Abstract
Toxin-antitoxin (TA) systems are found on both bacterial plasmids and chromosomes, but in most cases their functional role is unclear. Gene knockouts often yield limited insights into functions of individual TA systems because of their redundancy. The well-characterized F-plasmid–based CcdAB TA system is important for F-plasmid maintenance. We have isolated several point mutants of the toxin CcdB that fail to bind to its cellular target, DNA gyrase, but retain binding to the antitoxin, CcdA. Expression of such mutants is shown to result in release of the WT toxin from a functional preexisting TA complex as well as derepression of the TA operon. One such inactive, active-site mutant of CcdB was used to demonstrate the contribution of CcdB to antibiotic persistence. Transient activation of WT CcdB either by coexpression of the mutant or by antibiotic/heat stress was shown to enhance the generation of drug-tolerant persisters in a process dependent on Lon protease and RecA. An F-plasmid containing a ccd locus can, therefore, function as a transmissible persistence factor.
Keywords: CcdA-CcdB, conditional regulation, protein—protein interaction
Many bacterial genomes harbor a class of genes referred to as toxin-antitoxin (TA) genes. These genes were initially discovered on low copy-number plasmids of Escherichia coli and appear to be involved in plasmid maintenance in the bacterial population (1). TA systems comprise of a pair of genes organized in an operon encoding a stable toxin and a labile antitoxin that antagonizes it (1). Plasmid-based TA systems are also known as addiction modules and selectively eliminate daughter cells that do not inherit a plasmid copy during cell division (1). This mechanism, also called postsegregational killing, occurs in daughter cells devoid of a plasmid copy. The unstable antitoxin counterpart is degraded more rapidly by host proteases than the toxin. The toxin is released from the TA complex and interacts with an essential host target. This interaction often results in cell death but in some cases, as shown below, may also result in growth inhibition (2).
Homologs of plasmid-based TA systems have recently been discovered on the chromosomes of a large number of bacteria, many of which are pathogenic (3). The biological role of these TA systems in bacteria still remains controversial. A number of different models have been proposed to explain their presence on the chromosome (4). TA systems are difficult to study because overexpression of the active toxin component typically leads to cell death. Because many bacteria contain multiple homologous TA systems with redundant functions, multiple TA systems may need to be knocked out before there is an observable phenotype (5). In the present work, we describe a methodology to conditionally regulate expression of a toxin gene in a dose-dependent fashion. The method involves the release of the WT toxin from the TA complex by an overproduced mutant toxin that has a high affinity for the antitoxin but a low affinity for the cellular target of WT toxin (Fig. S1). This approach was validated using the well-established plasmid-based CcdAB TA system and used to demonstrate that this system plays a role in bacterial persistence. Bacterial persistence is a phenotype of dormant cells present at a low frequency in a growing population and characterized by tolerance to the presence of a variety of antibiotics, even in the absence of an active, specific resistance mechanism. Persisters are likely to be clinically important (6, 7). In the present work, we show that the F-plasmid derived ccd operon, whether located on a multicopy plasmid or in a single copy on the E. coli chromosome, plays a significant role in the generation of persisters. This finding is in addition to its well-studied role in plasmid maintenance (8). The methodology described here may also be used to probe the role of specific TA systems in other organisms where making knockouts are difficult or which have multiple homologous TA systems.
Results
Inactive, Active-Site Mutants of CcdB.
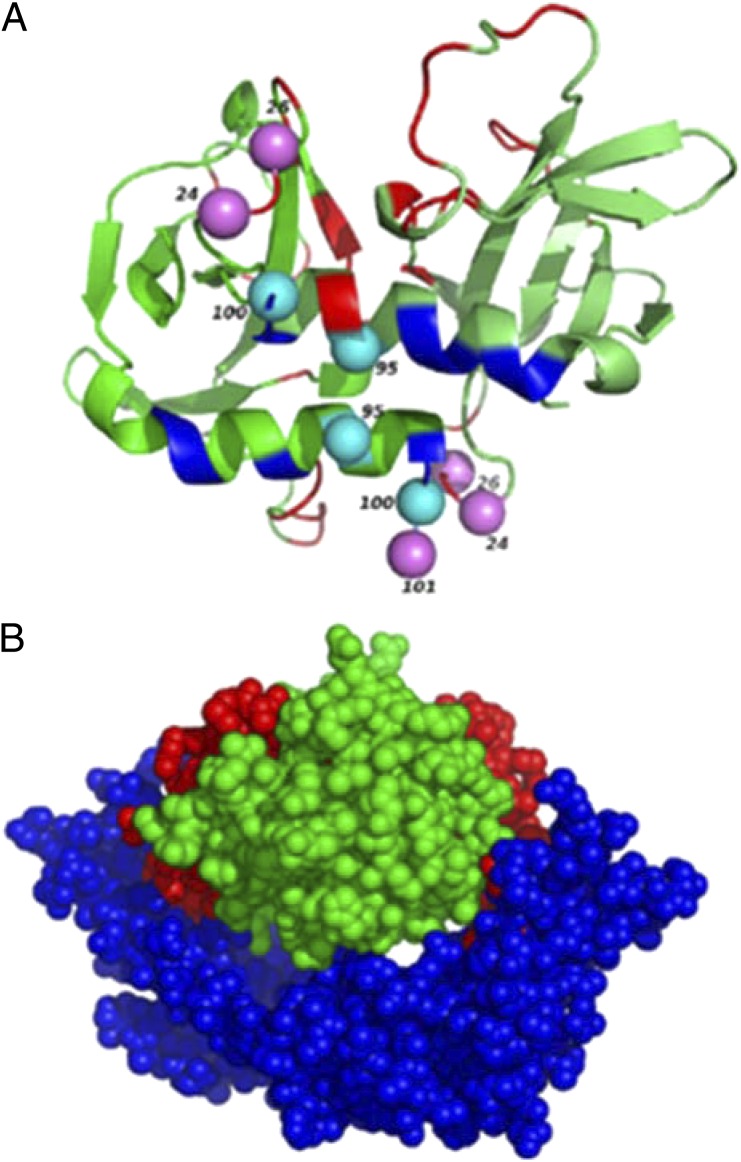
CcdAB is one of the most well-studied plasmid-based TA systems and is involved in maintenance of F-plasmid in E. coli (8). CcdB is a DNA gyrase poison that entraps a cleavage complex between gyrase and DNA (9). In the presence of its antagonist, CcdA, CcdB is sequestered in the form of a CcdAB complex. However, if the cell loses the F-plasmid, the labile CcdA is degraded by the ATP-dependent Lon protease (2), releasing CcdB from the complex to act on its target DNA gyrase, which eventually leads to cell death. The crystal structure of CcdB in complex with a fragment of DNA gyrase has been determined. The active-site residues of CcdB are defined as those that are involved in direct interaction with DNA gyrase, as determined by Ala and Asp scanning mutagenesis (10) and confirmed by X-ray crystallography of the CcdB:GyrA14 fragment complex (11). These comprise residues Ile24, Ile25, Asn95, Phe98, Trp99, and Ile101 (Fig. 1A). We have previously reported the construction and phenotypic characterization of a large library comprising of a total of 1,430 (75%) of the 1,900 possible single-site mutants of CcdB (12). The mutants were expressed under control of the arabinose inducible PBAD promoter in pBAD24ccdB (13). In this system, the level of expression of each mutant can be modulated by varying the arabinose concentration in the medium.
Fig. 1.
Interacting residues on CcdB. (A) CcdB2 structure (green) showing residues interacting with GyrA142 and CcdA2 in blue and red, respectively. Violet spheres correspond to residues which bind both to CcdA and GyrA14. Cyan spheres indicate Gyrase interacting residues, which are proximal (but not overlapping) to CcdA interacting residues (red). Residue numbers are shown adjacent to each sphere. (B) Overlap of CcdA and GyrA14 binding sites on CcdB. Model was generated by superposing CcdB2 (green) from complexes with GyrA142 [PDB ID 1X75, blue (11)] and CcdA2 [PDB ID 3G7Z, red (22)], respectively. This figure was created using PyMOL (http://www.pymol.org).
No structure was available for the CcdA:CcdB complex when this work was initiated. However, we hypothesized that there should be CcdB mutants that affect binding to DNA gyrase but not CcdA. Such mutants should be nontoxic when overexpressed in a CcdB-sensitive E. coli strain that lacks WT CcdB. However, when overexpressed in F-plasmid containing strains that express WT CcdB and its antitoxin CcdA, such mutants should cause cell death by titrating out CcdA, thereby permitting WT CcdB to bind to its cellular target. To validate this hypothesis, 10 inactive mutants previously isolated (12) at active-site residues involved in DNA gyrase binding were characterized. In initial experiments, mutants were overexpressed in two different E. coli strains, one containing an F-plasmid encoding WT CcdB and the other lacking the F-plasmid. The resulting phenotypes were characterized. The E. coli strains used were: Top10, which lacks the F-plasmid (and hence CcdA and CcdB), and either XL1-Blue or Top10F′, both of which carry the F-plasmid containing the ccd operon. The selected mutants (Table S1), when overexpressed in the Top10 strain with saturating inducer concentration (0.1% arabinose), showed an inactive phenotype. This finding demonstrates that in contrast to WT CcdB, these mutants lack the ability to bind and poison the DNA gyrase, and thus to cause cell death. When the same inactive CcdB mutants were overexpressed in either Top10F′ or XL1-Blue, 7 of the 10 showed complete absence of colonies (Fig. S2), comparable to the phenotype observed with overexpression of WT CcdB. This finding indicated that most inactive, active-site mutants of CcdB, when overexpressed, were able to titrate out WT CcdA from the WT CcdA-CcdB complex. The released WT CcdB likely caused cell death by binding to its cellular target, DNA gyrase. Although Top10F′ and XL1-Blue are recA−, similar results were obtained in the recA containing E. coli strain AB264. As a negative control, several inactive, buried-site mutants (Table S1) were also overexpressed in both Top10 and Top10F′ strains. These buried-site mutants are expected to have a distorted, destabilized, and aggregation-prone structure and thus are not expected to be able to release WT CcdB from the CcdAB complex. As expected, cells expressing these mutants did not show any growth defects and showed growth comparable to cells overexpressing the thioredoxin control protein, in both strains.
WT CcdB Causes Growth Arrest When Expressed at Low Levels.
To examine whether lower amounts of free WT CcdB cause reversible growth arrest or cell death in our system, Top10F′ and AB264 strains were transformed with inactive, active-site mutants and plated on LB/amp plates containing variable amounts of arabinose. Normal growth was observed in the range 0% to 1 × 10−4% arabinose. From 1 × 10−3 to 2 × 10−2% arabinose, colony size became progressively smaller, although the number of colonies was unchanged and at higher arabinose concentrations (1 × 10−1% and higher) no colonies were observed. To examine if the growth inhibition at low arabinose concentrations was reversible, the strain carrying F-plasmid was transformed with the inactive, active-site mutant 100TCcdB and grown in liquid culture. Log-phase cells (OD600 = 0.2) were induced with varying concentrations of arabinose for 2 h and plated on LB/amp plates containing 0.2% glucose to repress further expression of 100TCcdB from the PBAD promoter. It was found that cells induced with up to 0.01% arabinose showed reversible growth inhibition, but at higher arabinose concentrations there was significant cell death (Fig. S3). This finding demonstrates that exposure of cells to low levels of WT CcdB causes reversible growth inhibition but higher levels cause cell death.
In Vitro Studies of CcdB Mutants Binding to DNA Gyrase and CcdA.
To confirm that the inactive, active-site mutants of CcdB still bind CcdA but have diminished affinity for gyrase, four such mutants (24K, 24M, 95P, 100T) were purified and characterized by surface plasmon resonance (SPR). Purified GyrA14 (residues 363–494) or CcdA was immobilized on the surface of a CM5 chip. WT CcdB and all mutants were passed over the chip surface. Values of the measured parameters are summarized in Table S2. The data show that the mutants bind 5- to 50-fold more weakly to GyrA than WT CcdB, but bind CcdA with similar or higher affinity than WT CcdB.
Expression of Inactive, Active-Site Mutant Leads to Derepression of the ccd Promoter.
The phenotypic effects observed on overexpression of the inactive, active-site mutants in the presence of WT CcdAB could arise through two different mechanisms. The inactive mutant could displace WT CcdB from its complex with CcdA either directly or by binding to free CcdA. In the latter case, the consequent decrease in free CcdA would lead to dissociation of WT CcdB from its complex with CcdA to maintain equilibrium. In either event, the free WT CcdB could then bind to its cellular target, DNA gyrase, resulting in growth inhibition or cell death. Alternatively (or in addition), an increase in the CcdB:CcdA ratio as a result of mutant expression could lead to derepression of the ccd operon and fresh synthesis of CcdA and WT CcdB (14). The fresh CcdA would bind to the excess, inactive mutant of CcdB and the newly synthesized CcdB could bind to its cellular target, DNA gyrase. To determine whether expression of an inactive, active-site mutant of CcdB results in derepression of the ccd operon, two different approaches were used. In the first approach, the ability of excess 100TCcdB to decrease binding of WT CcdAB complex to promoter DNA was studied by SPR (Fig. S4) and EMSA (Fig. 2). In both cases, it was observed that DNA binding of the CcdAB complex was decreased in the presence of excess 100TCcdB, as described previously for WT CcdB (14). It can also be seen that the complex of 100TCcdB with CcdA (lanes 6–8, Fig. 2) binds ccdP/O DNA, although less well than the WT CcdB:CcdA complex. This finding is consistent with an earlier study (15) that showed that 100R and 100E CcdB mutants led to loss of toxin activity but retained the ability to bind the operator when complexed to CcdA. In an alternate approach, a GFP reporter was fused to the ccd promoter and cloned to yield the plasmid pccdP/O-gfp with a p15A origin. The effect of expression of the inactive mutant, 100TCcdB on GFP expression was monitored in both Top10F′ and Top10 strains. As can be seen in Fig. S5A, the ratio of GFP expression in induced to uninduced cells was appreciably higher for Top10F′ strain. This finding suggests that overexpression of 100TCcdB leads to derepression of the ccd operon by reducing the CcdA:CcdB ratio, resulting in fresh synthesis of WT CcdB.
Fig. 2.
Binding of CcdA-WTCcdB and CcdA-100TCcdB complex to ccdP/O monitored by EMSA. A radiolabeled DNA fragment containing the ccdP/O was incubated with increasing concentrations of CcdA-WTCcdB (lanes 1–5) or CcdA-100TCcdB complex (lanes 6–8) at a CcdA:CcdB ratio of 1.5:1. The data show that CcdA-100TCcdB complex binds less well to DNA than CcdA-WTCcdB (lanes 5 and 8) and also that addition of 100TCcdB to the preformed CcdA-WTCcdB complex, results in a decrease in the amount of DNA binding (lanes 9–11), presumably because of a decrease in the overall CcdA:CcdB ratio.
Overexpression Studies with 100TCcdB Suggest a Possible Role of CcdB in Generation of Persisters.
Plasmids expressing WT CcdB under control of the PBAD promoter are toxic to E. coli, even under highly repressed conditions (0.2% glucose) (12). However, expression of low levels of an inactive, active-site mutant (such as 100TCcdB) in cells containing the WT ccd operon, by the addition of 0.001% arabinose results in growth inhibition. Under these conditions, we examined the effects of the antibiotics ciprofloxacin, mitomycin C, cefotaxime, kanamycin, and tobramycin on cell viability. For this process, AB264/p100TccdB cells induced for 100TCcdB expression for 1 h were subsequently exposed to lethal doses of different drugs (ciprofloxacin, mitomycin C, cefotaxime, kanamycin, tobramycin) for 4 h. Cells were then washed, plated on 0.2% glucose containing medium to repress further expression of 100TCcdB, and the surviving colonies were counted. In comparison with the uninduced cells, induced cells showed ∼850-, 750-, and 350-fold more tolerance to ciprofloxacin, mitomycin C and cefotaxime, respectively, as well 250- and 150-fold increased tolerance against kanamycin and tobramycin, respectively (Fig. 3). Compared with cells lacking the 100TCcdB plasmid, levels of tolerance were even higher (Fig. S6). To rule out the possibility of development of antibiotic resistance because of mutation during the course of the experiment, persister-derived colonies were replica plated on LB plates containing antibiotics or without any antibiotic. Bacterial growth was found only in the absence of any antibiotic. This finding confirms the generation of true persisters at low levels of CcdB expression. These data show that in addition to its well-studied role in plasmid maintenance, the CcdAB system may also be involved in the generation of persisters.
Fig. 3.
Effect of Lon and RecA on CcdB mediated persister generation. AB264 (WT), AB264Δrec, AB264Δlon, and AB264ΔlonΔrec strains were induced for the expression of 100TCcdB mutant protein for 1 h at an OD600 of 0.2. Subsequently, the cells were challenged with different antibiotics at ∼10 times minimal inhibitory concentration for 4 h. Percent survival was calculated as the ratio (cfu after antibiotic exposure/cfu before antibiotic exposure) × 100. Survival ratio is defined as the ratio of percent survival of the E. coli strain induced for 100TCcdB to the corresponding uninduced strain. (A) The percent survival of the induced cells; (B) the survival ratio. Both values are shown in log scale. Error bars indicate the SE from three independent experiments.
CcdB-Induced Persister Formation Is Dependent on RecA and Lon Protease.
Poisoning of Gyrase:DNA complexes by CcdB in vivo, results in induction of a RecA mediated SOS response (16). The SOS response has previously been linked to persister formation via enhanced expression of TisB (17). To study the role of RecA on CcdB mediated persister formation, 100TCcdB was expressed in AB264 deleted for recA. Relative to the WT strain under identical conditions, the survival was over 100-fold reduced in the presence of ciprofloxacin and mitomycin C, respectively, and smaller reductions in survival were observed for the other antibiotics (Fig. 3). The involvement of RecA in DNA repair (18) explains why recA deletion makes bacteria particularly sensitive toward antibiotics that target DNA either directly (mitomycin C) or indirectly (ciprofloxacin).
Overexpression of inactive 100TCcdB results in an increase in the level of free WTCcdB, because of titration of any CcdA that is present as well as fresh synthesis of WT CcdB. The protease Lon is involved in degradation of free CcdA as well as activation of various genes during stress (19). In the present case, there should be little free CcdA present because of the presence of excess, inactive 100TCcdB. Nevertheless, to investigate the possible role of Lon in CcdB-mediated persister formation, the antibiotic sensitivity was examined in the presence and absence of overexpressed 100TCcdB in E. coli strain AB264:Δlon/p100TccdB, as described above. Deletion of lon led to about a 10-fold decrease in percentage survival relative to the WT strain and 50- to 200-fold decrease in survival ratio relative to the corresponding uninduced control (Fig. 3). This result clearly demonstrates that Lon also plays an important role in CcdB-mediated persister generation, apart from its role in antitoxin degradation (2, 5). Because both Lon and RecA are involved in CcdB-mediated persister generation, the above experiments were repeated in the AB264ΔlonΔrec double-knockout. In the double-knockout, persisters were further reduced to close to background levels, suggesting that the downstream pathways activated by Lon and RecA to generate persisters are at least partially independent.
Further Confirmation of CcdB’s Role in Persistence.
The above studies involving overexpression of 100TCcdB suggested a possible role for the ccd operon in the generation of persisters. Use of an inactive, active-site mutant, such as 100TCcdB, provides a powerful tool to conditionally regulate expression from the WT ccd operon. However, the WT CcdB levels generated using this system may not be physiologically relevant. Expression of the ccd operon is known to be triggered by various stresses, including heat (2, 20). We therefore compared the survival of cells with and without the ccd operon. Cells were first exposed to either heat or a sublethal dose of antibiotic prestress (21) (to derepress the operon) (Fig. S5B) followed by exposure to lethal concentrations of various different antibiotics (Fig. 4). Three different formats were used, namely: (i) Top10 cells containing F-plasmid, present at one to two copies, each molecule of plasmid contains one copy of the ccd operon but numerous other genes as well; (ii) Top10 cells transformed with the small multicopy pccd plasmid (10–12 copies per cell); and (iii) cells in which the ccd operon was integrated in single copy into the chromosome of the E. coli strain BW25113 (see SI Materials and Methods). This process was done not to simulate chromosomal ccd systems, but rather to quantitate the role of the F-plasmidic ccd operon in single copy on persister formation, in the absence of other F-plasmid genes. In each case, the number of persister cells were compared with the relevant control strain lacking the ccd operon (Fig. 4). In all cases, there was a significant increase in survival for cells containing the ccd operon, although the effects were not as large as those seen upon continuous expression of the inactive, active-site mutant (Fig. 3). This result is presumably because of lower activation of the ccd operon by transient heat or antibiotic prestress, than by continuous expression of an inactive, active-site mutant.
Fig. 4.
Role of CcdB in persister generation when present in multiple (A–C) or single copy (D–F). (A–C) Top10 strain containing either the multicopy plasmid pccd (Top10pccd) or F-plasmid (Top10F′) or lacking any plasmid (Top10) were grown to an OD600 of 0.2–0.3. Subsequently, cells were exposed to different prestresses, either a sublethal dose of ampicillin (1 μg/mL) for 1 h (A), heat at 48 °C for 20 min (B), or without any prestress (C). Subsequently, cultures were exposed to different antibiotics at ∼10 times the minimal inhibitory concentration for 4 h, washed, and then plated on LB agar media for cfu/mL determination. Survival ratio was calculated and is defined as the ratio of percent survival of the E. coli strain containing the ccd operon (either Top10pccd or Top10F′) to that lacking the ccd operon (Top10) after antibiotic exposure. (D–F) A similar experiment was done in strains with the ccd operon integrated into the chromosome. Normalized data for BWccd (containing ccd) E. coli strain with ampicillin (1 μg/mL) prestress (D), heat prestress (E), and without any prestress (F) are shown. Normalization was done with respect to two different reference strains, BWcat (containing insertion of cat marker at the same locus as ccd) and BW25113 (WT E. coli strain, lacking both cat and ccd), respectively. Error bars indicate the SE from three independent experiments.
Discussion
In this study we have used a method to conditionally regulate the expression of TA systems and to obtain new insights into the role of the ccd operon in persistence. Although we have specifically applied this method to a prokaryotic TA system, it can be used for any regulated system, including those within eukaryotic cells wherein an inactive mutant of the protein can titrate a cellular inhibitor. The present studies show that through appropriate point mutations, it is possible to selectively modulate some binding/catalytic functions without affecting others, even in compact, single-domain globular proteins. In many cases it should be possible to set up appropriate genetic screens to isolate such mutants. In a related approach, overexpression of a nontoxic mutant of RelE was used to titrate endogenous RelB and thereby activate endogenous RelE (5). In the relBE, mazEF and many other TA systems the toxin component is enzymatically active. It is therefore relatively straightforward to design mutations that lead to loss of enzyme activity without significantly affecting antitoxin binding. The CcdAB system is more challenging because CcdB has no enzymatic activity. Furthermore, CcdB has significantly overlapping binding sites for CcdA and DNA gyrase and binds both full-length proteins with KDs that are in the subnanomolar range (22) (Fig. 1B). Despite this challenge, it was possible to isolate CcdB mutants that abolish its Gyrase poisoning activity without affecting CcdA binding (Fig. S2 and Table S1), even at residue positions (such as 24I) that are involved in binding both proteins. This is a surprising result, which demonstrates that it should be possible to apply this methodology to many other TA systems, to conditionally activate a single toxin in a background of multiple other TA systems because of the specificity of TA interactions. A recent study has shown that the ccdF system is able to mediate postsegregational killing in an E. coli strain harboring the ccdO157 system on its chromosome. This finding shows that the plasmid ccdF system is functional even in the presence of its chromosomal counterpart (23). Inhibitors of TA interactions can be important leads for the development of new drugs in case of TA systems that are involved in killing cells. The present study validates the CcdAB system as a potential drug target because complete titration of WT CcdA by an excess of inactive, active-site mutants of CcdB results in cell death.
The factors responsible for generating bacterial persisters are poorly understood. Transcriptional profiling of isolated persisters indicated an overexpression of TA modules (24, 25). Although the ectopic expression of several different toxins, such as RelA, MazF, HipA, and YgiU resulted in increased persistence, strains deleted for individual TA modules do not show decreased persister formation until multiple TA modules are deleted (5). It was previously known that the ccd operon is involved in plasmid maintenance (8). In the present study, a putative role for the F-plasmidic CcdAB TA system in persistence was elucidated. Activation of endogenous CcdB either by CcdA titration or by transient heat or antibiotic stress lead to measurably higher amounts of persisters. Because the ccd operon occurs on the transmissible F-plasmid, it is also an example of a transmissible persistence factor. Homologs of F-plasmid ccd are also present on the chromosome of pathogenic strains of E. coli (E. coli O157:H7 str. EC4206, RefSeq accession no. ZP_03253116.1) and CcdB homologs are present in other human pathogens, such as Shigella dysenteriae (GenBank accession no. EGI89125.1) and Vibrio cholera (RefSeq accession no. ZP_01978533). The sequence of the chromosomal ccd systems have diverged significantly from the F-plasmid ccd operon studied here, although in ∼70% of cases both components of the chromosomal ccd have retained their toxic activity (26). Molecular evolutionary analysis of 47 isolates of ccdB0157 suggested that the chromosomally encoded ccdAB TA systems appear devoid of any biological function and are under neutral evolution (26). Using approaches similar to the ones described herein should clarify whether or not specific chromosomally encoded ccd systems also contribute to persistence.
The present studies also suggest that, in addition to its known role in antitoxin degradation (2, 5), the Lon protease has other roles in the generation of persisters that need to be elucidated. Like ciprofloxacin and other fluroquinolone antibiotics, CcdB produces DNA lesions by stabilizing a cleaved DNA-gyrase complex. This stabilization leads to induction of the RecA mediated SOS response. It is likely that this induction in turn leads activation of other TA loci, the promoters of which contain a Lex box. It has previously been shown that ciprofloxacin can induce formation of persisters by SOS-mediated activation of the tisAB, as well as other TA systems, which in turn may lead to dormancy (17). A similar mechanism may hold for CcdB also and this is outlined in Fig. 5. A stress that leads to elevated levels of Lon will lead to release of CcdB from its complex with CcdA and the resulting DNA lesion will result in an SOS response, ultimately triggering conversion of cells to a dormant state.
Fig. 5.
Schematic representation of CcdB induced persister formation. A stress activates Lon protease-mediated CcdA degradation that leads to release of CcdB from the CcdA-CcdB complex. Free CcdB poisons DNA Gyrase by forming a cleaved DNA-gyrase-CcdB ternary complex. This process leads to induction of the RecA mediated SOS response, which in turn activates other TA systems, ultimately leading to formation of multidrug-tolerant persister cells.
Materials and Methods
E. coli host strains and plasmids used for this study are listed in Table S3. The Top10 strain, which lacks the F-plasmid–encoded WT CcdA and CcdB, was used to select inactive mutants of CcdB. XL1-Blue, Top10F′ and AB264 strains have the F-plasmid that carries the ccd operon. All strains except AB264 are recA−. BW25113 is a WT E. coli K12 strain that was used for chromosomal insertions of the ccd operon and cat cassette. The ccdB gene was expressed under control of the arabinose PBAD promoter in plasmid pBAD24ccdB. The CcdB mutants for the study were taken from the library of CcdB mutants described previously (12). The GyrA14 fragment (residues 363–494) (11), CcdA (27), and CcdB (28) were expressed and purified in E. coli. Additional methodological details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Profs. John E. Cronan, Remy Loris, and Laurence Van Melderen for the wild GFP, GyrA14, and CcdA plasmids, respectively; Anusmita Sahoo and Bharat Adkar for preparing Fig. 1 and Table S1; and Dr. Pranveer Singh for the construction of the Top10F′ Escherichia coli strain. A.T. and P.C.D. are Council of Scientific and Industrial Research Fellows.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121217109/-/DCSupplemental.
References
- 1.Hayes F. Toxins-antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 2.Van Melderen L, Bernard P, Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994;11:1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 3.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: More than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci USA. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 7.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 8.Ogura T, Hiraga S. Partition mechanism of F plasmid: Two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 9.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 10.Bajaj K, Chakrabarti P, Varadarajan R. Mutagenesis-based definitions and probes of residue burial in proteins. Proc Natl Acad Sci USA. 2005;102:16221–16226. doi: 10.1073/pnas.0505089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dao-Thi MH, et al. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol. 2005;348:1091–1102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 12.Bajaj K, et al. Structural correlates of the temperature sensitive phenotype derived from saturation mutagenesis studies of CcdB. Biochemistry. 2008;47:12964–12973. doi: 10.1021/bi8014345. [DOI] [PubMed] [Google Scholar]
- 13.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afif H, Allali N, Couturier M, Van Melderen L. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol Microbiol. 2001;41:73–82. doi: 10.1046/j.1365-2958.2001.02492.x. [DOI] [PubMed] [Google Scholar]
- 15.Bahassi EM, Salmon MA, Van Melderen L, Bernard P, Couturier M. F plasmid CcdB killer protein: CcdB gene mutants coding for non-cytotoxic proteins which retain their regulatory functions. Mol Microbiol. 1995;15:1031–1037. doi: 10.1111/j.1365-2958.1995.tb02278.x. [DOI] [PubMed] [Google Scholar]
- 16.Couturier M, Bahassi el-M, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 17.Dörr T, Vulić M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 19.Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Aguirre-Ramírez M, Ramírez-Santos J, Van Melderen L, Gómez-Eichelmann MC. Expression of the F plasmid ccd toxin-antitoxin system in Escherichia coli cells under nutritional stress. Can J Microbiol. 2006;52:24–30. doi: 10.1139/w05-107. [DOI] [PubMed] [Google Scholar]
- 21.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Jonge N, et al. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol Cell. 2009;35:154–163. doi: 10.1016/j.molcel.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 23.Wilbaux M, Mine N, Guérout AM, Mazel D, Van Melderen L. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J Bacteriol. 2007;189:2712–2719. doi: 10.1128/JB.01679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah D, et al. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mine N, Guglielmini J, Wilbaux M, Van Melderen L. The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species. Genetics. 2009;181:1557–1566. doi: 10.1534/genetics.108.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madl T, et al. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J Mol Biol. 2006;364:170–185. doi: 10.1016/j.jmb.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 28.Bajaj K, Chakshusmathi G, Bachhawat-Sikder K, Surolia A, Varadarajan R. Thermodynamic characterization of monomeric and dimeric forms of CcdB (controller of cell division or death B protein) Biochem J. 2004;380:409–417. doi: 10.1042/BJ20031528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.