Abstract
Proapoptotic drugs are a mainstay of cancer drug treatment. These drugs stress cells and ultimately trigger the activation of caspases, cysteine-class proteases that cleave after aspartic acid and deconstruct the cell. It is well known that cells respond differently to proapoptotic cancer drug treatments. Here, using a global and unbiased quantitative N-terminomics technology, we show that ∼500 products of caspase cleavage and their kinetics vary dramatically between cell type and cytotoxic drug treatment. It is likely that variations arise from differences in baseline proteome composition of the cell type and the alterations induced by drug treatments to yield a unique cohort of proteins that caspases finally target. Many targets are specific to both drug treatment and cell type, providing candidate-specific biomarkers for apoptosis. For example, in multiple myeloma cells treated with the proteasome inhibitor bortezomib, levels of activating transcription factor-4 increase dramatically early in drug treatment and then decrease upon cleavage by activated caspases. Thus, caspase-derived cleavage products are a sensitive reflection of cell-type and drug-induced stress, and provide useful fingerprints for mechanisms of drug action and response.
Keywords: mass spectrometry, selected reaction monitoring, proteomics, degradomics
Apoptosis, programmed cell death, is a process conserved in all metazoans that is critical for removing unneeded or diseased cells. Blocking apoptosis is critical to cancer cell survival, and promoting apoptosis is key to most cancer chemotherapies. Apoptosis can be triggered principally by two pathways, depending on whether the inducing ligand interacts with membrane receptors (the extrinsic pathway) or by inducing internal cellular stress ultimately via the mitochondria (the intrinsic pathway). Many cancer drugs can induce apoptosis through different targets in these pathways; examples include blocking protein kinases or topoisomerases, or inhibiting the proteasome to induce cellular stress. Moreover, different cell lines show differential sensitivity to these drugs for reasons that are often poorly understood. Drug-induced cellular stresses initially cause changes in transcription and translation as cells struggle to reduce the insult (1). However, when this fails, apoptosis often ensues and ultimately converges on the activation of a family of cysteine-class aspartate specific proteases, known as caspases.
The apoptotic caspases are activated in two waves. First, the initiator caspases, such as caspases 8 and 9, sense the extrinsic or intrinsic pathways, respectively, and are activated. These caspases in turn cleave and activate the primary executioner caspases 3 and 7. Large-scale proteomics studies in cells have shown the caspases together cut hundreds of proteins generally at single sites (2–5). Recently, using a label-free quantitative proteomics approach called selected reaction monitoring (SRM) coupled with a positive enrichment N-terminomics approach, the substrate proteins have been shown to be cleaved at rates that vary more than 500-fold, suggesting an order to the process (6). These substrate proteins represent key nodes in maintaining cellular homeostasis and some are even known cancer drug targets (7).
Given the importance of understanding the common targets that are cleaved by caspases, and the mechanisms by which chemotherapeutic agents induce apoptosis in different cells, we used SRM analysis to follow the rates of caspase-cleaved products in three different hematopoietic cancer cell types treated with three different cytotoxic drugs that induce the intrinsic apoptotic pathway. We found much the same collection of cleaved proteins could be detected at some level in these cells, but their relative abundances varied dramatically. Moreover, different drugs caused huge alterations in the patterns and rates of apoptotic substrates that accumulated even in the same cell type. This finding suggested the nature of the cellular stress triggers differential production of proteins followed by activation of the apoptotic caspases that cut them. These studies further revealed common apoptotic substrates irrespective of cell type or drug, which is consistent with the hypothesis that some targets are critical nodes in cellular homeostasis. We also found cell-type and drug-specific pathways consistent with changes in substrate abundance and differential responses to cellular stresses. These studies begin to address the importance of critical targets for inducing apoptosis and exemplify sensitive mechanistic fingerprints for the action of cancer drug classes for inducing apoptosis.
Results
N-Terminal Proteome in Apoptotic Hematological Cells.
To selectively identify caspase-derived peptides in cells undergoing apoptosis, we used an approach that uses subtiligase to selectively label and enrich for N-terminal peptides before mass spectrometric analysis (Fig. S1) (4, 8). The advantage of this approach is that most α-amines derived from the translation start site are acetylated (9), so that the bulk of N termini detected are the result of either constitutive or regulated proteolysis. The subtiligase enrichment thus labels a small set of proteolyzed proteins, which greatly simplifies the sample complexity and is ideally suited for SRM analysis.
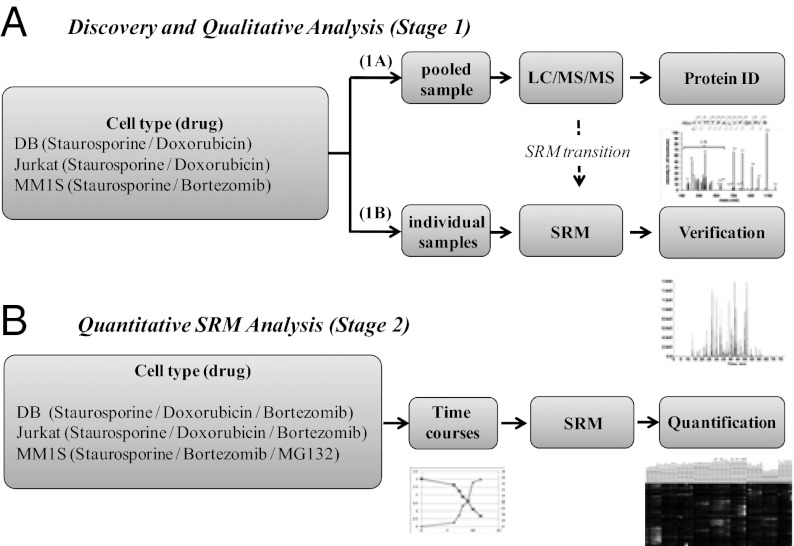
As shown in Fig. 1, the SRM process is divided into two stages: first, the identification of target peptides and generation of SRM transitions (stage 1) and second, the quantification of the relative abundance of those peptides using SRM (stage 2). To generate a comprehensive list of apoptotic-cleaved proteins in stage 1, we used three different hematopoietic cell lines: Jurkat, a T-cell line; DB, a B-cell line; and MM1S, a plasma cell line derived from multiple myeloma. These lines were induced to undergo apoptosis with two different drugs, staurosporine as the common inducer, and either doxorubicin or bortezomib as clinically relevant cytotoxic drugs. Cells were harvested at ∼50% and 75% cell death as monitored by cellular ATP levels. Following sample enrichment for free α-amine N termini (Fig. S1), aliquots from all conditions were pooled for discovery experiments using a QTOF LC-MS/MS.
Fig. 1.
Two-stage process for quantitative identification of caspase-derived peptides in three different cell lines each treated with three different drugs. (A) Discovery experiment to identify all possible α-amino termini in apoptotic cells. Three cell lines were treated with two apoptosis inducers per cell line to a level of ∼10% and 50% cell viability. The apoptotic cells were pooled for all conditions, lysed, and the free α-amino termini were biotinylated with subtiligase, and N-terminal tryptic peptides were isolated (Fig. S1). The N-terminal peptides were identified by LC-MS/MS using a QTOF instrument and the Protein Prospector informatics suite (stage 1A). The seven intense fragment ions from MS/MS spectra from each peptide were selected and collision energy for each transition was optimized for SRM analysis (stage 1B). (B) Having validated SRM transitions for caspase-derived peptides, SRM analysis was carried out for each of three different cell lines treated with three different cytotoxic drug as a function of time and degree of apoptosis (stage 2).
From these large-scale discovery runs, we identified a total of 2,083 α-amine–containing peptides from 1,256 proteins, with a false-discovery rate (FDR) of 2.0% (Table 1 and Dataset S1). To determine the number of putative endoproteolytic processing events, we subtracted away the 106 peptides with N termini within 3 aa of the translation initiation site (suggesting N-terminal exoproteolysis) and the 435 cut sites within the first 40 aa of the translation start site that contained a hydrophobic sequence typical of signal peptides or organelle transit peptides. This result left a total 1,542 peptides that are candidates for constitutive or regulated endoproteolysis. These sites are distributed among 916 proteins, and the ratio (1,542 cuts/916 proteins) indicates an average of 1.7 cuts per protein target within this general class of endoprotease substrates.
Table 1.
Summary of discovery experiments in pooled apoptotic cells (stage 1A)
| Total proteins identified | Sites | Proteins |
| Total α-amines | 2,083 | 1,256 |
| α-Amines from N-terminal protease* | 106 | 99 |
| α-Amines from signal or transit peptide removal† | 435 | 351 |
| α-Amines from endoprotease processing‡ | 1,542 | 916 |
| Endoprotease processing/protein | 1,542/916 = 1.68 | |
| Total Asp cleavage | 674 | 564 |
| α-Amines from endoproteolytic caspase cleavage‡ | 557 | 470 |
| Endoproteolytic Asp cleavage/protein | 557/470 = 1.19 | |
*α-Amine < 4 amino acids of translation and initiation.
†α-Amine 4 to 40 amino acids of translation and containing signal peptide.
‡α-Amine > 40 amino acids.
Of the 916 proteins processed by endoproteolysis, 674 sites were cut after aspartic acid. Removing the aspartic acid cuts within the first 40 residues of the start site, which eliminates those that could be cut by signal peptide processing proteases, leaves 557 sites as high-confidence caspase cut sites. These 557 cuts were distributed among 470 proteins, indicating an average of 1.2 caspase cuts per protein, which is considerably lower than the average cuts for the general endoprotease substrate class. Of the total number of 2,083 peptides identified in these pooled experiments, about one-third (674/2083) followed aspartic acid, representing a large fraction of the total proteolytic events. These 674 sites yielded a total of 846 identifiable peptide precursor ions (including different charge states, missed cleavages by trypsin, and methionine oxidations). We were interested in determining how the N-terminal dataset we found was affected by protein abundance. Representation of proteins by known abundance [using PaxDb (pax-db.org)] reveals broad coverage of proteins across five to six orders-of-magnitude (Fig. S2 A and B).
SRM for Quantitative Analysis.
To permit relative quantification of peptides across samples, we used SRM on a QTRAP mass spectrometer. SRM permits rapid and highly sensitive peptide quantification because it does not require acquisition of full mass spectra (10, 11). Instead, the presence of validated precursor-fragment ion pairs, or transitions, unique to each peptide provides positive identification. For the purpose of SRM, transitions were determined for the putative 846 identifiable caspase-derived peptide precursor ions identified in the pooled sample from stage 1A. These peptides correspond to the 674 aspartic acid cleavage sites. For each peptide of interest, the seven most intense fragment ions observed in stage 1A were selected for every precursor. Transitions were then verified by performing SRM analysis on the individual samples that comprised the pooled sample in stage 1A. The detected peptides were qualified by having at least five of seven coeluting transitions on HPLC present in one or more samples (see Methods). Using these criteria, 527 of the 637 unique peptides (81.9%) were detected by SRM analysis of the individual samples. The 110 peptides not detected were generally of low abundance or did not generate sufficient expected transitions to allow for high-quality identification. SRM analysis of apoptotic cell lysates allowed confident identification of 488, 505, and 520 peptides in DB, Jurkat, and MM1S cell lines, respectively. A common set of 472 peptides (89.6%) from multiple samplings could be detected at some level in all three cell lines indicating good coverage and sensitive detection (Fig. S2D, Table S1, and Datasets S2 and S3A).
To evaluate systematic variation within a given sample, we determined the average coefficient of variation for a sample run in triplicate. The averaged coefficient of variation was 12.3% for 246 peptides whose peak intensities were greater than 1.0 × 104 (Fig. S3A). Moreover, linearity in peptide intensities for these cell lysate samples were shown in sequentially diluted cell lysate samples (Fig. S3B). Furthermore, a strong correlation was observed in triplicate runs of Jurkat cells treated with doxorubicin; samples from four different time points clustered together by Spearman correlation (Fig. S3C). For further quantitative analysis, we calculated the rms error of peak intensity ratios (Fig. S3D). All of the calculated rms errors from decoy transitions (see Methods) were greater than 0.5 and therefore we only analyzed peptide data where the rms error was less than 0.4 for the quantification analysis of stage 2 (Dataset S2). To evaluate the biological reproducibility, Jurkat cells were treated individually with different concentrations of doxorubicin and compared at similar extents of cell death. The peptides detected showed good correlations for the peaks with highest intensities, even when treated with different drug concentrations (Fig. S3 E and F). To address the systematic error and reproducibility in the following SRM experiments, samples were analyzed in duplicate in random order and signal intensity was normalized to the intensities of internal standard proteins that were added at the time of cell lysis and normalized to total protein concentration.
Caspase-Derived Peptide Profiling in Apoptosis.
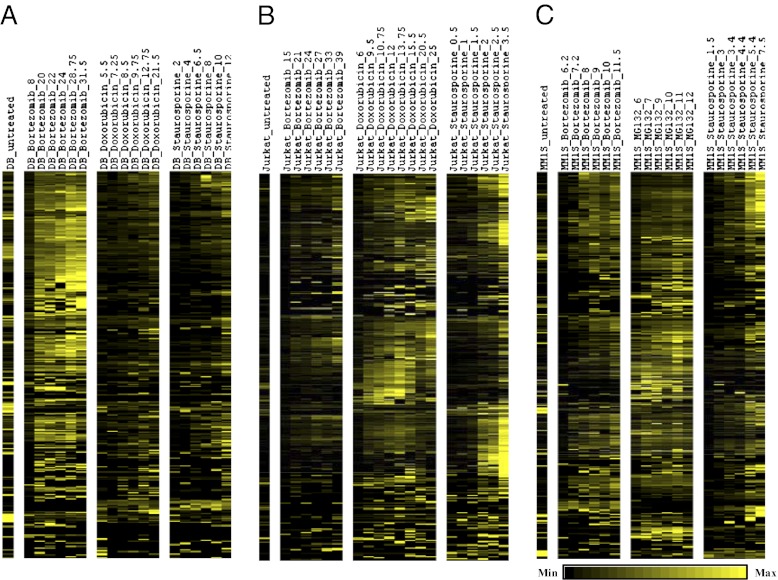
Caspase-derived peptide profiles were determined for Jurkat, DB, and MM1S cell lines treated with three different drugs at multiple time points over the entire cell death time course (Fig. 1) (stage 2). Each sample (cell line treated with drug and harvested at specific cell viabilities) was analyzed by SRM and we found 628 target peptides (including different charge states, missed cleavages by trypsin, and methionine oxidations). Although there is considerable overlap in peptides identified across cell lines and inducers, there is wide variation in peptide signal intensities across conditions and time (Fig. 2 and Dataset S3B). This finding suggests either a difference in abundance of the apoptotic proteins or a differential effect on amount or activation of specific caspases. To evaluate if there were differences in relative caspase activation rates, we tested the rates of activation of the major apoptotic caspases (3, 7–9) upon treating the MM1S cell line with bortezomib, MG132, and staurosporine using immunoblots (Fig. S4). We see that the executioner caspases (3, 7) are induced after a lag at similar rates and their activation relative to each other does not change with drug treatment. Similar effects were seen for the initiator caspases (8, 9). Thus, the differences in the MM1S cells treated with different drugs do not appear to be because of differential activation of different caspases.
Fig. 2.
Heat maps of the 628 selected caspase-derived peptides (stage 2). Three cell lines, (A) DB, (B) Jurkat, and (C) MM1S were treated with three different cytotoxic drugs. SRM permits relative quantification of each peptide. Across each time-course profile, the summed intensities of each peptide were normalized to 1.0 to show relative differences.
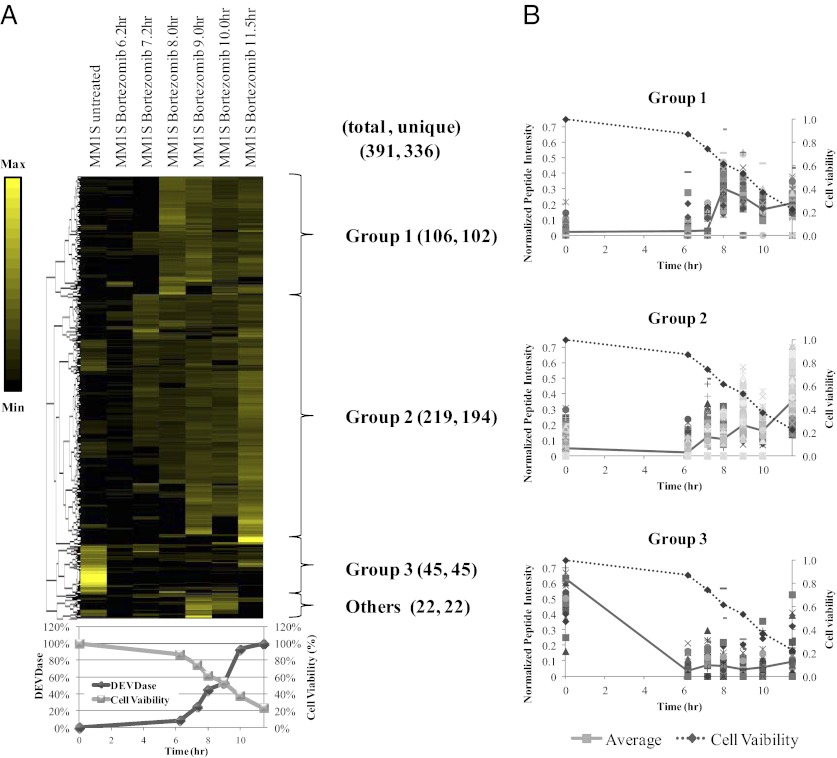
We next went on to dissect one of these cell lines and drug treatment in more detail. The time course of appearance for caspase-derived peptides of bortezomib-treated MM1S cells can be categorized into three groups using an unbiased complete-linkage hierarchical clustering (Fig. 3). Group 1 represents cleavage peptides whose abundance peaks at ∼50% cell death and then decreases or stabilizes over time. Group 2 represents peptides whose abundance continually increases as cell death proceeds. Group 3 represents peptides whose abundance decreases over time or is low throughout the time course. For most peptides, level of the caspase-derived peptides increases with the extent of cell death. Approximately 56% of caspase-derived peptides have their peak abundance at the last time point measured (group 2; i.e., the lowest cell viability). Another 33% have peak abundance at ∼50% cell viability and gradually decrease or stabilize with time (group 1) or remain at low but detectable levels throughout apoptosis (group 3).
Fig. 3.
Profile of caspase-derived peptides from MM1S cells treated with bortezomib. (A) Heat map view for unbiased hierarchically clustered peptides. The peptides were normalized so that the sum of intensities across all points in the time course equaled 1. Using unbiased complete-linkage clustering, peptides were classified into three major groups (groups 1, 2, and 3, described in the text). (B) Plot of cell viability (dashed line) and the normalized peptide intensities (solid line) of each group.
Caspase-Derived Peptide Profiling for Proapoptotic Drug and Cell-Types.
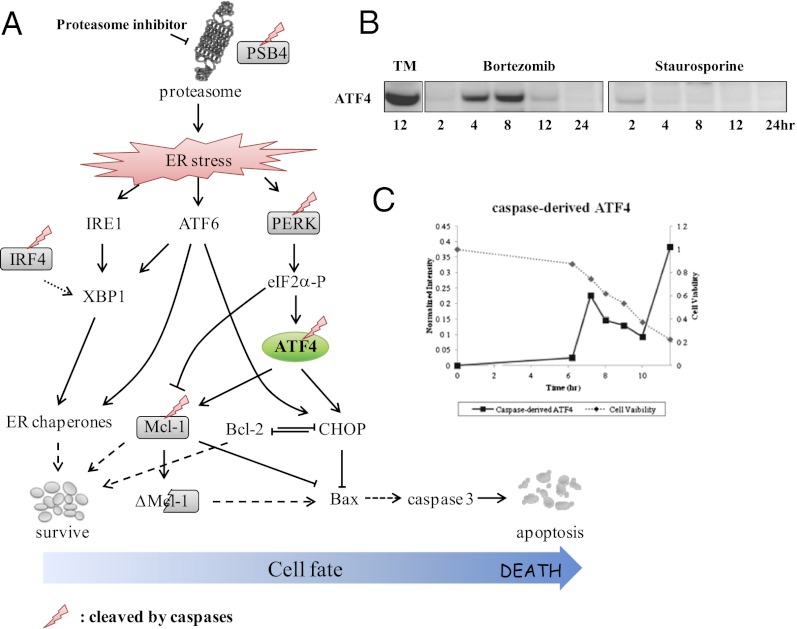
With the interest to develop cell-type and inducer-specific fingerprints, we determined if there are peptides that are specific to particular inducers or cell types. Peptides were stratified based on their proportional representation within a particular inducer or cell type (see Methods). There are clear cell-type and inducer-specific peptides listed in Table S2 and Table S3. Review of inducer-specific peptides also reveals proteins implicated in drug-specific mechanisms. For example, among the inducer-specific peptides identified is activating transcription factor-4 (ATF4), a transcription factor that is up-regulated in the unfolded protein response during endoplasmic reticulum (ER) stress (Fig. 4A). To further validate ER stress-mediated ATF4 as an inducer specific peptide, MM1S cells were treated with bortezomib or staurosporine over 24 h for immunoblotting. Only MM1S cells exposed to bortezomib showed induction of ATF4 (Fig. 4B). Levels of full-length ATF4 peak at 8 h following exposure to bortezomib. Caspase levels rise to about 50% activation at 8 h and levels of cleaved ATF4 then peak at 12 h when cell viability is less than 20% (Fig. 4C). Cleaved ATF4 is also specifically present in DB cells treated with bortezomib and in MM1S cells treated with MG132, but not in DB cells treated with other inducers (Dataset S3B). ATF4 represents a sterling example of a drug-specific target, which is induced upon celluar stress and subsequently cleaved by caspases.
Fig. 4.
ATF4 plays crucial roles in ER stress-induced apoptosis. (A) ER stress induced by the proteasome inhibitor bortezomib initiates the three different pathways in the unfolded protein response: IRE1, ATF6, and pERK. ATF4 is a member of the pERK pathway. (B) ATF4 is up-regulated in MM1S cells treated with bortezomib, but not with the pan-kinase inhibitor, staurosporine. Tunicamycin (TM) induces ER stress and is shown as a positive control. Total protein levels were adjusted by BCA assay. (C) Plot of normalized intensity of caspase-derived ATF4 detected in MM1S cell treated with bortezomib (time along x-axis) by SRM (solid line). MM1S cell viability is also shown (dashed line).
Discussion
Using quantitative mass spectrometry, we have characterized the degradomes of three cell lines representing hematologic malignancies treated with different intrinsic apoptotic drugs. The subtiligase N-terminome enrichment method provided highly reproducible results for both technical and biological replicates, and excellent coverage of the proteome. To detect low abundance peptides, we pooled the samples just before the discovery experiment and used high resolution 2D LC-MS/MS to reduce the complexity of the samples (Figs. S2 and S3). This process allowed us to broadly identify ∼900 proteins that are products of endoproteolysis in aggregate samples. Over half of these were caspase-derived, as evidenced by cuts between aspartic acid and a small amino acid residue (Fig. S2C). From these discovery data we developed and validated the SRM transitions, which provided a rapid and sensitive method for relative quantification to follow the kinetics for cleaving >500 caspase targets during apoptosis in a label-free fashion. Moreover, the sample coverage was excellent in that 89% of all peptides were detected at some level in all three cell lines. By following the level of caspase activation and corresponding cell death, it was possible to normalize cell-line and inducer-specific effects to comparable extents of apoptosis in the cultures.
One of the most striking results is that within any cell line, the kinetic patterns for production of caspase substrates differ widely depending on drug treatment, and far beyond stochastic variability in the data (Fig. 2). We have previously shown that caspases 3, 7, and 8 each have preferred sets of substrates and cleave their targets at rates that range >500-fold (6), so it was possible that the differences were because of differential caspase activation. However, we found no difference in the ratios of activation of the major apoptotic caspases by treatment of MM1S cells with either bortezomib, MG132, or staurosporine (Fig. S4). Cohen et al. used fluorescent tagging of ∼1,000 proteins in H1299 lung carcinoma cells to show that treatment with the cytotoxic camptothecin, a topoisomerase I inhibitor, caused dramatic changes in protein levels and localization; these effects even varied between cells (12). Hsieh et al. recently showed that treatment of PC3 cells with a mammalian target of rapamycin inhibitor caused significant changes in the proteome as assessed by ribosomal profiling (13). These drug-induced alterations to the proteome are superimposed on presumed differences in the baseline proteome of each cell-type, as suggested by transcriptome analyses (14, 15). The functional consequences of drug-induced alterations have been shown by Wolpaw et al. to permit classification of apoptotic drugs based on these “modulatory profiles” (16). We hypothesize that the large cell-type and drug-specific differences in kinetic patterns we observe for caspase-derived products is because of the drug treatment causing specific alterations in the proteome before activation of the caspases.
Previous proteomic studies have studied the kinetics for cleavage of caspase substrates in Jurkat cells with the single inducer, staurosporine (3, 6). These studies showed a wide variation in rate at which substrates are cleaved, and that the bulk of products persist in some form after they are cleaved. The data presented here with a broader panel of cells and inducers (Fig. 2) are fully consistent with these findings. As a more detailed example, an unbiased clustering analysis of ∼400 caspase targets in MM1S cells treated with bortezomib over extended times shows three broad categories: group 1 (∼33% of targets) is cleaved and ultimately degraded; group 2 (∼50% of targets) is cleaved and levels are sustained; and group 3 (∼10% of targets), the levels of which are persistently low; the remaining 15% could not be clustered (Fig. 3). Boisvert et al. have shown a correlation between protein abundance and turnover rates by using pulsed stable isotope labeling by amino acids in cell culture methods in homeostatic HeLa cells (17). Protein stability indices have also been measured by Yen et al. by measuring the half-lives of EGFP fusion proteins (18). We did not see a significant correlation between the group 1 and group 2 peptides based on these expected half-lives or abundances, which may be because of differences in protein stabilities in homeostatic compared with apoptotic cells. Alternatively, correlation may not have been observed because of further modification or cleavage to other stable peptide forms.
To identify common targets across all cell lines that may be fundamental to apoptosis, we correlated peptide intensity with the degree of cell death across all inducers and all cell types (Table S4). The top 15 targets had a correlation coefficient across the nine cell and treatment conditions ranging from R values of 0.87–0.76. Gene ontology (GO) term analysis for the aggregate distribution of the caspase-cleaved proteins is not distinctly different from the whole proteome. However, enrichment in GO terms is observed for this small common set, particularly with RNA splicing and mRNA metabolism. Specific components of spliceosome core proteins have previously been identified as targets of caspase cleavage during FAS-induced apoptosis (19) and 9 of 15 proteins are reported as the proteins with high catalytic efficiencies for caspases (6) (Table S4). For example, perilipin-3 (also known as TIP47) is a 47-kDa mannose 6-phosphate receptor-binding protein that returns mannose 6-phosphate receptors from the late endosomes back to the trans-Golgi network. Overexpression of TIP47 has been shown to protect NIH 3T3 cells from taxol-induced cell death and siRNA suppression of TIP47 facilitates cell death (20). Given its antiapoptotic function, cleavage of TIP47 and other antiapoptotic proteins by caspases is one mechanism by which apoptosis ensues. These are small numbers to draw firm conclusions but these begin to highlight common and aggressive targets that caspases attack. Time will tell if these make for useful cancer drug targets.
The cell type and inducer-specific nature of caspase cleavage profiles presents opportunities to dissect the pathways and proteins that define a cell-type and its response to drugs that induce apoptosis. An example of a cell-type–specific peptide detected with near exclusivity in a single cell-type in the present work is IFN regulatory factor 4 (IRF4), which is required for lymphocyte activation and the generation of Ig-secreting plasma cells (Table S2). The exclusivity observed here corroborates previous work that has shown IRF4 expression levels in MM1S cells to be higher than any other cell line (21–24). Shaffer et al. (25) have also shown that IRF4 activity is dysregulated in multiple myeloma cell lines. Clinically, IRF4 transcript levels in plasma cells inversely correlate with survival and IRF4 has therefore been proposed as a prognostic biomarker and therapeutic target (25).
It is intriguing that many of the caspase products are highly drug-induced and suggest that caspases may be attacking the very stress-response proteins the cells are using to survive. A clear example of this is ATF4, the expression of which is drug-specific; it is induced upon treatment with bortezomib, a proteasome inhibitor, but not staurosporine a general kinase inhibitor. ATF4 levels peak after 8 h of bortezomib treatment. Here we show ATF4 is a caspase substrate, with a cleavage site at Asp65 and that full-length ATF4 levels fall after caspases are induced. A fragment corresponding to the C-terminal end of ATF4 were not observed; as the functional domains of ATF4 are near the C terminus, the absence of a stable fragment suggests that ATF4 is inactive after caspase cleavage. ATF4 is known to be an important part of the pERK pathway and up-regulates antiapoptotic protein Mcl-1 (26) (Fig. 4A). Thus, a loss-of-function cleavage of Mcl-1 at Asp157 may redundantly enhance cell death. There may be many more examples of this kind of regulation in our dataset and further detailed analysis, which is beyond the scope of this article, should reveal them.
In sum, these studies reveal the remarkable sensitivity of caspase-cleavage profiles for drug and cell-type. Further studies will reveal how these drugs induce differences in protein-expression profiles and whether these are preferred targets of caspase cleavage. These data may aid in the development of specific apoptotic biomarkers for determining drug action or cell-type–specific cell death. Clinically, malignancy and drug-specific caspase-derived peptides may serve as early readouts of drug efficacy both before and during therapy, and possibly lead to identification of new targets and pathways for cancer drug treatments.
Methods
Cell Lines.
Jurkat clone A3 (CRL-2570), DB (CRL-2289), and MM1S (CRL-2974) were purchased from the ATCC. All of the media and supplements for cultivation were obtained from the University of California at San Francisco Cell Culture Facility.
Jurkat cells were grown in RPMI-1640 supplemented with 10% (vol/vol) FBS, 25 mM Hepes, 1 mM sodium pyruvate, and antibiotics [0.0064% (wt/vol) penicillin, and 0.01% (wt/vol) streptomycin]; DB cells were grown in RPMI-1640 supplemented with 10% FBS, 10 mM Hepes, 1 mM sodium pyruvate, and antibiotics; and MM1S was grown in media supplemented with 10% FBS, 2 mM glutamine, and antibiotics. Cells were cultured at a density of 1 × 106 cells/mL and treated with appropriate drug [bortezomib, doxorubicin, staurosporine (LC Laboratories), or MG132 (Tocris)]. The caspase activities and cell viabilities were monitored with Ac-DEVD-AFC caspase substrate and Cell Titer-Glo (Promega) to monitor ATP levels, respectively. The harvested cells were washed with phosphate buffer saline and the pellets were stored in a –80 °C freezer until use.
N-Terminal Isolation.
The pellets were lysed by sonication in 4.0% SDS, 400 mMBicine (pH8.0), containing the protease inhibitors; 0.1 mM z-VAD-fmk, 0.1 mM E-64, 1 mM AEBSF, 1mM PMSF, and 5 mM EDTA. Proteins were reduced by 2 mM TCEP at 90 °C for 15 min, and alkylated with 4 mM iodoacetamide (IAM) in dark for 1 h at room temperature. The IAM reaction was then quenched with 10 mM DTT. One-percent Triton-X was added followed by water [final concentrations 1% (wt/vol) SDS, 1% (wt/vol) Triton X-100, 100 mM Bicine]. Before biotinylation, 2 nM of human adrenocorticotropic hormone (fragment 1–17) (ACTH) and 2 nM of recombinant human somatotropin (rhGH) were added to the lysates as internal standards. For the biotinylation, 1 mM TEVest4 (10 mM in DMSO) (Fig. S1B), and 1 μM subtiligase were added to the lysate and allowed to react for 1 h at room temperature. Biotinylated peptides were precipitated in acetonitrile and resolved in 8 M guanidine hydrochloride before a second precipitation in cold ethanol at –80 °C for 16 h. The precipitates were resolved in 5.3 M guanidine hydrochloride and were captured on immobilized NeutrAvidin agarose beads (Thermo Fisher Scientific). The beads were washed with 5 M guanidine hydrochloride and on-bead digestion was performed overnight using sequence-grade trypsin [Promega, in 100 mM Bicine (pH 8.0), 200 mM sodium chloride, 20 mM calcium chloride, 1 M guanidine chloride]. N-terminal peptides were isolated by TEV protease [in 0.4 μM in 100 mM ammonium bicarbonate (ABC), 2 mM DTT and 1 mM EDTA] for 4 h. Peptides were desalted with ZipTip Pipette Tips (Millipore), evaporated and stored at −80 °C.
LC-MS/MS.
For discovery experiments, peptides were analyzed via off-line 2D reverse-phase LC-MS/MS. Samples underwent gradient elution HPLC fractionation on an XBridge C18 column (1.0 × 100 mm, 3.5 μm; Waters), using the following conditions: acetonitrile and water in a 70-min linear gradient, with 100 mM ABC (pH 11). Fractions were analyzed by nanoAcquity UPLC System (Waters) coupled with a QSTAR Elite System (Applied Biosystems; current AB/Sciex), using the following conditions: 0.1% formic acid in water (eluent A) and 0.1% formic acid in acetonitorile (eluent B) in a 90-min linear gradient. Injected samples were trapped on a Symmetry C18 Column (0.18 × 20 mm, 5 μm; Waters) for 5 min at 1% of eluent B before start the gradient; a BEH130 C18 column (0.075 × 200 mm column, 1.7 μm; Waters), with a flow rate of 1 μL/min was used for analysis. A QTRAP 5500 LC-MS/MS system (AB/Sciex) was used for a SRM experiments. Before quantitative experiments by SRM, a collision energy scan was performed to optimize the collision energy. The FDR for SRM analysis was evaluated by analyzing the sample with randomized sequenced peptides with increasing precursor m/z by 10 for decoy transitions (27).
Peptide Identification and Generation of SRM Transitions.
Protein Prospector (version 5.6.2 or later) (University of California at San Francisco) was used to identify the N-termini Abu-labeled peptides from the Swiss-Prot database (2011.01.11) from the discovery experiment. The FDR was calculated from decoy database of random sequenced proteins. The SRM transition were generated using experimental data derived from discovery experiments with in-house perl scripts (SI Text). For details about data analysis, bioinformatics, immunoblotting of caspase cleavages, and cell viability asays, see SI Methods.
Supplementary Material
Acknowledgments
We thank Nicholas Agard, Julie Zorn, and Emily Crawford for their critical suggestions; Carmela Sidrauski and Diego Acosta-Alvear for providing antibodies for immunoblotting; Peter Baker, Aenoch Lynn, Robert Chalkley, and Takahiko Muramatsu for Protein prospector and perl script; and members of the J.A.W. laboratory for assistance with reagents. This research was supported by National Institutes of Health Grant R01 GM081051 (to J.A.W.); the Rogers Foundation; and the Stephen and Nancy Grand Multiple Myeloma Translational Initiative. Mass spectrometry was performed at the Bio-Organic Biomedical Mass Spectrometry Resource at University of California at San Francisco (A.L.B., Director), supported by the Biomedical Research Technology Program of the National Institutes of Health National Center for Research Resources, Grants P41RR001614 and 1S10RR026662.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208616109/-/DCSupplemental.
References
- 1.Geva-Zatorsky N, et al. Protein dynamics in drug combinations: A linear superposition of individual-drug responses. Cell. 2010;140:643–651. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Agard NJ, Maltby D, Wells JA. Inflammatory stimuli regulate caspase substrate profiles. Mol Cell Proteomics. 2010;9:880–893. doi: 10.1074/mcp.M900528-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wildes D, Wells JA. Sampling the N-terminal proteome of human blood. Proc Natl Acad Sci USA. 2010;107:4561–4566. doi: 10.1073/pnas.0914495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agard NJ, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 8.Agard NJ, Wells JA. Methods for the proteomic identification of protease substrates. Curr Opin Chem Biol. 2009;13:503–509. doi: 10.1016/j.cbpa.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picotti P, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 12.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitsiades CS, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: Biological and clinical implications. Proc Natl Acad Sci USA. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shringarpure R, et al. Gene expression analysis of B-lymphoma cells resistant and sensitive to bortezomib. Br J Haematol. 2006;134:145–156. doi: 10.1111/j.1365-2141.2006.06132.x. [DOI] [PubMed] [Google Scholar]
- 16.Wolpaw AJ, et al. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc Natl Acad Sci USA. 2011;108:E771–E780. doi: 10.1073/pnas.1106149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisvert F-M, et al. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics. 2011;11 doi: 10.1074/mcp.M111.011429. M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 19.Van Damme P, et al. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- 20.Hocsak E, et al. TIP47 protects mitochondrial membrane integrity and inhibits oxidative-stress-induced cell death. FEBS Lett. 2010;584:2953–2960. doi: 10.1016/j.febslet.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 21.Heintel D, et al. Expression of MUM1/IRF4 mRNA as a prognostic marker in patients with multiple myeloma. Leukemia. 2008;22:441–445. doi: 10.1038/sj.leu.2404895. [DOI] [PubMed] [Google Scholar]
- 22.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 23.Mittrücker H-W, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 24.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, et al. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood. 2012;119:826–837. doi: 10.1182/blood-2011-07-366492. [DOI] [PubMed] [Google Scholar]
- 27.Reiter L, et al. mProphet: Automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.