Abstract
HLA-DM (DM) plays a critical role in Ag presentation to CD4 T cells by catalyzing the exchange of peptides bound to MHC class II molecules. Large lateral surfaces involved in the DM:HLA-DR (DR) interaction have been defined, but the mechanism of catalysis is not understood. In this study, we describe four small molecules that accelerate DM-catalyzed peptide exchange. Mechanistic studies demonstrate that these small molecules substantially enhance the catalytic efficiency of DM, indicating that they make the transition state of the DM:DR/peptide complex energetically more favorable. These compounds fall into two functional classes: two compounds are active only in the presence of DM, and binding data for one show a direct interaction with DM. The remaining two compounds have partial activity in the absence of DM, suggesting that they may act at the interface between DM and DR/peptide. A hydrophobic ridge in the DMβ1 domain was implicated in the catalysis of peptide exchange because the activity of three of these enhancers was substantially reduced by point mutations in this area.
HLA-DM (DM)3 was discovered by analysis of mutant B cell lines in which Ag presentation by MHC class II molecules to CD4 T cells is defective (1, 2). MHC class II molecules assemble in the endoplasmic reticulum with invariant chain, which protects the hydrophobic peptide binding site before MHC class II molecules reach the endosomal compartment (3, 4), where invariant chain is proteolytically degraded such that only the CLIP fragment remains in the peptide binding site (5). DM substantially accelerates the dissociation of CLIP and thereby facilitates binding of peptides created by limited proteolysis of foreign and self Ags (6–8). DM also accelerates the dissociation of other MHC class II-bound peptides, and this editing function favors the presentation of high-affinity peptides, an aspect that is important in antimicrobial T cell responses (7–11). High-affinity peptides are bound to MHC class II molecules with half-lives of several days or even weeks, and display of such peptides over extended periods of time increases the sensitivity of pathogen detection by T cells (12, 13). DM accelerates the peptide exchange reaction by several orders of magnitude so that replacement of CLIP by other peptides as well as peptide editing can take place while HLA-DR (DR) molecules pass through this compartment en route to the cell surface (8). Recent work has demonstrated that DM primarily acts as an enzyme that accelerates the forward and reverse reaction from unstable to stable DR/peptide complexes (14, 15), the rate-limiting steps in peptide dissociation and peptide binding, respectively.
DM is a membrane-anchored heterodimer that belongs to the extended family of proteins with a MHC fold. Within this family it has the closest sequence similarity to MHC class II molecules, but the crystal structures of DM and its murine homologue H2-M demonstrated that these molecules lack a peptide-binding groove because the α1 and β1 helices are in closer proximity than in MHC class II molecules (16, 17). A critical question in the field is the mechanism by which DM catalyzes the exchange of MHC class II-bound peptides. Mutagenesis experiments have identified lateral surfaces on DM and DR molecules that are involved in the interaction between the two proteins. On the DR side, these mutations span the entire length of the ectodomain and are localized to the α1 and β2 domains (E40 and F51 in the α1 domain, as well as D152, L184, V186, E187, and S197 in the β2 domain) (18). On the DM side, an extended interaction surface has been mapped that also spans the entire length of the ectodomain (E8, D31, E47, and A55 in the DM β1 domain; R110 in the DM β2 domain; F100 and I173 in the DM α2 domain) (19). Several of these mutations involve acidic residues (E8, D31, and E47 of DMβ) that are located in the vicinity of a hydrophobic ridge in the DM β1 domain. The lateral interaction model is also supported by experiments in which a peptide was covalently attached to a surface-exposed cysteine located in the vicinity of the hydrophobic ridge (DM βC46) (20). When DM was attached to the peptide N terminus, it induced rapid dissociation of the peptide from DR, whereas it could not catalyze dissociation of the complex when attached to the peptide C terminus. These data support a model in which lateral interactions between DM and DR molecules induce a conformational change that destabilizes the DR/peptide interaction, but it is not known which DM residues are only involved in binding and which catalyze peptide exchange. It has been proposed that DM disrupts one or several hydrogen bonds between the peptide backbone and conserved DR residues, which could account for the fact that DM accelerates dissociation of peptides that are highly diverse in sequence (8, 17). However, recent work has demonstrated that DR/peptide complexes vary substantially in susceptibility to DM and that peptide and DR residues along the entire length of the binding site can affect susceptibility to DM (21). These data suggest that DM causes a more global conformational change of the peptide binding site. We have approached this question by searching for small molecules that modify the kinetics of DM-catalyzed peptide exchange and report the identification of four compounds that substantially increase the catalytic efficiency of the enzyme.
Materials and Methods
Fluorescence polarization (FP) assay
Binding of an Alexa-488-labeled myelin basic protein (MBP) (85–99) peptide to DR2/CLIP was examined by FP. Based on the crystal structure of the DR2/MBP peptide complex (22), the solvent-exposed P5 lysine of MBP (85–99) was mutated to cysteine (ENPVVHFFC(Alexa-488)NIVTPR) for labeling with a maleimide derivative of Alexa-488 (Molecular Probes). The peptide was synthesized by Jerini, reverse-phase HPLC purified, and analyzed by mass spectrometry to confirm fluorophore attachment. Soluble DR2/CLIP (DRA, DRB1*1501) was prepared from Chinese hamster ovary cell transfectants, as described previously (23). DM was purified as described from a S2 cell transfectant provided by Dr. D. Zaller (Merck) (7). Peptide binding assays were performed in 384-well plates (Corning) in 40-μl volumes. Reactions were typically set up in a 150 mM sodium chloride, 50 mM sodium citrate (pH 5.2) buffer (except for pH profiling). Compounds in DMSO or DMSO controls were added to empty wells of 384-well plates, usually in a 1-μl volume. DR2/CLIP was then added (typically in a 34-μl volume) to a final concentration of 100 nM (100 nM to 3.2 μM for Vmax and KM calculations) to compounds and mixed well to dissolve compounds from DMSO stocks into an aqueous environment. DM and MBP were added together in a 5-μl volume to final concentrations of 20 nM and 30 nM, respectively (unless indicated otherwise in the figure legends). FP measurements were made using a LJL Biosystems Analyst HT plate reader (Molecular Devices) with a 485/20 bandpass, 505 DRLP dichroic, 530/30 bandpass filter set for excitation and emission. FP values were read at 25°C every minute for the first 20 min and every 10 min thereafter for up to 1000 min.
High-throughput screening
Compounds were screened for the ability to modify DM-catalyzed peptide exchange in reactions containing 100 nM DR2/CLIP, 20 nM DM, and 10 nM fluorescent MBP peptide. A Beckman Biomek FX pipette station with a Sagian Core system controlled by SAMI software (Beckman Coulter) was used to add 34.6 μl of 114 nM DR2 in 150 mM NaCl, 50 mM sodium citrate (pH 5.2) to wells of 384-well plates containing 0.4 μl of library compound in DMSO. DR2/CLIP and library compound were allowed to preincubate for 30 min at room temperature. During this time, the fluorescence intensity of each well was measured to determine whether compounds were fluorescent at the wavelengths used to read FP. DM and MBP were premixed at concentrations of 160 nM and 240 nM, respectively, and added in a 5-μl volume to the DR2/CLIP-compound mixture to start the reaction. Reading of each plate required 5 min, and plates were therefore set up 5 min apart. FP values were read at 30, 120, and 360 min following addition of DM and MBP peptide. Finally, the overall fluorescence of each well was read to ensure that each well had received equal quantities of labeled MBP peptide. Compounds of interest were identified from the data using several criteria. Fluorescent compounds were excluded based on readings taken before addition of the labeled MBP peptide. In addition, the total fluorescence signal from a well had to be within an acceptable range to indicate that it received an appropriate amount of DM and fluorescent peptide. Finally, the FP measurements had to be within the range of free peptide and fully bound peptide at each time point. Candidate compounds were then further tested in dose-response experiments.
Mutagenesis of DM
The baculovirus system (pAcDB3 plasmid with BaculoGold Baculovirus; BD Biosciences) was used to generate DM mutants with the α- and β-chains under the control of separate P10 promoters. “Wild-type” DM consisted of the α-chain (signal peptide, residues α1–204 of the mature polypeptide followed by a FLAG tag) and β-chain (signal peptide, residues β1–210 of the mature polypeptide and a protein C tag) with two of the three glycosylation sites mutated (αN165D and βN92D mutations). The remaining glycosylation site (αN15) was left intact because it was ordered in the DM structure and located close to the face of DM that is thought to interact with DR. This DM glycosylation mutant had comparable activity to DM expressed in S2 insect cells and was used when comparisons between mutant DM and wild-type DM were made. The mutations βL51D, βL51N, βD31N, βD31A, βE47A, βE47Q, βD31N/βE47Q, and βD31A/βE47A were introduced by overlapping PCR. Recombinant Baculoviruses were made according to the BD BaculoGold Transfection kit instructions (BD Biosciences). The titer of Baculovirus supernatants was determined by real-time PCR using a supernatant of known titer as a standard (24). Sf9 cells were infected with a multiplicity of infection >5, and the protein was allowed to accumulate for 3 days. Culture supernatants were harvested and concentrated 10-fold before purification on an anti-protein C epitope tag column (Roche Applied Science), as described by the manufacturer. Bound DM was eluted with EDTA (5 mM), concentrated and stored at −80°C. All mutants were analyzed by SDS-PAGE to confirm the presence of both DM chains and protein concentration measurements.
Calculation of Vmax and KM
To determine the Vmax and KM of DM at different pH values or in the presence of small molecules, the peptide binding curves measured by FP were analyzed. FP and anisotropy are mathematically related ways of expressing parallel:perpendicular emission ratios and are easily interconverted. Although FP is approximately linear with respect to the ratio of free:bound peptide, FP was converted to anisotropy (which is exactly linear) by the following equation so as not to introduce unnecessary errors.
A = anisotropy
FP = fluorescence polarization
Time points of the peptide binding reaction were fit to a single phase exponential association curve that was modified to account for the fact that at t = 0 anisotropy = 83.3 mA.
At = (Aequilibrium · (1 − e−Kob · t)) + At = 0
t = Time in minutes
At = Anisotropy (mA) at a given time
Aequilibrium = Anisotropy (mA) at equilibrium
At = 0 = Anisotropy (mA) of free peptide at room temperature (83.3 mA)
kob = Observed rate constant (min−1) (kon[peptide] + koff)
The slope of the first 10% of the curve was defined as the initial velocity of the reaction (v) in mA/min. The initial velocities of reactions in the absence of DM were subtracted from initial velocities in the presence of DM to define the DM contribution. These values were plotted against the DR2/CLIP concentration (μM). This curve could be fit to the following equation to determine the Vmax (mA min−1 ) and KM (μM) by nonlinear regression. All curve fitting was preformed in Prism (GraphPad), and each curve was visually inspected for fit.
The Vmax/KM was determined by dividing the Vmax by the KM, giving the relative Vmax/KM values in the following units: mA min−1 μM−1 to allow for comparison between DM mutants at equivalent concentrations.
pH dependence of DM mutants
The pH dependence of DM mutants was determined by setting up 2-fold serial dilutions of DR2/CLIP (0.162–5.12 μM) in 10 mM sodium citrate, sodium phosphate buffer (pH 7.13). Twenty-five microliters of each dilution was added to different wells to screen a range of pH values from 4.79 to 7.13. Five microliters of a 160 nM stock solution of DM (or mock) in 10 mM citrate phosphate (pH 7.13) was added to wells containing DR2/CLIP. To adjust each well to the desired pH value, 5 μl of a 500 mM stock solution of citrate/phosphate buffer at the appropriate pH was added. The reaction was started by addition of 5 μl of a 240 nM solution of fluorescent MBP peptide and vigorous mixing. The plate was then quickly transferred to the plate reader, and FP was measured over 1000 min to track the progress of peptide binding reactions.
Microdialysis
Solutions of DR/CLIP, DM, and DR:DM (all at 5 mg/ml) in 150 mM NaCl, 50 mM sodium citrate (pH 5.2) were aliquoted into one side of a Micro-Equilibrium Dialyzer (model SE0200, Harvard Apparatus; Amika) with 100 μl of dialysis chambers. The other side was filled with aldolase (5 mg/ml) as a control protein in the same buffer. The two chambers were separated by a dialysis membrane with a 10-kDa molecular mass cutoff, which allowed small molecules but not proteins to pass between chambers. Before the chambers were closed, 1 μl of 1 mM M19 or F15 dissolved in DMSO was added to each side to a final concentration of 10 μM. The chambers were left to equilibrate overnight at 4°C on a rotator to ensure mixing. After equilibrium had been reached, the concentration of M19 in each chamber was measured by absorbance at 380 nm using a series of M19 standards. Because F15 was not colored, it was quantitated using a C8 reverse-phase HPLC column using 90 μl of the dialyzed material. The peak area (at 254 nm) was determined and converted into a concentration by comparison with a standard curve of F15 compiled from reverse-phase runs with known quantities of F15.
Structure determination
DM was purified from a stable S2 cell transfectant provided by Dr. D. Zaller (Merck) (7), and crystals were grown by the hanging drop method using conditions similar to those reported by Mosyak et al. (17) in 1 M lithium sulfate, 300 mM ammonium sulfate, and 100 mM sodium citrate (pH 5.4). Crystals were looped out of drops and equilibrated for 30 min in 1 μl of a solution containing 30% polyethylene glycol 4000, 100 mM ammonium sulfate, 100 mM sodium citrate (pH 5.4), and 20% glycerol. Small molecules were added from stock solutions in DMSO to the crystal soaking solution to final concentrations of 1 mM for M19, 300 μM for F15, and 100 μM for F12, and 9 μl of compound in the soaking solution was added to the 1-μl drop containing the equilibrated crystal and allowed to soak for 30 min. The crystals were harvested in cryo loops and frozen in liquid nitrogen. The structure presented here was from a crystal soaked with F15 that diffracted to 2.28 Å. Data was collected at beamline X6A (National Synchrotron Light Source at Brookhaven National Laboratories, Upton, NY) and processed with HKL2000 (25). The published DM crystal structure (Brookhaven Protein Data Bank (PDB) accession no. 1HDM) was solved with the C2221 space group, but our crystals had a P212121 space group even though the crystallization conditions were similar. The structure was solved by molecular replacement with the DM coordinates (1HDM residues truncated to alanine) in MolRep 7.3.03 (26). A single rotation solution was found with two translation functions. The LSQKAB program of the CCP4 suite (27) was used to rotate 1HDM and apply the two translations forming a PDB file with two DM molecules. This PDB file was then put through rigid body refinement in the crystallography and nuclear magnetic resonance system (CNS) and the Rfree refined from 52 to 42%. This model was used in rounds of refinement with CNS (28) and model building in O (29). Strict noncrystallographic symmetry was used for initial refinement until extra DM residues at the C terminus could be built into the density. In one molecule, electron density for residues α13 to 200 and β3 to 193 could be seen, whereas in the other DM heterodimer residues α12 to 206 and β3 to 199 were resolved. Noncrystallographic symmetry restraints were used for the portions of DM that each heterodimer had in common. These differences and small differences in the packing probably explain why the space group P212121 refined better than C2221. Water molecules (420 shared between two DM molecules) were added after the Rfree could no longer be lowered by refinement alone and this reduced the Rfree from 28.6 to 26.9%. The number of waters present is in close agreement to the previously reported structure, which identified 198 water molecules per DM molecule. For statistics on data collection and refinement see Table I.
Table I.
Statistics for data collection and refinement
| Data collection | |
| Space group | P212121 |
| Unit cell (Å) | a = 98.1; b = 108.4; c = 110.2 |
| Molecules/asymmetric unit | 2 |
| Resolution limit (Å)a | 30–2.28 (2.37 – 2.28) |
| Unique reflections | 59,443 |
| Multiplicity | 5.6 |
| Average IIσ (I) | 12.3 (5.02) |
| Completeness (%) | 99.7 (99.6) |
| Rsym (%)b | 5.7 (30.9) |
| Wilson plot B (Å2) | 32.9 |
| Model refinement | |
| Rcryst (%)c | 22.9 |
| Rfree (%) | 26.9 |
| Root mean square deviations from ideality |
|
| Bond lengths (Å) | 0.0063 |
| Bond angles (°) | 1.41 |
| Dihedrals (°) | 25.6 |
| Impropers (°) | 0.81 |
| Ramachandran plot statistics (%) | |
| Favored | 86.2 |
| Allowed | 12.9 |
| Generously allowed | 0.6 |
| Disallowed | 0 |
Values in parentheses refer to the highest resolution shell.
Rsym = Σ∣I − 〈I〉∣/ΣI, where I is the measured intensity of a reflection and 〈I〉 is the average intensity of that reflection.
R = Σ‖Fobs∣ − ∣Fcalc‖/Σ∣Fobs∣, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
Rcryst is calculated from 97.3% of data;
Rfree is used for cross-validation and based on the remaining 2.7% of data.
Results
Identification of small molecules that accelerate DM-catalyzed peptide exchange using a FP assay
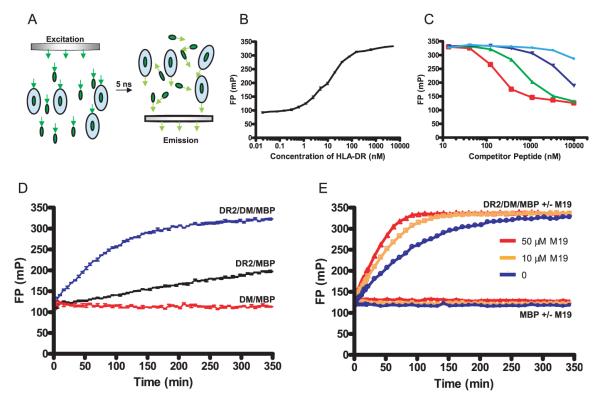
Kinetic measurements of the peptide binding reaction are critical for the identification of small molecules that modulate the activity of DM. Most investigators use endpoint assays to measure peptide binding, but it is difficult to capture the kinetics of the reaction with such an approach. We developed a real-time peptide binding assay in which progression of the reaction is monitored by FP without any additional manipulation. Monitoring of peptide binding by FP is based on the fact that a small fluorescent peptide tumbles significantly faster than the peptide-receptor complex. Polarized fluorescent light is used to excite the fluorophore, and following a 5-nanosecond delay the emitted light is measured both parallel (polarization retained) and perpendicular (polarization lost) to the incident light (Fig. 1A) (30, 31). Binding of the fluorescent peptide to DR2 increases the fraction of emitted fluorescent light that has retained polarization, as illustrated in Fig. 1B. Peptide binding was monitored using an Alexa-488 fluorescently labeled version of a MBP (residues 85–99) peptide that binds to DR2 (the DRA/DRB1*1501 allele). DR2/CLIP complexes expressed in Chinese hamster ovary cells were used in these experiments because they represent the natural substrate for peptide loading in APCs (23). The experiment in Fig. 1B shows how FP values increase when an increasing fraction of the labeled peptide is bound to DR2. The labeled MBP peptide (10 nM) was added to increasing concentrations of DR2/CLIP (20 pM to 5 μM final concentrations), and FP measurements were made following overnight incubation at 37°C. Specificity of binding was demonstrated using unlabeled competitor peptides with substitutions at key DR2 anchor residues that occupy the P1 and P4 pockets of the binding site (residues 89 and 92 of the MBP peptide, respectively) (Fig. 1C). Substitution of the P1 and P4 anchors by alanine reduced the binding affinity of the peptide, and substitution of the P4 anchor by aspartic acid abrogated binding, in agreement with previous studies The results from these experiments were in close agreement with data from radio-labeled peptide binding assays (see Fig. 1C) (32, 33). A real-time peptide binding assay involving fluorescence resonance energy transfer from tryptophan residues of the MHC to a fluorophore attached to the peptide has also been reported (34). An advantage of the FP method is that it does not require excitation in the UV range, conditions under which many small molecules are highly fluorescent.
FIGURE 1.
Identification of small molecules that accelerate DM-catalyzed peptide exchange using a FP-based peptide binding assay. A, Principle of monitoring the binding of a fluorescently labeled peptide to DR by FP. The rate at which molecules tumble in solution is directly related to their molecular mass, and a small fluorescently labeled peptide tumbles significantly faster than the complex of such a peptide bound to a DR molecule. The light used for excitation of the fluorophore passes through a polarization filter (left), and the fraction of the emitted fluorescence that has retained polarization is measured following a 5-nanosecond delay (right). The relative intensities of polarized vs scattered emission depend on the ratio of DR-bound vs free fluorescent peptide. B, Analysis of peptide binding to DR2 by FP. An Alexa-488-labeled MBP peptide (10 nM) was incubated with DR2/CLIP at a wide range of concentrations (ranging from 20 nM to 5000 nM) overnight at 37°C. Free peptide yielded FP values of ~100 mA; FP values increased to ~320 mA when the majority of fluorescent peptide molecules were bound to DR2. Reactions were set up in triplicates, and the average and SDs are shown. FP was plotted as millipolarization units (mP), with larger numbers indicating that a larger fraction of emitted fluorescence has retained polarization. C, FP binding measurements are specific. Competitor peptides of different affinities were used to compete for binding of 10 nM fluorescently labeled MBP to 100 nM DR2. Competitor peptides tested were MBP (85–99) (red open squares) as well as single amino acid analogues with substitutions at key DR2 anchor residues: alanine substitution of position 89 (green triangles), alanine substitution of position 92 (dark blue triangles), or aspartic acid substitution of position 92 (light blue circles). D, Real-time analysis of DM-catalyzed and noncatalyzed peptide binding to DR2/CLIP. DR2/CLIP (100 nM) was incubated with labeled MBP peptide (10 nM) in the presence (blue line) or absence (black line) of DM (20 nM) at 25°C, and progression of the binding reaction was monitored by FP. As a control, DM (20 nM) and MBP peptide (10 nM) were incubated in the absence of DR2 (red line). E, A small molecule (M19) that significantly accelerates DM-catalyzed peptide binding. Screening of libraries of small molecules using this assay yielded four compounds that substantially accelerate the peptide binding reaction. The kinetics of reactions involving DR2, DM, and MBP peptide were compared in the absence of M19 (blue line) and in the presence of M19 (orange, 10 μM; red, 50 μM). M19 had no effect on FP measurements for reactions involving only the peptide (labeled MBP alone ± M19; blue: no M19, orange: 10 μM M19, red: 50 μM M19). MBP peptide, DM, and DR were used at 10 nM, 20 nM, and 100 nM, respectively.
The major advantage of this technique is that the reaction can be followed in real time so that many time points can be analyzed without the need to withdraw samples for analysis. In Fig. 1D, the kinetics of the peptide binding reaction were followed in the presence and absence of DM (blue and black lines, respectively). In the absence of DM, peptide binding proceeded slowly with a half-life of association of 587 min. Inclusion of DM at 20 nM substantially accelerated the reaction, with a half-life of association of 62 min. With extended incubation times (18 h), the same FP values were reached in the presence and absence of DM (data not shown), confirming that DM acts as a catalyst but does not change the equilibrium of the reaction. In a control reaction, DM and the MBP peptide were incubated in the absence of DR2 (red line). The graphs show the average and SDs of triplicate measurements, which were highly reproducible due to the ratiometric nature of the readout (Fig. 1, B–D).
Using this assay, we screened several libraries of small molecules (~48,500 compounds) with the aim of identifying compounds that modulate DM-catalyzed peptide exchange. The screening led to the identification of four small molecules, M19 (LDN-111299), F15 (LDN-9224), F12 (LDN-8609), and E03 (LDN-5176) that substantially accelerated the rate of DM-catalyzed peptide binding. In Fig. 1E, the effect of M19 on DM-catalyzed peptide binding is shown. This small molecule substantially increased the rate of the reaction and thus significantly shortened the time required to reach equilibrium. The halftimes of association in the presence of 50 and 10 μM of M19 and no M19 were 25, 35, and 62 min, respectively.
Targets of the small molecules that accelerate peptide exchange
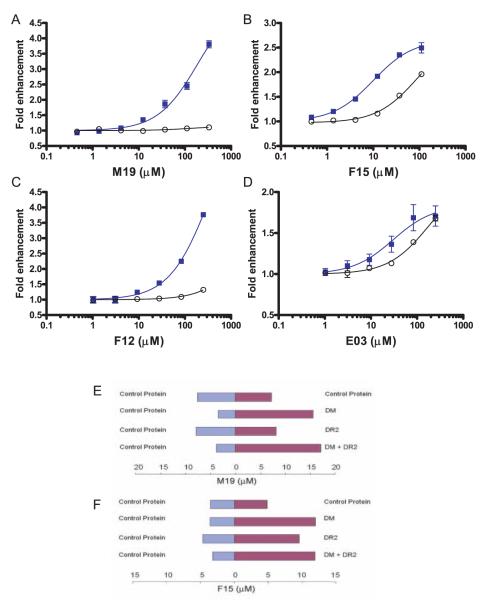
The mechanisms by which the small molecules accelerate peptide exchange (CLIP to MBP) were investigated in peptide binding experiments in the presence and absence of DM. The fold-enhancement of the rate of MBP peptide binding by the small molecules was calculated based on the slope of the initial linear component of the reaction. These experiments demonstrated that M19 and F12 (Fig. 2, A and C) were only active in the presence of DM. The enhancement of the initial rate of the reaction was substantial, ~3.5–4-fold compared with reactions without the small molecules. No plateau was observed even at relatively high concentrations of these compounds, indicating that they induce a striking acceleration of the reaction despite a low affinity. The other two small molecules (F15 and E03; Fig. 2, B and D) accelerated peptide exchange even in the absence of DM, but yielded a more substantial enhancement in the presence of the enzyme. The enhancer effects in the presence of DM could not be attributed to their action on DR alone, because this would not be expected to significantly affect the fold enhancement of the reaction rate in the presence of DM. These results demonstrate that F15 and E03 act directly on the DR/peptide complex.
FIGURE 2.
Targets of the small molecules that accelerate peptide exchange. A–D, The kinetics of the peptide binding reaction were measured over a wide range of small molecule concentrations, both in the presence and absence of DM (■ and ○, respectively). The initial rate of the reactions was determined, and the fold enhancement over reactions without small molecules was plotted as a function of small molecule concentration. Such graphs are shown in A–D for each of the small molecules (M19, F15, F12, and E03). The M19 and F12 enhancers (A and C) only accelerated the peptide binding reaction in the presence of DM. For the F15 and E03 enhancers (B and D), an acceleration of the peptide binding reaction was also observed in the absence of DM, but these molecules were most effective in the presence of DM. E–F, Binding of small molecules was examined in microdialysis experiments in which two compartments carrying a control protein (aldolase, blue bars) or an experimental protein (red bars) were separated by a dialysis membrane. Following overnight incubation at 4°C, the concentration of the small molecule in both compartments was measured. E, M19 was found to bind to DM but not to DR2 because it was only enriched when DM was present in the experimental compartment. F, The F15 enhancer was found to bind to both DM and DR2, consistent with the functional data shown in B. Microdialysis experiments were conducted three times with similar results, and a representative experiment in each case is shown.
Binding of small molecules to their protein targets can be studied in microdialysis experiments in which two compartments that contain a test protein or a control protein are separated by a dialysis membrane. The small molecule but not the target protein can cross the dialysis membrane, and binding of the small molecule to its target thus leads to an enrichment of the small molecule in the compartment containing the receptor and depletion from the compartment containing the control protein provided that the small molecule is at a lower concentration than the target (final concentration of 10 μM for small molecules, 100 μM for DM, and 80 μM for DR). After the reaction had reached equilibrium, we determined the concentration of M19 in both compartments by absorbance measurements. M19 was substantially enriched when DM or both DM and DR were present in the experimental chamber, indicating that M19 binds to DM. No enrichment was observed in the presence of DR (Fig. 2E). Similar experiments were performed with F15, and F15 was quantitated at the end of the reaction by reverse-phase HPLC. In the presence of control proteins that did not bind to F15 there was some nonspecific loss of the compound. F15 was strongly enriched when DM or both DM and DR were present in the experimental chamber. Binding of F15 to DR could also be demonstrated, but the enrichment was not as strong as with DM (Fig. 2F). These results demonstrated that F15 binds to both DM and DR, explaining the functional results described above. The small molecules that accelerate peptide exchange thus fell into two groups: M19 and F12 were only active in the presence of DM, and microdialysis experiments demonstrated binding of M19 to DM, but not DR. In contrast, F15 and E03 were active in the absence of DM but showed the most substantial enhancement in the presence of DM. F15 bound to both DM and DR, and may thus localize to the interface between the two proteins in the DM:DR/peptide complex.
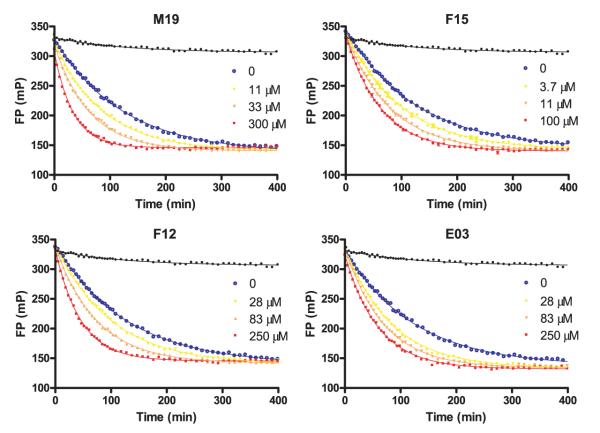
We also examined whether these small molecules accelerated DM-catalyzed dissociation of a high-affinity peptide from DR2. For that purpose, DR2 molecules were loaded with the labeled MBP peptide, and the complex was isolated using gel filtration to remove free peptide. Dissociation of the labeled peptide was examined by FP in the presence of a large molar excess of unlabeled MBP peptide to prevent rebinding of labeled peptide. DM significantly accelerated dissociation of the MBP peptide from DR2, and the rate of peptide dissociation was substantially increased by each small molecule tested (see Fig. 3). Taken together, these data demonstrated that these small molecules substantially accelerate peptide association and dissociation in the presence of DM.
FIGURE 3.
Small molecules also accelerate dissociation of the high-affinity peptide MBP (85–99) from DR2. Dissociation of fluorescently labeled MBP peptide from DR2 (150 nM) catalyzed by DM (150 nM) was followed by FP in the absence (blue line) and in the presence of different concentrations of small molecules (yellow, orange, and red lines; concentrations indicated in the figure); a molar excess of unlabeled MBP peptide was added to prevent rebinding of the fluorescent peptide. MBP peptide dissociation in the absence of DM and small molecules is also shown in each graph (black line). Peptide dissociation was substantially accelerated by all of the small molecules.
Direct effect of small molecules on catalysis by DM
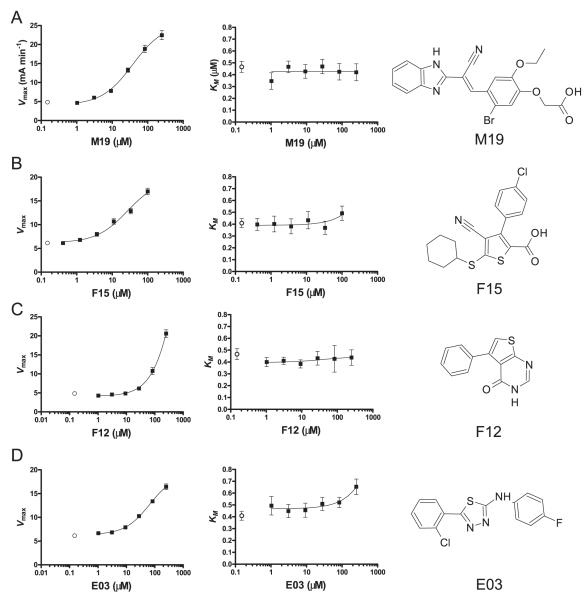
The four small molecules were quite distinct in their structure, but M19 and F15 shared a carboxyl group (Fig. 4). Nevertheless, detailed analysis of enzyme kinetics demonstrated significant functional similarities. The influence of each of the compounds on the key parameters Vmax, KM, and Vmax/KM of DM-catalyzed peptide exchange were analyzed. In simple Michaelis-Menten enzyme kinetics, the Vmax relates to the maximum number of substrate molecules that can be converted to product during a specified unit of time at a defined enzyme concentration, whereas the KM (the substrate concentration required for half-maximal velocity) is dependent on the affinity of the enzyme for the substrate. In a more complex enzymatic reaction, other aspects of the reaction can also contribute to the KM. These parameters were determined for all four small molecules by analyzing reactions in a grid of 6 × 6 DR/CLIP and small molecule concentrations with and without DM (described in Fig. 5). The initial rate of reactions in the absence of DM were subtracted from the initial rate in the presence of DM such that the enzymatic parameters are specific to DM in the presence of the compound and do not include components attributable to the effects of small molecules on DR alone. These experiments demonstrated that M19 and F12 had no effect on the KM and that F15 and E03 had only subtle effects on the KM at the highest concentration. All small molecules substantially increased the Vmax and the Vmax/KM, indicating that all four compounds increase the catalytic efficiency of DM.
FIGURE 4.
The small molecules increase the efficiency of catalysis by DM. A–D, The functional effects of the small molecules on the kinetics of DM-catalyzed peptide exchange were examined by measuring the initial rate of the reaction at six different concentrations of the substrate (DR2/CLIP, 0.1–3.2 μM; 30 nM MBP) at each of six enhancer concentrations, as illustrated in Fig. 5. For reasons described in the experimental procedures, FP was converted to anisotropy to calculate these initial rates. From these titration curves, the maximal rate of the reaction (Vmax) and the substrate concentration (DR2/CLIP) required for half-maximal activity (KM) were calculated. The resulting Vmax and KM values were plotted relative to the concentration of the small molecule. The open black circle in each graph represents the Vmax or KM in the absence of small molecule. All four molecules substantially increased the Vmax but had only minor effects on the KM, indicating that they increased the rate at which the enzyme-substrate complex was converted to enzyme and product. The structures of the four small molecules (M19, F15, F12, and E03) that accelerate DM-catalyzed peptide exchange are shown on the right. M19 and F15 share a carboxyl and a nitrene group, but the structures are otherwise quite distinct.
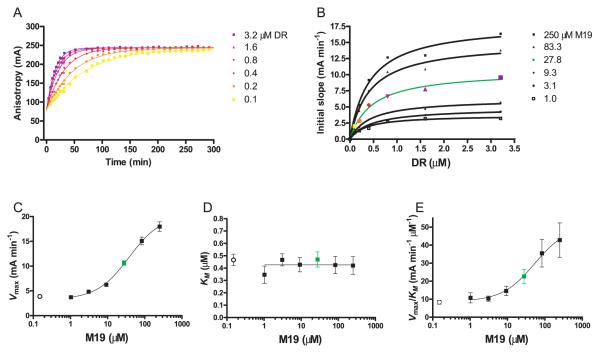
FIGURE 5.
Calculation of Vmax and KM values for DM in the presence and absence of the M19 small molecule. A, Peptide binding reactions were set up with a grid of six different substrate concentrations (DR2/CLIP, 0.1–3.2 μM; 30 nM MBP) and six concentrations of the M19 enhancer (1–250 μM). Shown is a set of six reactions with a fixed M19 concentration (27.8 μM) and six different DR2/CLIP concentrations. The initial slopes were calculated from anisotropy instead of FP for reasons described in Materials and Methods. B, For each concentration of M19, the initial slope of the reaction was plotted relative to the DR2/CLIP concentration. Each curve from A yielded one data point in B. From each of these curves, the maximum rate of the reaction (Vmax) and the substrate concentration (DR2/CLIP) required for half-maximal activity (KM) was calculated. C–E, The Vmax (C), KM (D), and Vmax/KM (E) values were then plotted as a function of the M19 concentration. The ○ in both graphs show Vmax (C), KM (D), and Vmax/KM (E) values in the absence of the small molecule.
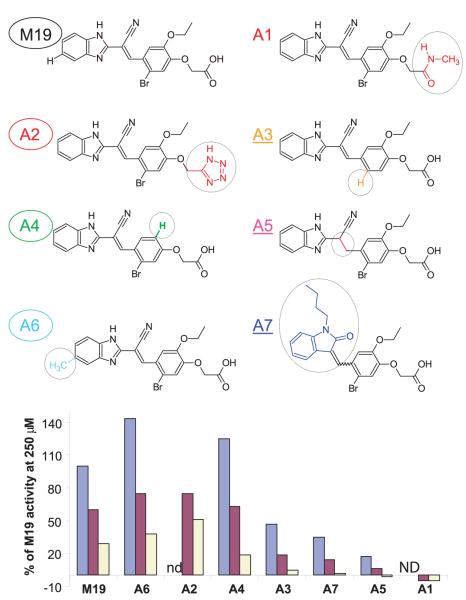
Structure-activity relationship of M19
Medicinal chemistry was used to identify regions of M19 important for activity (Fig. 6). Both M19 and F15 share a carboxylic acid group, and the importance of this group was demonstrated by replacement with either an amide (analog A1) or a tetrazole (analog A2). Substitution with the amide resulted in a loss of activity (this compound could only be tested at 10 μM and 50 μM because of limited solubility). Tetrazole is commonly used as a bioisostere for carboxylic acids in medicinal chemistry and retains a similar pKa, but delocalizes the negative charge around the ring and is somewhat larger than its carboxylic acid counterpart. The tetrazole analog had slightly higher activity than M19 at concentrations of 10 μM and 50 μM. Taken together, these results demonstrate the importance of the carboxylic acid group for M19 activity. In contrast, the neighboring ethyl-ether group could be replaced by a hydrogen without loss of activity (analog A4). Removal of the aryl bromide (analog A3) resulted in a partial loss of activity.
FIGURE 6.
Structural features of M19 that are important for activity. A series of M19 analogues with changes at defined sites (circled in each of the analogues) were tested at three concentrations (250, 50, and 10 μM); analogues A1 and A2 had limited solubility and were therefore tested only at 50 and 10 μM, but not at 250 μM (ND). Activity is expressed as percentage relative to M19 at a concentration of 250 μM. Analogues that have activity similar to or higher than M19 are circled (A2, A4, and A6), and analogues with reduced activity are underlined (A3, A5, and A7). The carboxylate was critical for activity because replacement with an amide (A1) resulted in a complete loss of activity, whereas substitution with a tetrazole (a carboxylic acid bioisostere) did not (A2). The ethyl-ether was not required, as shown by analog A4, and removal of the aryl bromide (A3) reduced but did not abrogate activity. Significant changes to the left portion of the molecule could be made as shown by analog A7, and addition of a methyl group to the benzimidazole ring (A6) yielded a compound with activity higher than M19.
Replacement of the double bond linking the two ring structures with a single bond significantly reduced activity (analog A5) to <20%, relative to M19 at the highest concentration tested (250 μM). This change has large effects on the rigidity of M19, suggesting that the arrangement of the two ring structures relative to each other is important for activity. An increase in activity was observed with the addition of a single methyl group to the benzimidazole ring (analog A6). A dose-response curve indicated that the EC50 of this analog was 2- to 5-fold lower than M19 (data not shown). This suggested that the benzimidazole ring may not be optimal and that binding might be significantly increased by changes to this part of the molecule. In analog A7, the entire left portion of the molecule was replaced with a substituted oxindole ring. This analog had partial activity (~30% relative to M19), indicating that substantial changes to this part of the molecule can be made without complete loss of activity. These small molecules are thus structurally and functionally distinct from previously reported compounds (phenolic compounds with a single −OH group) that accelerate release of low-affinity peptides from DR in the absence of DM, presumably through disruption of hydrogen bonds between the peptide and the MHC molecule (35, 36).
Determination of the binding site for enhancers on DM
Our first attempt to define the binding site of the enhancers involved x-ray studies of DM crystals soaked with M19, F15 or F12. The structure was solved, and extra electron density was sought to account for the soaked small molecules, but unambiguous density representing bound enhancers could not be identified (the nonphysiological conditions required for crystal growth and soaking may have interfered with small molecule binding). However, this effort yielded a more complete structure of DM compared with the deposited PDB structure 1HDM (17) (see Table I for statistics). The conformation of the extra residues is unlikely to be altered by soaking of crystals with the small molecules because comparisons of five structures soaked with three different small molecules yielded similar electron density maps. The resolution of the five structures was in the range of 2.3–2.7 Å (Rfrees of 0.269, 0.274, 0.276, 0.287, and 0.335).
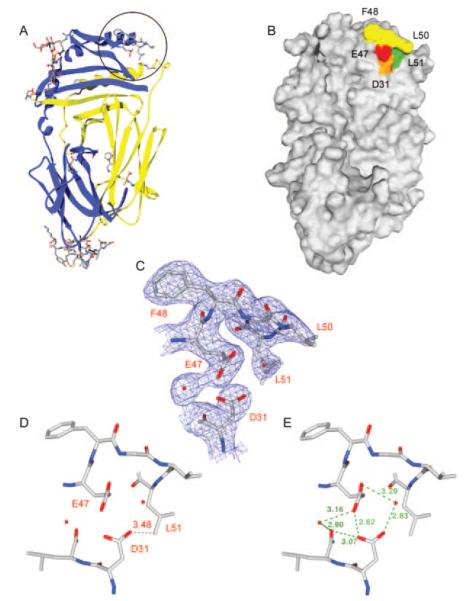
Of the regions not present in the 1HDM structure, a 5-aa loop (DM β47–51) that leads into the β1 helix of DM could be defined (Fig. 7, A–C). This region of DM is of interest because previous mutagenesis experiments had shown that substitutions of the neighboring DM βE47 and βD31 residues affect the DM:DR interaction (19). The higher resolution structure showed that these two acidic residues are within hydrogen bonding distance from each other and that they coordinate two water molecules (Fig. 7, C–E). These two acidic residues are in close proximity to a hydrophobic pocket formed by DM βL50 and βL51.
FIGURE 7.
Structural analysis of DM resolves the electron density for a hydrophobic ridge located in the DM β1 domain. A, A number of DM residues that were absent in PDB file 1HDM could be resolved in a higher resolution crystal structure and are shown as a stick representation on a ribbon diagram of DM. Five amino acids (DMβ 47–51, EFGLL) that form a hydrophobic ridge are circled. B, Molecular surface representation of DM in the same orientation as in A. Residues F48, G49, and L50 of the hydrophobic ridge are colored yellow, and residues βL51 and βE47 are colored green and red, respectively. A neighboring acidic residue (βD31) is colored orange. C, Electron density calculated from a 2Fobs-Fcalc map at 1σ shows that the DM β47–51 region is clearly resolved. Two neighboring water molecules and βD31 are also shown. The view is the same as in A and B. D–E, The distance between βD31 and βL51 as well as the hydrogen bonding network between the two acidic residues and the surrounding environment is shown. The view is tilted back slightly compared with A–C to clearly show the hydrogen bond distances.
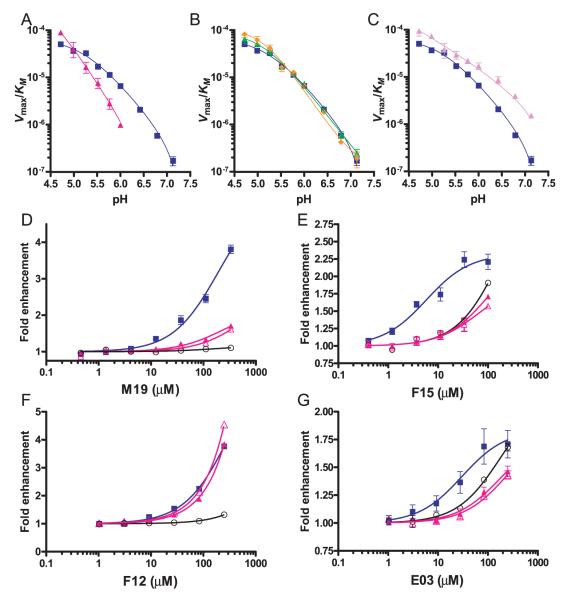
We investigated this area as a potential binding site for the four enhancers, and aimed to introduce mutations that would maintain DM function but abrogate the activity of the small molecules. Because all four small molecules shared hydrophobic ring structures, we mutated DM βL51 as well as the neighboring βE47 and βD31 residues. The catalytic activity of the DM βL51D mutant was similar to wild-type DM at pH 4.7 to 5.2, but activity was reduced substantially at higher pH levels (Fig. 8A, red and blue lines, respectively). Activity could not be accurately determined at a pH lower than pH 4.5 because both DR2 and DM were less stable under these conditions. Given the proximity of position 51 to βE47 and βD31, introduction of an acidic residue may affect the protonation state and the hydrogen-bonding pattern of βE47 and/or βD31.
FIGURE 8.
Three of the four small molecule enhancers bind in the vicinity of the hydrophobic ridge of DM. A–C, Mutations were introduced at positions βL51, βE47, or βD31 of DMβ, and the activity of these DM mutants was examined as a function of pH in the absence of small molecule enhancers. Graphs show the activity of these mutants as Vmax/KM from pH 4.7 to 7.3. A, The βL51D mutation (red line) narrows the pH range of the enzyme relative to wildtype DM (blue). B–C, The individual βD31N (green) or βE47Q (orange) mutations (B) did not affect DM activity, whereas the βD31N-βE47Q double substitution (C, purple) increased the activity of the enzyme ~2-fold at a low pH (pH 4.7 and 5.0) and ~9-fold at a neutral pH (pH 7.3) compared with wild-type DM (blue). D–G, The effect of mutation of DM residue βL51 on the activity of the four small molecules. The dose response for each enhancer is shown in the presence of wild-type DM (blue line), the βL51D mutant (red, closed triangles), the βL51N mutant (red, open triangles), and in the absence of DM (black, open circles). Both βL51 mutations greatly reduced or abrogated the activity of M19 (D), F15 (E), and E03 (G).
Importantly, the βL51D mutation abrogated or substantially reduced the activity of three of the four small molecules, M19, F15, and E03 (Fig. 8, D–G, closed red triangles). Only F12 retained the ability to accelerate the reaction in the presence of the βL51D mutant. This result was surprising because these compounds belonged to the two functional groups described above. We therefore extended these experiments with a substitution that did not introduce a negative charge at this site. The results with this βL51N mutation were strikingly similar, confirming the importance of this residue for the activity of three of the small molecules (Figure 8, D–G, open red triangles). These results show that βL51 forms part of the binding site of M19, F15, and E03 on DM.
Individual mutations of βD31 or βE47 to alanine or the corresponding carboxamide did not affect the activity of DM at pH 4.8, 5.5, and 6.8. A more detailed pH profiling of the βD31N and βE47Q mutations also showed no measurable effect on catalysis over a wide pH range (data not shown and Fig. 8B). However, the double βD31N-βE47Q mutant in which both residues were substituted by the corresponding carboxamide had increased activity. At an acidic pH, the activity of the mutant was ~2-fold higher than wild-type DM. This mutant was also active at a neutral pH at which wild-type DM had little activity; the difference in activity at this pH was ~9-fold (Fig. 8C). Mutations in this area of DM can thus substantially affect the pH range within which the enzyme is active. The βL51D mutation narrowed the pH range, whereas mutation of the neighboring βD31-βE47 residues extended it into the neutral pH area.
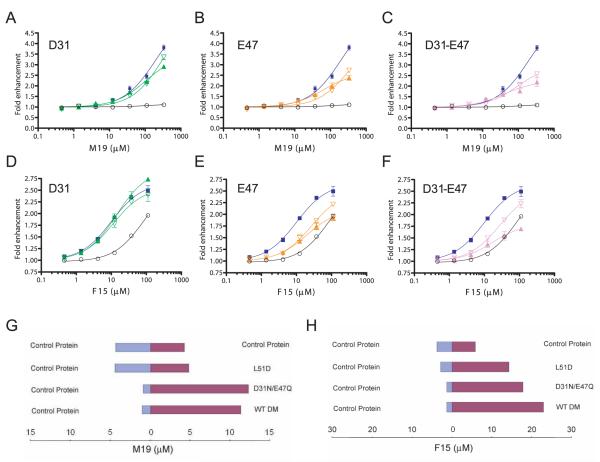
Analysis of the βE47 mutants strengthened the conclusion that this site is important for the activity of the small molecules. The activity of both M19 and F15 was significantly reduced by the βE47N and βE47A mutations (Fig. 9, B and E), whereas mutations of the neighboring βD31 residue had little effect (Fig. 9, A and D). The βD31-βE47 double mutants had properties very similar to the βE47 single mutants (Fig. 9, C and F).
FIGURE 9.
Mutation of DM βE47 reduces the activity of M19 and F15. A–C, The dose response of M19 is shown for DM molecules with mutations at positions βD31 and/or βE47. Graphs show the M19 dose response with (A) βD31N (green, closed triangles) and βD31A (green, open triangles) (A); βE47Q (orange, closed triangles) and βE47A (orange, open triangles) (B), and E31N-βE47Q (purple, closed triangles) and βD31A-βE47A double mutations (purple, open triangles) (C). The dose response for reactions with wild-type DM (blue) and without DM (black) is shown in each graph. D–F, The corresponding experiments are shown for F15 with the set of mutants described in A–C. G, The βL51D mutation abrogates M19 binding, whereas the βD31N-βE47Q double mutation only affects activity. Microdialysis experiments were performed with wild-type DM and two DM mutants; the concentration of M19 in the two chambers was measured when equilibrium had been reached. H, Similar microdialysis experiments as described in G were performed with F15.
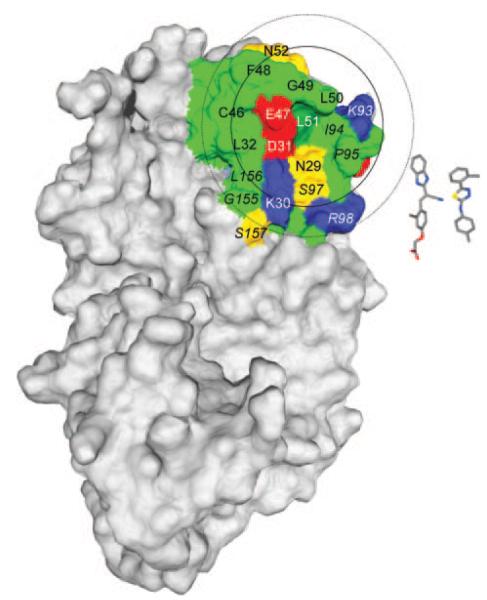
Microdialysis experiments demonstrated that the βL51D mutation interfered with M19 binding (Fig. 9G). In contrast, the βD31N-βE47Q mutant retained the ability to bind M19, despite the lower activity of M19 with this mutant. This may be because other residues provide most of the binding energy required for M19 binding, whereas this region is relevant for the catalytic activity of the DM-enhancer complex. The βL51D mutation reduced binding of F15 to an intermediate level (Fig. 9H), indicating that βL51 makes a larger contribution to binding of M19 compared with F15. Taken together, these results demonstrated that βL51 and βE47 located within the vicinity of the hydrophobic ridge of DM are important for the activity of these DM enhancers. It should be noted that the residues contacted by these small molecules may not be limited to these acidic and hydrophobic ridge residues. The largest of the small molecules is M19 (~15.6 Å in length), and a sphere with a radius of 15.6 Å that is centered on βL51 has been drawn onto the surface of DM in Fig. 10. The functional properties of the small molecules that bind to this site implicate this area in the catalysis of peptide exchange by DM.
FIGURE 10.
Surface area of DM surrounding DMβ L51 and E47. A surface representation of DM as well as the structures of the M19 (left) and E03 (right) small molecule enhancers are shown. The two circles are centered on the DMββL51 residue that is critical for the activity of three of the four small molecules. The larger circle was drawn using the longest dimension of M19 (15.6 Å) as the radius; the smaller circle represents E03. Hydrophobic residues are colored green, basic residues blue, acidic residues red, and other polar residues yellow. Residues from the DMα-chain are italicized and residues from the DMβ-chain are indicated by a regular font.
Discussion
These results demonstrate that small molecules can substantially accelerate DM-catalyzed peptide exchange. The four structurally distinct compounds that we have identified fall into two functional groups. The compounds in the first group (F15 and E03) accelerate peptide release even in the absence of DM and thus act directly on the DR/peptide complex. These small molecules have the highest activity in the presence of DM, indicating that they may bind to the DM:DR/peptide complex. This conclusion is supported by microdialysis experiments that demonstrated specific binding of F15 to DM and DR. The second set of compounds (M19 and F12) is only active in the presence of DM, and microdialysis experiments demonstrated that M19 binds to DM but not DR. M19 may only act on the DM component of the complex and induce a conformational change in the enzyme, but it is also possible that a functional group of DM-bound M19 directly modulates the conformation of the DR/peptide complex and that its mechanism of action is thus related to F15.
Enzymatic reactions proceed in two stages, the formation of the enzyme-substrate complex and conversion of the enzyme-substrate complex to enzyme and product. Formation of the enzyme-substrate complex is dependent on the affinity of the enzyme for its substrate, and the initial interactions between enzyme and substrate contribute to the KM (36). All small molecules had striking effects on the Vmax and the Vmax/KM but little or no effect on the KM, demonstrating that the small molecules modulate the efficiency of catalysis rather than early steps in the enzyme-substrate interaction.
Enzymes accelerate chemical reactions by binding with a higher affinity to the transition state than the initial state of the substrate (37). In the peptide exchange reaction, the transition state is likely to represent the complex of DM and an unstable DR/peptide species. Formation of the DM:DR/peptide transition state may require conformational changes in both the DM and the DR/peptide component, and may therefore involve several discrete steps. All small molecules directly affect the rate of catalysis, as reflected by the Vmax/KM, indicating that they make a transition state energetically more favorable. The four small molecules share this functional property, even though they are structurally quite diverse and may promote different facets of the conformational changes within the DM:DR/peptide complex that lead to peptide release. Structural studies on DM and the DM:DR/peptide complex with these small molecules are required to precisely define the structural changes induced by the small molecules.
Given the structural diversity among these small molecules, it was surprising that a point mutation in DM (substitution of DM βL51 by D or N) greatly reduced the activity of three of the four small molecules. Microdialysis experiments confirmed that the βL51D mutation abrogated M19 binding to DM and that it reduced binding by F15. These three small molecules thus bind to a similar site of DM, even though they belong to the two different functional groups described above. DM βL51 is part of the hydrophobic ridge that had been first noted as an unusual feature in the crystal structure of H2-M (16), and the lateral interaction model that has been developed based on extensive mutagenesis of DM and DR positions this region of DM within proximity of the peptide binding groove (18, 19). Binding of the small molecules to this site is also supported by mutational analysis of the neighboring DM βE47 residue. The activity of both M19 and F15 was reduced by the βE47N and βE47A single mutations as well as the corresponding βE47-βD31 double substitutions. The functional properties of the small molecules that bind to this DM site therefore implicate the DMβ1 domain in the catalysis of peptide exchange. This area has an extended, semicircular hydrophobic surface as well as a number of polar residues (Fig. 10) that may be relevant for binding of DM to DR or for catalysis. A potential caveat of any mutagenesis experiment is that the effects of the mutation may not be strictly local, but result in more substantive conformational changes. The point mutations studied here in terms of small molecule function all yielded proteins that were catalytically active, excluding the possibility of misfolding. Four different mutations involving two DM residues located at this site had similar effects on small molecule function, providing strong experimental support for the conclusion that three of these molecules bind to or within close proximity of this site. Nevertheless, we cannot exclude the possibility that the small molecules induce changes in DM conformation that extend beyond the site identified in this mutagenesis experiments.
The compounds identified in this study are the first small molecules identified that modulate the catalytic efficiency of DM and have distinct properties from other compounds such as the detergents identified by Avva and Cresswell (38), which promote the release of low-affinity peptides from DR in the absence of DM and have no effect on the catalysis of peptide exchange by DM.
We identified DM mutants (βL51D, βD31N-βE47Q) with substantially altered pH activity profiles, suggesting that this site is involved in defining the pH optimum of DM. The protonation state of βE47 and βD31 is relevant for DM activity because substitution of both residues by the corresponding carboxamides (βD31N-βE47Q double mutant) broadened the pH range within which DM was active. At a neutral pH, these residues are likely to be deprotonated and thus carry a negative charge that may interfere with binding of DM to DR. Although wild-type DM had no activity at a neutral pH, this double mutant was catalytically active at a neutral pH. The βL51D mutation may alter the protonation state of the neighboring βD31 and βE47 residues and thus alter the pH profile. These data and the previous mutagenesis experiments involving DM βD31 and βE47 suggest that these residues are located at or close to the interface with DR/peptide in the DM:DR/peptide complex (19).
These results demonstrate that the catalytic efficiency of DM can be substantially increased by several different small molecules, raising the possibility that the editing function of DM could be increased to modify the peptide repertoire presented to CD4 T cells. Biasing the repertoire toward increased representation of high-affinity peptides may be useful in the induction of antimicrobial or antitumor T cell responses mediated by T cells that recognize peptides bound with high affinity to MHC class II molecules. Self-peptides with an intermediate or low affinity have been implicated in animal models of T cell-mediated autoimmune diseases, and presentation of such peptides may be reduced when the editing function of DM is increased (39). The compounds that are currently available have limited solubility and also have shown toxicity in preliminary cellular studies, indicating that they need to be modified for evaluation of these potential applications. The small molecules may also be valuable in conjunction with structural studies for further elucidating the mechanism by which DM catalyzes peptide exchange.
Acknowledgments
We thank Dr. Dennis Zaller for providing the transfectant that produces soluble DM, Drs. Steve DeWall and Brian DeDecker for discussions on the use of FP for monitoring of peptide binding, and Dr. Li-An Yeh and Jake Ni for their expertise and help in setting up the high-throughput screen.
Footnotes
This work was supported by National Institutes of Health Grants R01 NS044914 (to K.W.W. and R.L.S.), R01 AI057493 (to K.W.W.), and K01 DK068383 (to N.P.S.), and a fellowship from the HCNR (to M.J.N.).
Abbreviations used in this paper: DM, HLA-DM; DR, HLA-DR; FP, fluorescence polarization; MBP, myelin basic protein.
Disclosures The authors have no financial conflict of interest.
References
- 1.Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. Defective processing and presentation of exogenous antigens in mutants with normal HLA class II genes. Nature. 1990;343:71–74. doi: 10.1038/343071a0. [DOI] [PubMed] [Google Scholar]
- 2.Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, Monaco JJ, Mellins E. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- 3.Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- 4.Germain RN, Rinker AG., Jr. Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 5.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 6.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II αβ dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 7.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 8.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 9.Katz JF, Stebbins C, Appella E, Sant AJ. Invariant chain and DM edit self-peptide presentation by major histocompatibility complex (MHC) class II molecules. J. Exp. Med. 1996;184:1747–1753. doi: 10.1084/jem.184.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovitch SB, Petzold SJ, Unanue ER. Cutting edge: H-2DM is responsible for the large differences in presentation among peptides selected by I-Ak during antigen processing. J. Immunol. 2003;171:2183–2186. doi: 10.4049/jimmunol.171.5.2183. [DOI] [PubMed] [Google Scholar]
- 11.Nanda NK, Sant AJ. DM determines the cryptic and immunodominant fate of T cell epitopes. J. Exp. Med. 2000;192:781–788. doi: 10.1084/jem.192.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen PE. Long-lived complexes between peptide and class II major histocompatibility complex are formed at low pH with no requirement for pH neutralization. J. Exp. Med. 1992;176:793–798. doi: 10.1084/jem.176.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanzavecchia A, Reid PA, Watts C. Irreversible association of peptides with class II MHC molecules in living cells. Nature. 1992;357:249–252. doi: 10.1038/357249a0. [DOI] [PubMed] [Google Scholar]
- 14.Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc. Natl. Acad. Sci. USA. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J. Exp. Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fremont DH, Crawford F, Marrack P, Hendrickson WA, Kappler J. Crystal structure of mouse H2-M. Immunity. 1998;9:385–393. doi: 10.1016/s1074-7613(00)80621-4. [DOI] [PubMed] [Google Scholar]
- 17.Mosyak L, Zaller DM, Wiley DC. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- 18.Doebele CR, Busch R, Scott MH, Pashine A, Mellins DE. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 19.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 20.Stratikos E, Mosyak L, Zaller DM, Wiley DC. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-DM catalysis. J. Exp. Med. 2002;196:173–183. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J. Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 22.Smith KJ, Pyrdol J, Gauthier L, Wiley DC, Wucherpfennig KW. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. J. Exp. Med. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosinski M, Reid S, Nielsen LK. Kinetics of baculovirus replication and release using real-time quantitative polymerase chain reaction. Biotechnol. Bioeng. 2002;77:476–480. [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Methods in Enzymology. Vol. 276. Academic; London: 1997. Processing of x-ray diffraction data collected in oscillation mode; pp. 307–326. Macromolecular Crystallography Part A. [DOI] [PubMed] [Google Scholar]
- 26.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 27.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A. 1976;32:922–923. [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt. 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt. 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Owicki JC. Fluorescence polarization and anisotropy in high throughput screening: perspectives and primer. J. Biomol. Screen. 2000;5:297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 31.Pin SS, Kariv I, Graciani NR, Oldenburg KR. Analysis of protein-peptide interaction by a miniaturized fluorescence polarization assay using cyclin-dependent kinase 2/cyclin E as a model system. Anal. Biochem. 1999;275:156–161. doi: 10.1006/abio.1999.4331. [DOI] [PubMed] [Google Scholar]
- 32.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL, Hafler DA. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J. Exp. Med. 1994;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogsgaard M, Wucherpfennig KW, Cannella B, Hansen BE, Svejgaard A, Pyrdol J, Ditzel H, Raine C, Engberg J, Fugger L. Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA)-DR2-MBP 85–99 complex. J. Exp. Med. 2000;191:1395–1412. doi: 10.1084/jem.191.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joshi RV, Zarutskie JA, Stern LJ. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry. 2000;39:3751–3762. doi: 10.1021/bi9923656. [DOI] [PubMed] [Google Scholar]
- 35.Falk K, Lau JM, Santambrogio L, Esteban VM, Puentes F, Rotzschke O, Strominger JL. Ligand exchange of major histocompatibility complex class II proteins is triggered by H-bond donor groups of small molecules. J. Biol. Chem. 2002;277:2709–2715. doi: 10.1074/jbc.M109098200. [DOI] [PubMed] [Google Scholar]
- 36.Marin-Esteban V, Falk K, Rotzschke O. Small-molecular compounds enhance the loading of APC with encephalitogenic MBP protein. J. Autoimmun. 2003;20:63–69. doi: 10.1016/s0896-8411(02)00107-5. [DOI] [PubMed] [Google Scholar]
- 37.Fersht A. Enzyme structure and mechanism. 2nd Ed W. H. Freeman; New York: 1984. [Google Scholar]
- 38.Avva RR, Cresswell P. In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity. 1994;1:763–774. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 39.Harrington CJ, Paez A, Hunkapiller T, Mannikko V, Brabb T, Ahearn M, Beeson C, Goverman J. Differential tolerance is induced in T cells recognizing distinct epitopes of myelin basic protein. Immunity. 1998;8:571–580. doi: 10.1016/s1074-7613(00)80562-2. [DOI] [PubMed] [Google Scholar]