficacy, which was comparable to or even better than that observed using clone 201B6 on day 7 (Fig. 3C). An immunostaining analysis performed on day 7 also detected many cells from both 201B7 and 201B6 clones that were positive for SOX17, an endoderm marker (Fig. 3D).
The modified protocol allowed both 201B6 and 201B7 clones to effectively differentiate into CXCR4- and Sox17-positive endodermal cells, but only the former can further differentiate into hepatic lineage cells.
Efficient Hepatic Differentiation from Blood-Derived hiPSCs.
With the modified protocol allowing efficient differentiation into CXCR4-positive cells, we compared the endodermal and hepatic differentiation propensities among various hiPSC lines derived from different origins and using different methods. We examined hiPSC lines derived from aHDFs (aHDF-iPSCs), dental pulp cells (DP-iPSCs), peripheral blood cells (PB-iPSCs), and cord blood cells (CB-iPSCs). We also tested seven hESC lines. Detailed cell information and passage numbers in this experiment are indicated in Table S1. We used the albumin secretion levels into the culture media on day 21 as a representative marker to monitor the hepatic differentiation, on the basis of the results described in Fig. 2. We thought that the NaB requirement might be different for various clones (Fig. 3 B and C), so we applied 0.5 mM NaB for three different durations during the first 7 d of endodermal differentiation (0 d, no administration; 3 d, NaB treatment from day 1 to day 3; or 6 d, treatment from day 1 to day 6).
PB-iPSCs and CB-iPSCs showed efficient generation of CXCR4-positive cells on day 7, although NaB administration caused significant cell death in some clones. aHDF-iPSCs and hESCs exhibited marked variations in their propensity to differentiate into CXCR4-positive cells. DP-iPSC lines did not respond to this differentiation protocol due to poor cell growth or cell death and failed to generate CXCR4-positive cells on day 7 (Fig. 4A). On day 21, the PB-iPSCs showed significantly higher levels of albumin secretion than did the aHDF-iPSC, DP-iPSC, and hESC lines (Fig. 4 B and C), suggesting that peripheral blood cells can be suitable donor cell candidates for generating hiPSCs for hepatocyte differentiation.
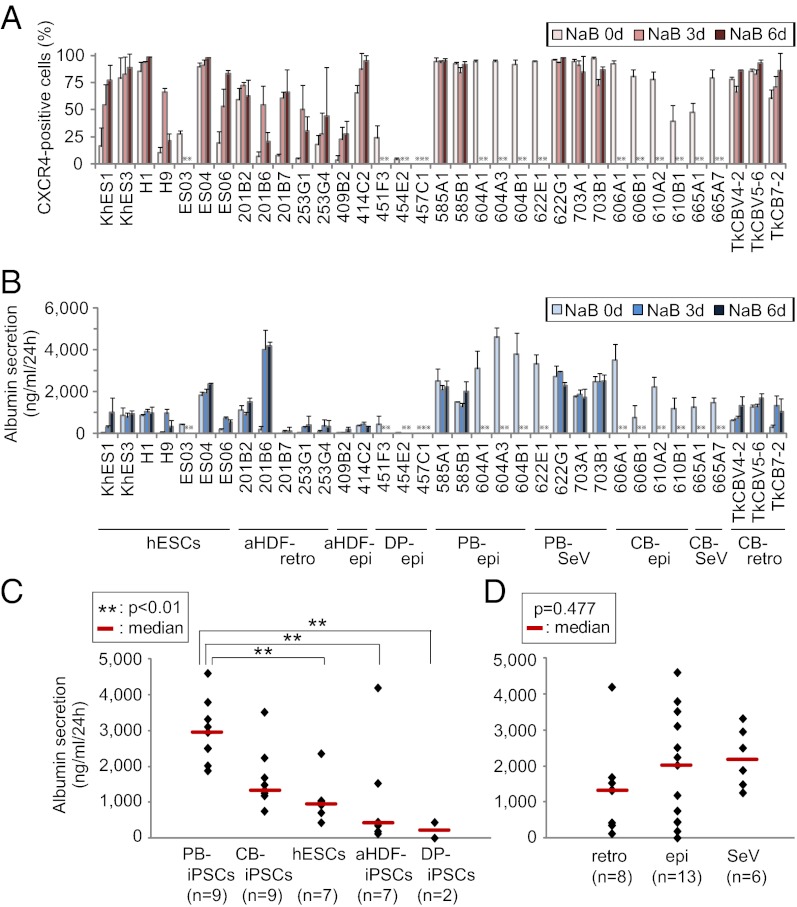
Fig. 4.
Comparison of the propensities for endodermal and hepatic differentiation among various hiPS/ESC lines. (A) Percentage of CXCR4-positive cells on day 7 was determined by flow cytometric analysis. (B) Albumin secretion potential of the hepatic differentiated hiPS/ESCs on day 21 was determined by an ELISA. For each clone, data were obtained after different durations of NaB administration (0, 3, and 6 d during the first 7 d of endodermal differentiation; x axis). Error bars indicate the SD (n = 3). epi, episomal vector; retro, retrovirus vector; SeV, Sendai virus vector.  : Data were not obtained due to significant cell death or poor cell growth. (C and D) Statistical analysis of the differences in the albumin secretion potential of hepatic differentiated hiPS/ESCs on day 21. For each clone, the highest level among the different NaB administration periods (0, 3, or 6 d) was used for the analysis. HiPSCs were grouped by the original cell types (C) and derivation methods (D).
: Data were not obtained due to significant cell death or poor cell growth. (C and D) Statistical analysis of the differences in the albumin secretion potential of hepatic differentiated hiPS/ESCs on day 21. For each clone, the highest level among the different NaB administration periods (0, 3, or 6 d) was used for the analysis. HiPSCs were grouped by the original cell types (C) and derivation methods (D).
PB-iPSCs, as well as aHDF-iPSCs and CB-iPSCs, were generated by three methods, including retrovirus vectors (1), Sendai virus vectors (23), and episomal plasmid vectors (24). However, we did not find any differences in the propensity for hepatic differentiation among the hiPSCs derived using different induction methods (Fig. 4D).
Donor-Dependent Variations in Hepatic Differentiation.
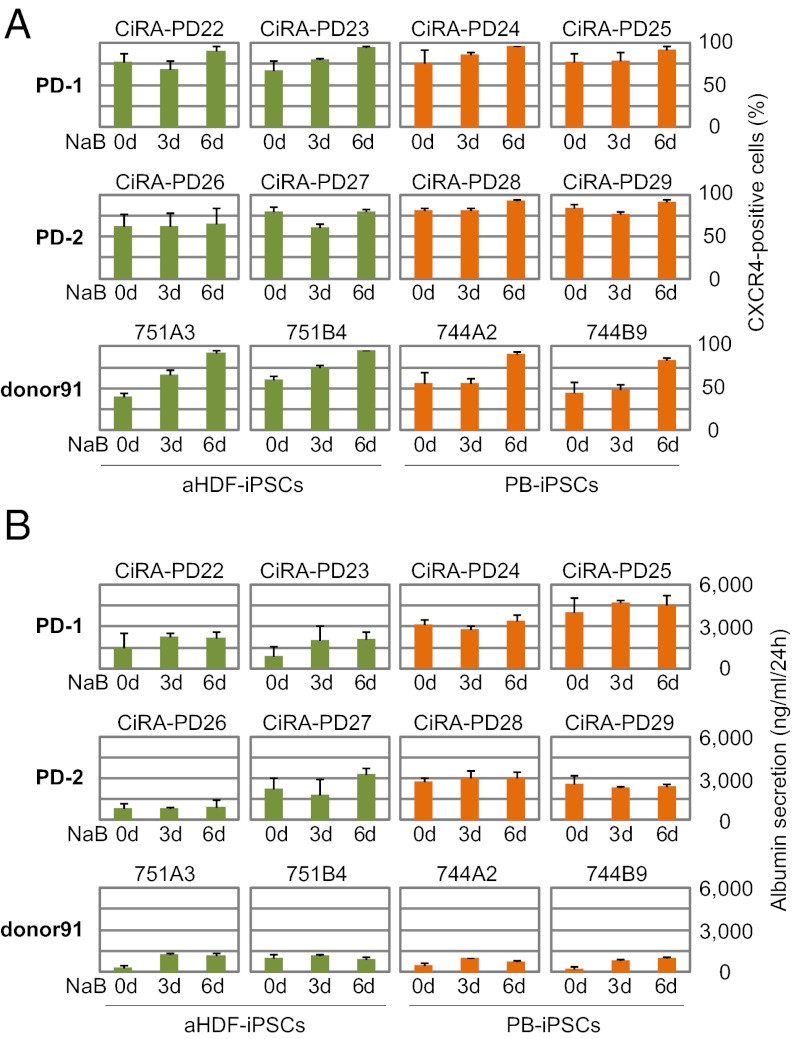
In the previous experiments, all aHDF-iPSC clones were derived from a single donor, whereas the PB-iPSC clones were derived from two donors, who were different from the fibroblast donor (Table S1). Thus, the observed differences in the propensities for hepatic differentiation may be attributable to the differences in donors, rather than to the differences in the types of somatic cells. To investigate this possibility, we directly compared the propensity for hepatic differentiation of aHDF-iPSCs and PB-iPSCs derived from the same individuals, two Parkinson disease patients (PD-1 and PD-2) and one healthy adult individual (donor91), using episomal plasmids. We analyzed two aHDF-iPSC clones and two PB-iPSC clones from each donor (Table S1). Again, we applied 0.5 mM NaB for three different durations during the first 7 d of endodermal differentiation (0 d, no administration; 3 d, NaB treatment from day 1 to day 3; or 6 d, treatment from day 1 to day 6).
The modified procedure effectively induced the differentiation of all hiPSC clones into CXCR4-positive cells by day 7, with at least 50% efficacy (Fig. 5A). On day 21, the PB-iPSCs tended to have higher albumin secretion than did the aHDF-iPSCs in the PD clones (Fig. 5B). In the donor91 clones, both aHDF-iPSCs and PB-iPSCs showed low but similar albumin secretion potential. These data suggest that, although the original cell types from which the hiPSCs are derived may influence the propensity of the cells for hepatic differentiation, the donor specificity may have a stronger impact on the hepatic differentiation. The NaB requirement was closely consistent within each individual, thereby supporting the importance of the donor-specific influence.
Fig. 5.
Comparison of the propensities of hiPSCs from the same individuals for endodermal and hepatic differentiation. Two aHDF-iPSC and PB-iPSC clones derived from the same individual (two Parkinson disease patients, PD-1 and PD-2, and one adult healthy donor, donor91) were differentiated into the hepatic lineage. (A) Percentage of CXCR4-positive cells on day 7 as determined by flow cytometric analysis. (B) Albumin secretion of hepatic differentiated hiPSCs on day 21 was analyzed by an ELISA. For each clone, the data were obtained for different NaB administration periods (0, 3, and 6 d during the first 7 d of endodermal differentiation; x axis). The green and orange bars indicate aHDF-iPSCs and PB-iPSCs, respectively. Error bars indicate the SD (n = 3).
Gene Expression or DNA Methylation Cannot Predict the Propensity for Hepatic Differentiation.
We then tried to understand the molecular mechanisms underlying the observed variations in hepatic differentiation. We first examined the global gene expression profile of sibling hiPSC clones 201B6 and 201B7 (derived from the same donor) by microarray analyses. These two clones effectively differentiated into CXCR4-positive cells, but only 201B6 hiPSCs were able to differentiate into hepatic cells. The two clones showed similar global expression patterns in the undifferentiated state (Fig. S3A). Surprisingly, the CXCR4-positive cells of these sibling hiPSC clones sorted by flow cytometry on day 7 were also very similar in terms of their global gene expression (Fig. S3B), despite the marked difference in their propensities for hepatic differentiation. In particular, the expression levels of 10 transcription factors related to liver development [HNF1A, HNF1B, FOXA2 (HNF3B), HNF4A, HNF6, SOX17, GATA4, GATA6, HHEX, and CEBPA] (25, 26) were comparable between the two hiPSC clones (Fig. S3 A and B).
We also examined the global gene expression patters of the undifferentiated aHDF-iPSC and PB-iPSC clones from the same individuals in Fig. 5. Again, we did not observe any correlation between the gene expression patterns and the propensities for hepatic differentiation (Fig. 5 and Fig. S4 A and B). Moreover, a similar gene expression pattern was observed in both the intraindividual comparisons (Fig. S4C) and the interindividual comparisons in PB-iPSCs (Fig. S4D).
We next examined the DNA methylation status of the promoter regions of the 10 liver-related transcription factors in five sibling hiPSC clones (201B2, 201B6, 201B7, 253G1, and 253G4) and four hESC clones (KhES1, KhES3, H1, and H9). A pyrosequencing analysis of undifferentiated cells showed no significant differences among the sibling hiPSC clones with various propensities for hepatic differentiation. The promoter regions of HNF1A and HNF4A were highly methylated, whereas the promoters for the other 8 liver-related transcription factors were unmethylated in all of the hiPS/ESCs (Fig. S5). Next, we analyzed the DNA methylation status of the 10 liver-related transcription factors in CXCR4-positive cells derived from clones 201B6 and 201B7 on day 7. Again, we did not find any significant differences between these clones, even within the CXCR4-positive cell populations (Fig. S6).
Discussion
In this report, we observed marked differences in the propensity for hepatic differentiation among hiPSC lines generated from various origins and using various methods. Our results suggest that the genetic background of individual donors has a strong impact on the hepatic differentiation of hiPSC clones. In addition, most PB-iPSC lines we tested showed favorable results in terms of their hepatic differentiation. In contrast, the methods used to generate hiPSCs did not show a significant impact on hepatic differentiation.
In previous studies that compared the differentiation propensities of hiPSC clones from different origins, the genetic backgrounds of the donors were not considered (18, 27). In these studies, one type of somatic cell was generally obtained from companies or repositories, and another type of somatic cell was obtained from a different source. Therefore, the observed differences in these analyses may have been attributable to different donors, rather than to the different original somatic cells.
In fact, in our own analyses, we initially concluded that PB-iPSC clones were much better than aHDF-iPSC clones in terms of their hepatic differentiation, on the basis of the comparison between hiPSC clones derived from a single purchased aHDF line and those from PB samples from two Japanese donors. However, when we compared PB-iPSCs and aHDF-iPSCs from the same donors, we found that the differences in the hepatic differentiation between PB-iPSCs and aHDF-iPSCs were small. Rather, the variations in hepatic differentiation were largely attributable to differences in the donors.
In two mouse studies (16, 17) and one human study (28), iPSC clones were generated from different types of cells from single donors. These studies showed that iPSCs at early passages retained epigenetic memories of the original cells and thus could efficiently redifferentiate back into the same lineage. However, at late passages, these epigenetic memories seem to be lost. These studies did not focus on the genetic backgrounds of different donors and thus did not show their impact on the differentiation propensities. As most of our experiments were performed using hiPSCs with passage numbers 20–30, it is unlikely that tissue-specific donor memory has significant impact on hepatic differentiation.
These previous studies also showed that the epigenetic memory skewed the ability of iPSCs to differentiate into cell lineages different from the original cell type. For example, Kim et al. (18) showed that hiPSC clones from blood cells have limited abilities to differentiate into keratinocytes. Our results, however, showed that most of the PB-iPSC clones effectively differentiated into hepatocyte-like cells, which have a different origin from that of blood cells.
The mechanisms underlying the propensity for efficient hepatic differentiation remain unclear. PB-iPSCs and aHDF-iPSCs from the same individuals were randomly located in a hierarchical clustering analysis of expression array data. In addition, the global gene expression patterns in sibling hiPS clones, 201B6 and 201B7, which showed opposing hepatic differentiation potentials, were similar not only in the undifferentiated state, but also in CXCR4-positive cell populations. We also investigated whether the epigenetic modification pattern of clone 201B6 resembled that of human liver. However, the targeted analyses of DNA methylation did not show any resemblance between clone 201B6 and adult liver. It is still possible that the diversity in the propensity for hepatic differentiation is due to differences in histone modifications, which still need to be evaluated.
We found that differences in the propensity for hepatic differentiation are largely derived from the process of changing from CXCR4-positive cells to hepatic cells. The earlier process from pluripotent cells to endodermal cells was comparable among most hiPSC clones. Thus, most hiPSC clones seem to have relatively uniform responsiveness to activin A and Wnt3a. In contrast, each clone seems to have a different level of responsiveness to hepatocyte growth factor (HGF) and oncostatin M (OSM), which are required for hepatic differentiation from endodermal cells. There may be single-nucleotide polymorphisms (SNPs) or other sequence variations in cytokine receptors or downstream molecules in each individual that affect this process. In addition, we have to consider the possibility that different protocols may yield different hepatic differentiation propensities.
We also found that CB-iPSCs are similar to PB-iPSCs in their hepatic differentiation potential. In general, cord blood cells are considered to be more plastic compared with peripheral blood cells and can be obtained noninvasively. They are also free from postnatal genetic mutations. As a result, cord blood cells may be a useful source for hiPSC generation (29–31).
In summary, our present data showed that the genetic background of donors is an important determinant of the propensity of hiPSC clones for hepatic differentiation. We also showed that peripheral blood and cord blood can be good sources for clinically useful hiPSC generation.
Materials and Methods
Cell Culture.
HiPS/ESCs were maintained on feeder layers of mitomycin C-treated STO/Neo resistant/LIF (SNL) feeder cells in Primate ES medium (ReproCELL) supplemented with 4 ng/mL recombinant human basic fibroblast growth factor (bFGF; Wako) as described previously (1). Detailed information about the hiPS/ESC lines is described in Table S1. The experiments using hiPSCs of healthy individuals and Parkinson disease patients were approved by the ethical committee of the Department of Medicine and Graduate School of Medicine, Kyoto University, and informed consent was obtained from the donors. HepG2 and HuH7 (human hepatoma cell lines) were cultured in DMEM (Nacalai Tesque) containing 10% FBS (vol/vol).
RNA Isolation and PCR.
Total RNA was purified using TRIzol reagent (Invitrogen). One microgram of total RNA was used for each reverse-transcription reaction with ReverTraAce-a (Toyobo) and dT20 primer, according to the manufacturer’s instructions. Reverse-transcription (RT)-PCR was performed with ExTaq (Takara). The real-time PCR analysis was carried out with SYBR Premix Ex Taq ll (Takara) and run on a StepOne Real-Time PCR System (Applied Biosystems). The means of duplicate measurements were normalized against the level of a housekeeping gene (GAPDH) for the same sample. The primer sequences used are shown in Tables S2 and S3.
DNA Methylation Analysis.
Bisulfite treatment of genomic DNA was carried out using an EZ DNA Methylation kit (Zymo) according to the manufacturer’s protocol. For pyrosequencing, bisulfate-treated DNA was amplified by the PyroMark PCR kit (QIAGEN). Pyrosequencing analysis was performed by the PyroMarkQ96 ID system (QIAGEN) according to standard procedures. Genomic DNA of human heart, liver, and brain was purchased from BioChain. The primer sequences used are shown in Table S4.
Additional Methods.
Detailed descriptions of methods for hepatic differentiation in vitro, immunohistochemical staining, flow cytometric analysis, functional analysis of hepatic differentiated hiPS/ESCs in vitro, DNA microarray analysis, and statistical analysis are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Yoshinori Yoshida, Masato Nakagawa, Kazutoshi Takahashi, Michiyo Koyanagi, and Tomonori Nakamura, and other members of the Yamanaka research group for their valuable scientific discussions; Megumi Narita and Akiko Oishi for technical assistance; and Drs. Tomohisa Kato and Akira Nasu (Center for Induced Pluripotent Stem Cell Research and Application) for the donor91 hiPSC lines. We also thank Naoki Nagata, Yoko Miyake, and Rie Kato for their valuable administrative support. This work was supported in part by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science; and the Ministry of Education, Culture, Sports, Science, and Technology; by a grant from the Leading Project of the Ministry of Education, Culture, Sports, Science, and Technology; and by a grant from the Funding Program for World-Leading Innovative Research and Development on Science and Technology (First Program) of the Japanese Society for the Promotion of Science. This work was also supported by research grants from the Ministry of Health and Labour (to R.T. and H.I.). M.K. was a Research Fellow of the Japanese Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: The DNA microarray analysis data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38155).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209979109/-/DCSupplemental.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.D’Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 4.Cai J, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 5.Hay DC, et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 2008;26:894–902. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- 7.Song Z, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan GJ, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Si-Tayeb K, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayama K, et al. Efficient generation of functional hepatocytes from human embryonic stem cells and induced pluripotent stem cells by HNF4α transduction. Mol Ther. 2012;20:127–137. doi: 10.1038/mt.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 13.Ramos-Mejia V, et al. Nodal/Activin signaling predicts human pluripotent stem cell lines prone to differentiate toward the hematopoietic lineage. Mol Ther. 2010;18:2173–2181. doi: 10.1038/mt.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miura K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji O, et al. Therapeutic potential of appropriately evaluated safe-induced pluripotent stem cells for spinal cord injury. Proc Natl Acad Sci USA. 2010;107:12704–12709. doi: 10.1073/pnas.0910106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 20.Suemori H, et al. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochem Biophys Res Commun. 2006;345:926–932. doi: 10.1016/j.bbrc.2006.04.135. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 22.Yasunaga M, et al. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 23.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 25.Zaret KS. Regulatory phases of early liver development: Paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 26.Zaret KS. Genetic programming of liver and pancreas progenitors: Lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–340. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- 27.Ohi Y, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Ye Z, et al. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giorgetti A, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5:353–357. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haase A, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.