Abstract
RNA interference is a fundamental gene regulatory mechanism that is mediated by the RNA-induced silencing complex (RISC). Here we report that an artificial nanoparticle complex can effectively mimic the function of the cellular RISC machinery for inducing target RNA cleavage. Our results show that a specifically designed nanozyme for the treatment of hepatitis C virus (HCV) can actively cleave HCV RNA in a sequence specific manner. This nanozyme is less susceptible to degradation by proteinase activity, can be effectively taken up by cultured human hepatoma cells, is nontoxic to the cultured cells and a xenotransplantation mouse model under the conditions studied, and does not trigger detectable cellular interferon response, but shows potent antiviral activity against HCV in cultured cells and in the mouse model. We have observed a more than 99% decrease in HCV RNA levels in mice treated with the nanozyme. These results show that this nanozyme approach has the potential to become a useful tool for functional genomics, as well as for combating protein-expression-related diseases such as viral infections and cancers.
Keywords: endoribonuclease, nanomedicine, gene therapy
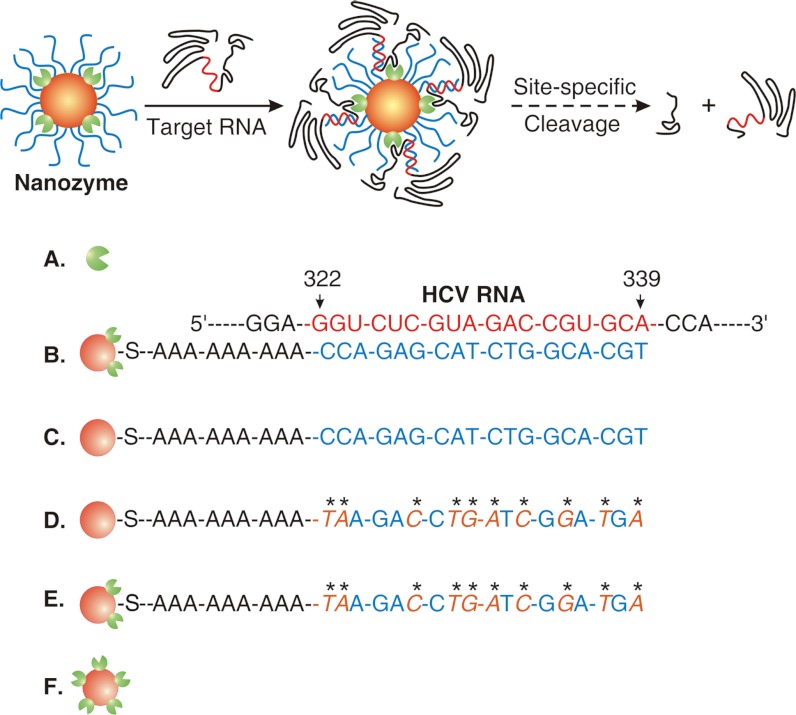
Owing to their size-dependent properties, nanoparticles have been extensively explored for biomedical applications (1–4). Their superior electronic, optical, magnetic, and photothermal properties have been used to develop new methods to sense and detect the state of living cells and organisms and to diagnose and treat diseases (2–9). Nanoparticle-based drug carriers—which can target cancer cells actively through the guiding moieties on their surface or passively through the enhanced permeation and retention (EPR) effect—have shown the remarkable ability to increase the selectivity of therapeutic agents against cancer cells while reducing their toxicity to normal cells (1, 10, 11). Such nanoparticle carriers also enable detailed tracking of drug locations within a patient’s body via biomedical imaging techniques such as MRI (1, 11, 12). In addition, self-assembled DNA nanoparticles having precisely controlled sizes and structural conformations have emerged as a new class of nanoparticle-based therapeutic agents (13, 14). Here, we report the use of nanoparticles as building blocks to construct multicomponent macromolecular complexes (called nanozymes, Fig. 1) for mimicking the RNA-cleavage function of the active RNA-induced silencing complex (RISC)—the cellular machinery—that mediates RNA interference (RNAi) pathways (15).
Fig. 1.
Schematic representation describing the design and function of a nanozyme: (A) an endoribonuclease, (B) a nanozyme with DNA oligonucleotides complementary to the sequence at the HCV RNA position (322–339 nucleotides), (C) a DNA-NP: a nanozyme counterpart that does not bear endoribonucleases, (D) a control DNA-NP, (E) a control nanozyme, and (F) an RNase-NP.
In cellular RNAi pathways, the RISC is an endonuclease-containing multiprotein complex that incorporates one strand of a small interfering RNA (siRNA) or microRNA; it uses this RNA strand to recognize and capture a complementary messenger RNA (mRNA) and then cleaves it into two pieces (15). Inspired by the structure and function of the RISC machinery, we designed a nanozyme that consists of a nanoparticle, sequence nonspecific endoribonucleases, and single-stranded DNA oligonucleotides (Fig. 1). The nanoparticle is the backbone of this nanozyme, providing a large surface area to hold endoribonucleases and DNA oligonucleotides at close proximity. Endoribonucleases are the catalytically active components of the nanozyme, whereas DNA oligonucleotides function as the components responsible for target RNA recognition via Watson–Crick base pairing and direct the neighboring endoribonucleases to cleave target RNAs that contain complementary sequences (Fig. 1). Because of their low toxicity and unique surface chemical properties for alkylthiol functionalization (2, 16–18), gold nanoparticles were chosen as the backbone to synthesize nanozymes. RNase A was used as the endoribonuclease component because it does not degrade the DNA-based recognition component of nanozymes, and it is one of the most robust and active ribonucleases for sequence nonspecific degradation of single-stranded RNAs, which have routinely been used for the removal of RNA contamination from DNA preparations as well as the removal of unhybridized regions of RNA from DNA/RNA or RNA/RNA hybrids (19). In addition, it is well documented that RNase A can effectively bind onto the surface of gold nanoparticles through noncovalent adsorption (20).
Hepatitis C virus (HCV) was chosen as a model system to evaluate the function and efficacy of the nanozyme for silencing gene expression and suppressing viral replication. HCV is a major cause of liver diseases such as chronic hepatitis, cirrhosis, and liver cancers (21). More than 170 million people are infected by HCV worldwide (22). Current interferon-based therapy results in sustained virus clearance in only around 50% patients; the therapy is not HCV-virus-specific and has significant side effects. In the absence of an effective vaccine, more specific antiviral therapies are urgently needed (22, 23).
HCV is a positive-strand RNA virus and has six major genotypes and numerous subtypes (24). The 5′ nontranslated region (5′ NTR) in the HCV genome is highly conserved among the six major genotypes, and this region contains an important structure known as the internal ribosome entry site that controls the initiation of HCV RNA translation (25). Previous reports have shown that, by targeting this viral genomic region, siRNA 331 can effectively inhibit the replication of HCV in cultured cells (25). The sequence of siRNA 331 was used in the design of an alkylthiol-terminated DNA oligonucleotide as the recognition component to synthesize anti-HCV nanozymes. This oligonucleotide consists of an A9 tether and an 18-nt-long fragment with sequence complementary to that of the region (nucleotides 322–339) in the HCV genome; this A9 tether was used as a spacer between the nanoparticle surface and the 18-nt recognition sequence for increasing the efficiency of the hybridization between a nanozyme and its complementary target (26). A mutant version of the alkylthiol-terminated oligonucleotide with nine mismatches was synthesized as a control DNA (Fig. 1).
Our results show that anti-HCV nanozymes are capable of actively cleaving a 5′ NTR-containing HCV RNA segment in a sequence specific manner, demonstrating that the sequence nonspecific RNase A and oligonucleotide components on the nanozymes can act cooperatively in target RNA cleavage. In addition, the nanozymes exhibit excellent stability against the degradation by proteinase, can be effectively internalized by human hepatoma cells in culture, are nontoxic to the cultured cells and a xenotransplantation mouse model under the conditions studied, and do not trigger detectable cellular interferon (IFN) response, but display a potent antiviral activity against HCV in cultured cells and in the mouse model.
Results and Discussion
Nanozymes were synthesized using a two-step method. In a typical experiment, gold nanoparticles (13 ± 1 nm in diameter) were first functionalized with RNase A in a carbonate buffered solution (pH 9.6) and then modified with the anti-HCV oligonucleotide (or the control DNA). This oligonucleotide-loading process was promoted by stepwise addition of a concentrated NaCl solution, which yielded anti-HCV nanozymes (or control nanozymes, Fig. 1). The resulting nanozymes were highly dispersible and stable in phosphate buffered saline (PBS) at pH 7.4, and they can be indefinitely suspended in 0.6 M PBS solutions. Transmission electron microscopy (TEM) shows that the surface functionalization with RNase A and alkylthiol-terminated DNA oligonucleotides did not affect the size and size distribution of gold nanoparticles (Fig. S1), but the surface functionalization led to a 6-nm red shift of the surface plasmon band of the gold nanoparticles (Fig. S2), suggesting a change in the dielectric constant of the surrounding environment of the nanoparticles due to the surface functionalization of DNA oligonucleotides (27).
The width of the surface plasmon band of the resulting nanozymes remained nearly unchanged as compared to that of unmodified gold nanoparticles, indicating that the nanozymes were not aggregated (27). Indeed, dynamic light scattering measurements further show that the nanozymes were discrete colloidal particles with a diameter of 48 ± 1.9 nm (Fig. S3); this size is in excellent agreement with a core/shell structure consisting of a 13-nm gold nanoparticle core and a shell of 27-nt single-stranded DNA oligonucleotides (Fig. 1). In addition, the nanozymes exhibited a less negative zeta potential than their counterparts without RNase A—gold nanoparticle-oligonucleotide conjugates (DNA-NPs)—at pH 7.4 (-14 ± 1.1 mV vs. -28 ± 1.7 mV; see Table S1), which suggests that the positively charged RNases were indeed functionalized on the surface of the nanozymes.
The average numbers of RNase A and oligonucleotide on each nanozyme were determined using an RNase activity assay and the OliGreen assay, respectively (SI Materials and Methods). Our results show that these average loading numbers were tunable by varying the amounts of RNase and oligonucleotide used in nanozyme syntheses, whereas the sequence difference in the oligonucleotides did not lead to a measurable change in their loading numbers on the nanozymes (Tables S2 and S3). In this study, the highest oligonucleotide loading number on the nanozymes with 12.1 ± 0.5 RNase A was found to be 78 ± 4, and these nanozymes were used as a model for evaluating nanozyme function in the following in vitro and in vivo experiments. To examine the oligonucleotide density effects on the target specificity of nanozymes, we synthesized nanozymes with 12.3 ± 0.8 RNase A and 29 ± 3 oligonucleotide (denoted as NZ-Ls). As controls, we prepared gold nanoparticle-RNase conjugates (RNase-NPs) and DNA-NPs with the anti-HCV or control sequences (Fig. 1).
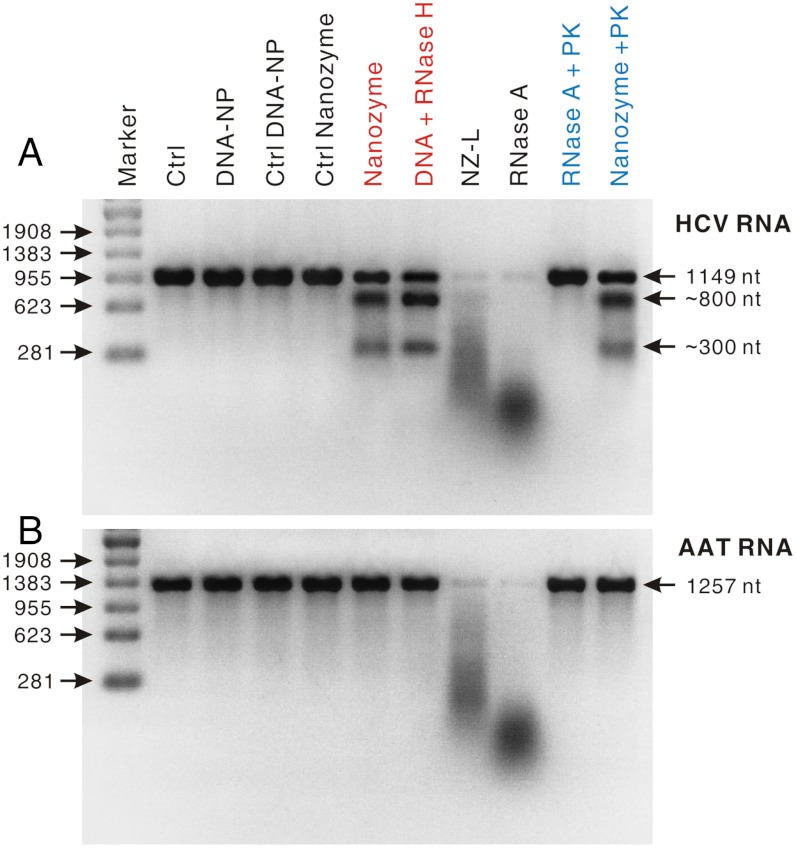
To assess the target specificity of the anti-HCV nanozyme, we performed an in vitro RNase activity assay with DNA-NPs, control DNA-NPs, control nanozymes as negative controls, and particle-free RNase A as a positive control. The target substrate was an HCV RNA segment (nucleotides 1–1149) that contains the entire 5′ NTR region of the HCV RNA genome of the HCV JFH-1 strain (see SI Materials and Methods). The control substrate was a 1,257-nt RNA segment of human alpha-1 antitrypsin (AAT) gene that does not contain complementary sequences to the nanozyme-bearing oligonucleotides (see SI Materials and Methods). In a typical RNase activity assay, the HCV (or AAT) RNA (0.12 μM) was incubated with the anti-HCV nanozymes or a control in a phosphate-buffered saline solution (PBS, 11 μL; pH 7.4) at 37 °C for 15 min, and the corresponding products were analyzed by electrophoresis in a denaturing agarose gel.
Electrophoresis analyses show that, although the anti-HCV nanozyme displayed no measurable cleavage activity on the AAT RNA, it did cleave the HCV RNA target into two major fragments with sizes of about 300 nt and 800 nt (Fig. 2A). This result corresponds to a RNA cleavage site fully matching the predicted position where the HCV RNA binds to the nanozyme via DNA/RNA hybridization (Fig. 1). In addition, the sizes of these two RNA fragments almost perfectly match the sizes of those corresponding HCV RNA fragments cut by RNase H—an endonuclease that specifically degrades the RNA of RNA-DNA hybrids (28)—in the presence of the free anti-HCV DNA oligonucleotide (Fig. 2A); this result further suggests that the anti-HCV nanozyme induced site-specific RNA cleavage (Fig. 1). Moreover, RNase H showed no cleavage activity towards the mixture of AAT RNA and the anti-HCV oligonucleotide (Fig. 2B), which experimentally shows that AAT RNA does not have complementary sequences to the anti-HCV oligonucleotide (28). The remarkable target specificity of nanozymes is further consistent with the results that the negative controls showed no cleavage activity against either the HCV or AAT RNAs (Fig. 2). On the contrary, unbound RNase A nonspecifically degraded both RNA substrates into short fragments, which appeared as broad smears (Fig. 2). Together, these results demonstrate that, like the RISC machinery, the anti-HCV nanozyme was capable of cleaving its target RNAs in a sequence- and site-specific manner (Fig. 1).
Fig. 2.
Ribonuclease activity tests for assessing the target selectivity of anti-HCV nanozyme and its ability to resist the degradation of proteinase activities. In these tests, the concentrations of anti-HCV nanozyme (or a control: DNA-NP, control DNA-NP, control nanozyme, and NZ-L) were 0.034 nM and that of unbound RNase A was 0.408 nM. The products of these tests were analyzed by using electrophoresis in a 2% formaldehyde agarose gel, and RNA bands were stained by using SYBR Green II. (A) HCV RNA segment (nucleotides 1–1149) as the substrate. (B) 1,257-nt AAT RNA segment as the substrate. In a typical proteinase K resistance test, nanozymes (0.034 nM) or particle-free RNase A (0.408 nM) were first incubated with proteinase K (10 nM) in a PBS buffer (pH 7.4) at 37 °C for 1 h. Then the product of this proteinase K treatment was divided into two parts and further incubated with the HCV (or AAT) RNA segment (0.12 μM) in a PBS buffer (11 μL; NaCl, 0.138 M; KCl, 0.027 M; pH 7.4) at 37 °C for 15 min. s: Ctrl, blank control; PK, proteinase K.
We attribute this site-specific RNA cleavage to the cooperative coupling between the RNase and DNA-oligonucleotide components of the nanozyme (Fig. 1). On one hand, the access of noncomplementary RNAs to the nanozyme-bearing RNase molecules is blocked by the densely packed oligonucleotides through steric hindrance and repulsive Coulomb interactions. On the other hand, these DNA oligonucleotides can also bind to target RNAs via base pairing and bring them to the RNase molecules on the nanozyme, resulting in the endonucleolytic cleavage of these RNAs into two fragments at positions close to the binding site (Figs. 1 and 2A). Therefore, the DNA oligonucleotide surface density should be critical for the RNA target specificity of nanozymes. Indeed, the RNase activity assay shows that the anti-HCV nanozymes with a low oligonucleotide surface coverage (i.e., NZ-Ls) did not exhibit target specificity and they cut both the HCV and AAT RNAs in a sequence nonspecific manner (Fig. 2).
Given the potential for RNase A degradation by proteinases in the cell or in vivo (29), we next examined the in vitro resistance of the anti-HCV nanozyme against proteinase K compared with particle-free RNase A (Fig. 2). RNase activity tests show that unbound RNase A lost its activity almost completely after 1 h incubation with proteinase K in a PBS buffer (pH 7.4) at 37 °C. In contrast, nearly no measurable change was observed in the nanozyme activity after an identical proteinase K treatment (Fig. 2). We attribute the resistance to proteinase degradation to the fact that the RNase molecules on the nanozyme were protected by the densely packed oligonucleotides through steric hindrance (Fig. 1). The ability to resist proteinase degradation should enhance the stability of these nanozymes in the cell and in vivo (see below).
To examine the intracellular activity of the nanozyme against HCV replication, we used an HCV replicon cell culture system, an FL-Neo cell line, which is a stable human hepatoma Huh7-derived cell line (30). This cell line harbors autonomously replicating genomic length genotype 1b HCV replicons and is an excellent system to evaluate anti-HCV agents in cell culture (30). We first evaluated the cellular uptake and cytotoxicity of anti-HCV nanozymes (see SI Mateirals and Methods). The assay based on inductively coupled plasma mass spectrometry shows that the nanozymes were effectively internalized by cultured FL-Neo cells. Uptake was nearly proportional to the concentration of nanozymes added to the cell media at low concentrations; an average number of nanozymes found in each cell was (4.6 ± 0.2) × 104 at a concentration of 0.54 nM (Table S4). Cell viability tests show that the nanozymes displayed no detectable toxicity to FL-Neo cells at concentrations ranging from 0.034 to 0.54 nM (Fig. S4).
We then examined the intracellular activity of the anti-HCV nanozyme with respect to gene knockdown for suppressing the replication of HCV RNA. FL-Neo cells were treated once with the nanozyme (or a control) at varying concentrations, incubated at 37 °C for 72 h, and then harvested and processed for a viral RNA assay using a quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR), with the endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as an internal standard (see SI Materials and Methods). Our results show that the treatments using anti-HCV nanozymes, control nanozymes, RNase-NPs, DNA-NPs, or control DNA-NPs did not lead to a measurable change in the GAPDH-RNA levels in FL-Neo cells (Fig. S5), indicating that these treatments did not induce toxicity effects on cell proliferation, which is in agreement with the results from our cell viability tests (Fig. S4).
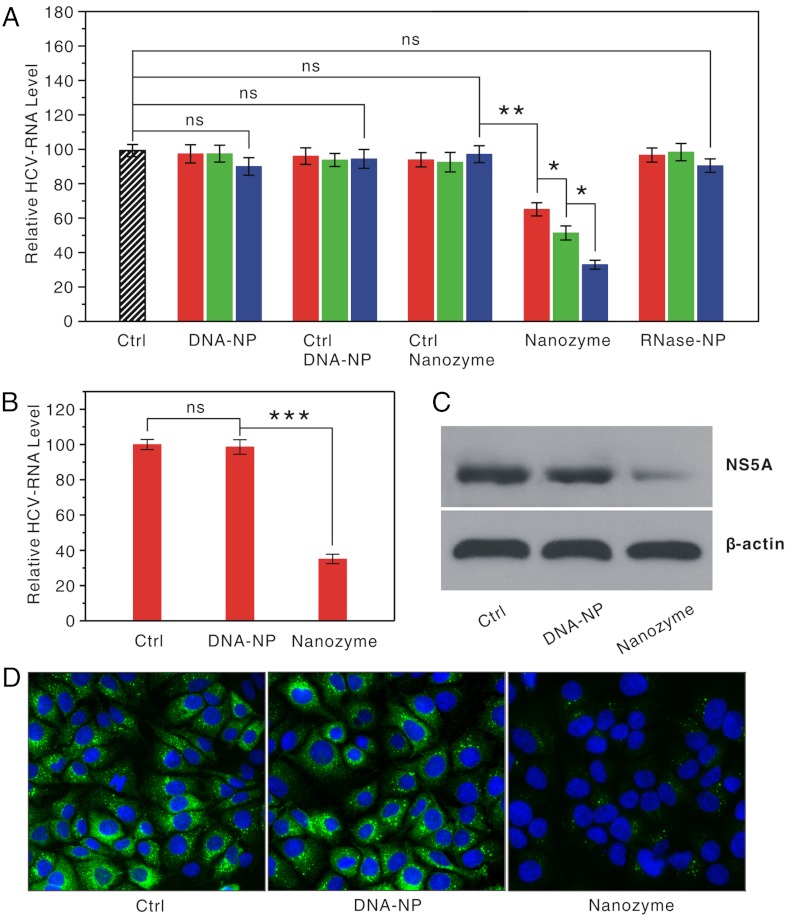
No measurable reduction in the HCV RNA levels was observed in the treatments using DNA-NPs at concentrations of 0.034, 0.14, and 0.54 nM (Fig. 3A). The inability of these conjugates to cause significant antisense effects on HCV replication is likely due to their low concentrations in our experiments (18). In contrast, when treated respectively with the corresponding nanozyme dosages, HCV replication in the cells dramatically decreased, and the inhibitory effect was dose dependent (Fig. 3A). As additional controls, neither control nanozymes nor control DNA-NPs induced detectable antiviral effects (Fig. 3A), which further confirms that these nanozymes exhibit excellent intracellular target specificity.
Fig. 3.
Anti-HCV effects of the anti-HCV nanozyme in FL-Neo cells. (A) qRT-PCR analyses of HCV RNA expression in the FL-Neo cells treated with anti-HCV nanozymes, DNA-NPs, control DNA-NPs, control nanozymes, and NZ-Ls at varying doses: 0.034 nM (red), 0.14 nM (green), and 0.54 nM (blue). Each bar presents the mean and standard deviation derived from three independent experiments; Student’s t test: ns, nonsignificance: P > 0.14, * for P < 0.01, and ** for P = 0.00053. Ctrl, blank control. (B) qRT-PCR analysis of HCV RNA expression in the FL-Neo cells treated with nanozymes or DNA-NPs three times during 7 d at concentration of 0.067 nM (see SI Materials and Methods). ns: P = 0.58, *** for P = 0.00024. (C) Western blot analysis of NS5A expression in the FL-Neo cells from the treatment in B, which was probed with anti-NS5A antibody and anti-β-actin antibody, respectively. The band intensity (NS5A/β-actin) relative to the control was found to be 0.25 ± 0.02 in the cells treated with nanozymes, whereas it was 0.97 ± 0.07 in the cells treated with DNA-NPs. (D) Immunofluorescence analysis of HCV NS5A expression in the individual FL-Neo cells from the treatment in B. The cells were fixed and the HCV NS5A expression level was examined by using fluorescent immunostaining with anti-HCV NS5A antibody and secondary antibody [fluorescein isothiocyanate (FITC)-labeled goat anti-mouse immunoglobulin G antibody]. The nuclei of the cells were stained with DAPI (4′,6-diamidino-2-phenylindole) as an internal reference. Mean and standard deviation were derived from three independent experiments.
In addition, the treatments with RNase-NPs did not lead to measurable effects on HCV RNA replication. This result is likely associated with the RNase deactivation/inhibition by intracellular proteinases and ribonuclease inhibitors (29, 31), and it indicates that the densely packed oligonucleotides are important for the nanozyme activity in cell and in vivo (vide supra). Moreover, the nanozyme-induced antiviral effects observed herein might also be caused by IFN activation (32). To evaluate this possibility, we examined the expression of gene 6–16 (G1P3), a downstream gene in the IFN signaling pathway (33). No changes were observed in the mRNA level of this gene in FL-Neo cells treated with anti-HCV nanozymes at the concentrations tested (Fig. S6), and this result essentially rules out the possibility of IFN pathway induction by these nanoparticles (33).
To further assess the intracellular antiviral activity of the anti-HCV nanozyme, we examined whether the nanozyme-mediated HCV RNA reduction is associated with suppression of viral protein synthesis. The nonstructured 5A (NS5A) protein of HCV—which plays key roles in both viral RNA replication and modulation of the physiology of the host cell—was used to evaluate HCV protein levels in FL-Neo cells. In a typical experiment, FL-Neo cells were treated with the anti-HCV nanozyme (0.068 nM) or a control on days 1, 3, and 5, and then harvested on day 7 (see SI Mateirals and Methods). The results from qRT-PCR analyses show that the nanozyme treatment resulted in a 65% decrease in HCV RNA levels in FL-Neo cells, whereas the treatment using the same amount of DNA-NPs did not induce measurable effects on HCV RNA replication (Fig. 3B).
Western blot analyses show fairly consistent results regarding the protein level: Barely any effect was observed in the FL-Neo cells treated with DNA-NPs, whereas the FL-Neo cells displayed a significant decrease in NS5A protein level of approximately 75% upon nanozyme treatment (Fig. 3C). NS5A protein reductions slightly exceeded the reduction levels obtained from HCV RNA, and this could be due to posttranscriptional mechanisms that have been observed previously (34). In addition, the results from these ensemble measurements are consistent with those from single-cell level observations on the basis of fluorescent immunohistochemical staining for NS5A protein (Fig. 3D). After the nanozyme treatment, more than 99% of FL-Neo cells displayed a significant decrease in the level of NS5A protein expression when compared to the control treatments (Fig. 3D). Altogether, these results unambiguously demonstrate that the anti-HCV nanozyme was capable of inducing an HCV gene knockdown in both the RNA and protein levels.
To evaluate the in vivo antiviral activity of the anti-HCV nanozyme, we constructed a xenotransplantation mouse model via subcutaneous injection of HCV-JFH1 infected Huh7.5 cells into nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (see SI Materials and Methods). HCV JFH1 is a genotype 2a strain capable of establishing robust infections in NOD/SCID mice harboring Huh7.5 hepatoma xenograft tumors, providing a simple and effective in vivo model system for the assessment of anti-HCV drugs (23). The HCV-infected mice were randomly divided into four groups. The group of mice without treatment was used as blank controls, and the other three groups of mice were injected in the tumors with particle controls (i.e., DNA-NPs or the control nanozymes), or anti-HCV nanozymes (in a dose of 0.34 pmol for each mouse) on days 1, 3, and 5. Treatment ended on day 7, when the animals were sacrificed and processed for the analyses of the viral and G1P3 mRNA levels using qRT-PCR with the human GAPDH gene as an internal control. In this mouse model, the HCV protein level of expression is too low to be detected by common methods such as Western blot and immunofluorescence. Note that no sign of toxicity was observed in any treated animals with either nanozymes or DNA-NPs (see SI Materials and Methods).
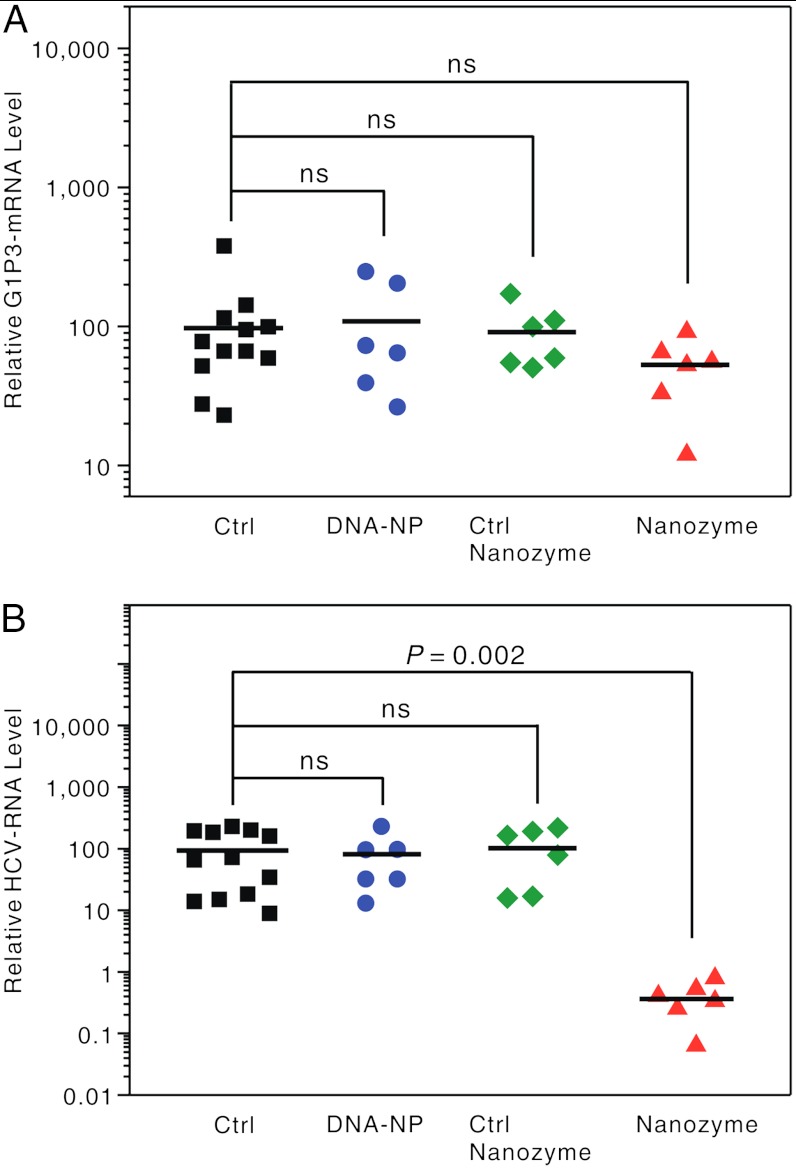
The analysis of G1P3 gene expression shows that none of the anti-HCV nanozymes, control nanozymes, or DNA-NPs led to IFN pathway activation in the mouse model (Fig. 4A, see ref. 32). The treatments with control nanozymes or DNA-NPs resulted in a statistically insignificant therapeutic effect on HCV RNA expression when compared with the control mice (Fig. 4B), which is consistent with those results from the in vitro experiments (Fig. 3). On the contrary, the nanozymes caused a potent therapeutic anti-HCV effect in the mouse model; the nanozyme-treated mice displayed an average decrease of 99.6% in HCV RNA levels. Remarkably, the in vivo HCV RNA reductions exceeded the reduction levels observed in the corresponding in vitro experiments (Fig. 4B). We attribute the improved antiviral therapeutic efficacy of the nanozyme to the fluidity and complex enzymatic characteristics of in vivo environments, which are much more dynamic systems than cell cultures (35).
Fig. 4.
In vivo antiviral effect of the nanozyme in xenotransplanted NOD/SCID mice. qRT-PCR analysis of (A) G1P3 mRNA and (B) HCV RNA expression in the xenograft tumors of the mice without treatment (Ctrl) shown as black squares, those treated with DNA-NPs shown as blue dots, control nanozyme shown as green diamonds, anti-HCV nanozyme shown as red triangles (the HCV RNA levels were relative to GAPDH). Each data point represents an individual mouse, and P-values were calculated using Student’s t test: ns, nonsignificance: P > 0.24.
Since the antiviral function of the nanozyme is independent of cellular RNAi machinery, this nanoparticle-based gene regulation approach complements RNAi-based approaches and has the potential to become a useful tool for controlling intracellular gene regulation. This platform will also allow one to add functionality that could direct nanozymes to specific tissues, organs, and even subcellular organelles that express target genes (1). In addition, this work presents a critical step toward creating a class of nanoparticle-based intracellular machineries with extraordinarily cooperative functions, remarkable target selectivity, and perhaps allosteric functions that enable these machineries to have an on/off switch in response to chosen allosteric effectors such as specific byproducts in disease-associated metabolic pathways (36), thus providing a powerful tool for studying and regulating a wide variety of biological pathways such as those in somatic cell reprogramming.
Materials and Methods
Nanozyme Synthesis.
In a typical synthesis, gold nanoparticles (10 nM, 13 ± 1 nm in diameter, see Fig. S1) were mixed with RNase A (0.5 μM) in a carbonate buffered solution (2 mL; carbonate, 10 mM; pH 9.6). After shaking for 30 min, alkylthiol-modified oligonucleotides with the anti-HCV or control sequence (6.4 nmol, see Fig. 1) and phosphate buffer (1.0 M, pH 7.4) were added to bring the mixture solution with 10 mM phosphate. After 8 h shaking, a sodium chloride solution (1.5 M solution in RNase-free water) was added to bring the NaCl concentration gradually to 0.3 M during a period of 32 h. The solution was further shaken for another 8 h. Then the resulting nanozyme particles were purified using centrifugation (13,000 rpm, 20 min), and were redispersed in RNase-free water for use. In addition, the average number of RNase A and oligonucleotide loaded onto individual nanozymes was tunable by varying the concentration of RNase A and oligonucleotides used in the synthesis (Tables S1– S3). Note that all the vials and tubes used herein were modified by silane for minimizing the nonspecific binding of RNase A onto the glass surface of these glass containers.
RNase A Activity Assay.
In a typical test, RNA substrates (0.5 μg) were incubated with nanozyme (0.034 nM), DNA-NPs (0.034 nM), control nanozyme (0.034 nM), control DNA-NPs (0.034 nM), or particle-free RNase A (0.408 nM) in a phosphate buffered saline solution (11 μL; phosphate, 10 mM; NaCl, 0.138 M; KCl, 0.027 M; pH 7.4) for 15 min. Then the formaldehyde loading buffer (11 μL, purchased from Londa Rockland, Inc.) was added to denature the RNA products, and the resulting solution was heated at 65 °C for 11 min and then immediately placed on ice for 2 min before loading onto a 2% agarose/formaldehyde denaturing gel (10×3-(N-morpholino) propanesulfonic acid (denoted as MOPS) buffer, 5 mL: RNase free water, 45 mL; agarose, molecular biology grade, 1.0 g; and 37% formaldehyde solution, 0.9 mL). Gel electrophoresis was performed at 60 V for approximately 90 min or until the front line of bromophenol blue dyes migrated about 6 cm in the gel. Afterwards, the gel was stained by SYBR® Green II for visualization.
Construction of a HCV-Infected Xenotransplantation Mouse Model.
Xenographed mice harboring HCV were constructed as follows. First, HCV human cells (Huh7.5 cells) were generated as follows. The pJFH1 plasmid was cut by Xba I, and the resulting linearized DNA was purified and used as the template for in vitro transcription. In vitro transcribed JFH1 RNA was delivered into Huh7.5 cells by electroporation. HCV replication in transfected cells was confirmed by NS5A immunostaining. The tumorigenicity of Huh7.5 HCV transfected cells was performed by inoculating 5 × 106 cells, resuspended in PBS (100 μL, pH 7.4), subcutaneously injected into the back of the nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (8–10 wk old). The tumor volume of the mice was evaluated twice a week.
Evaluation of in Vitro and in Vivo Nanozyme Activities.
Analysis of the HCV and G1P3 RNAs were performed using qRT-PCR. RNA samples were extracted from FL-Neo cells or mouse tumor tissues using an RNA isolation reagent (TRIzol; Invitrogen). To prevent DNA contamination, total RNA was treated with RNase-free DNase II (Invitrogen). Total RNA samples (2 μg per reaction) were reversely transcribed into cDNAs by RT II reverse transcriptase (Invitrogen). Then, the cDNAs were used as templates in quantitative real-time PCR with HCV 3′ NTR gene-specific primers, (i.e., forward primer (FP): 5′-CCTTCTTTAATGGTGGCTCCAT-3′, nucleotides 9538–9559; reverse primer (RP): 5′-GGCTCACGGACCTTTCACA-3′, nucleotides 9582–9600; Probe 5′-TTAGCCCTAGTCACGGCT-3′, nucleotides 9561–9578). The amplification reactions were performed using TaqMan RT-PCR on a StepOne Plus real-time PCR system (Applied Biosystems). The primers for G1P3 gene were 5′-CAAGCTTAACCG-TTTACTCGCTGCTGT-3′ (Forward) and 5′-TGCGGCCGCTGCTGGCTACTCCTCATCCT-3′ (Reverse, see ref. 32). The human GAPDH was used as an internal control in PCR amplification, and its primers were 5′-TCACCAGGGCTGCTTTTA-3′ (FP) and 5′-TTCACACCCATGACGAACA-3′ (RP). The PCR conditions were as follows: 10 min at 95 °C and 40 cycles of 15 s at 95 °C, 30 s at 58 °C (GAPDH or G1P3) or 60 °C (HCV), and 30 s at 72 °C, with an extension for 10 min at 72 °C.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Shen-hsiu Hung and Prof. Yiider Tseng for their assistance in microscope measurements. Y.C.C. and C.L. thank Research Opportunity Seed Fund of the University of Florida for partial support of this work. Y.C.C. acknowledges support from the Office of Naval Research (ONR: N00014-09-1-0441) and National Science Foundation (DMR-0645520). C.L. acknowledges support from National Institutes of Health (R01CA133086 and K26 RR023976). We acknowledge the Major Analytical Instrumentation Center (MAIC) at the University of Florida for TEM usage.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207766109/-/DCSupplemental.
References
- 1.Peer D, et al. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Giljohann DA, et al. Gold nanoparticles for biology and medicine. Angew Chem Int Edit. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alivisatos P. The use of nanocrystals in biological detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 4.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 5.Nederberg F, et al. Biodegradable nanostructures with selective lysis of microbial membranes. Nat Chem. 2011;3:409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 6.Qian XM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 7.Wu W, Li AD. Optically switchable nanoparticles for biological imaging. Nanomedicine (Lond) 2007;2:523–531. doi: 10.2217/17435889.2.4.523. [DOI] [PubMed] [Google Scholar]
- 8.Graham D, Thompson DG, Smith WE, Faulds K. Control of enhanced Raman scattering using a DNA-based assembly process of dye-coded nanoparticles. Nat Nanotechnol. 2008;3:548–551. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 9.Cho EC, Zhang Q, Xia YN. The effect of sedimentation and diffusion on cellular uptake of gold nanoparticles. Nat Nanotechnol. 2011;6:385–391. doi: 10.1038/nnano.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim B, et al. Tuning payload delivery in tumour cylindroids using gold nanoparticles. Nat Nanotechnol. 2010;5:465–472. doi: 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigger I, Dubernet C, Couvreur P. Nanoparticles in cancer therapy and diagnosis. Adv Drug Deliv Rev. 2002;54:631–651. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 12.Yezhelyev MV, Qi LF, O’Regan RM, Nie S, Gao XH. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J Am Chem Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khaled A, Guo SC, Li F, Guo PX. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu D, Shu Y, Haque F, Abdelmawla S, Guo PX. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 17.Pan Y, et al. Size-dependent cytotoxicity of gold nanoparticles. Small. 2007;3:1941–1949. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 18.Rosi NL, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 19.Alberts B. Molecular Biology of the Cell. 5th Ed. New York: Garland Science; 2008. [Google Scholar]
- 20.Bendayan M. In: “The Enzyme-Gold Cytochemical Approach: A Review”in Colloidal Gold Principles, Methods, and Applications. Hayat MA, editor. Vol. 2. San Diego: Academic Press; 1989. pp. 117–147. [Google Scholar]
- 21.Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309–2324. doi: 10.1038/onc.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploss A, Rice CM. Towards a small animal model for hepatitis C. EMBO Rep. 2009;10:1220–1227. doi: 10.1038/embor.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMullan LK, et al. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci USA. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokota T, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demers LM, Mucic RC, Reynolds RA, Mirkin CA, Letsinger RL. Fluorescence-based method for the determination of surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold nanoparticles. Abstr Pap Am Chem Soc. 2000;219:U870–U870. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 27.Lazarides AA, Schatz GC. DNA-linked metal nanosphere materials: Structural basis for the optical properties. J Phys Chem B. 2000;104:460–467. [Google Scholar]
- 28.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly BM, Yu CZ, Chang PL. Presence of a lysosomal-enzyme, arylsulfatase-a, in the prelysosome-endosome compartments of human cultured fibroblasts. Eur J Cell Biol. 1989;48:71–78. [PubMed] [Google Scholar]
- 30.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 32.Marques JT, Williams BRG. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 33.Zhu HZ, et al. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology. 2003;37:1180–1188. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]
- 34.Kanda T, Steele R, Ray R, Ray RB. Small interfering RNA targeted to hepatitis C virus 5′ nontranslated region exerts potent antiviral effect. J Virol. 2007;81:669–676. doi: 10.1128/JVI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 36.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.