Abstract
Uncharacterized open reading frames (ORFs) in human genomic sequence often show a high degree of evolutionary conservation, yet have little or no tissue EST or protein data suggestive of protein product function. The encoded proteins may have highly restricted expression in specialized cells, subcellular specializations, and/or narrow windows during development. One such highly specialized and minute subcellular compartment is the neuromuscular junction (NMJ), where motorneurons contact muscle fibers. The electric Torpedo ray has evolved to expand the NMJ structure to the size of a large organ (electroplax organ), and we hypothesized that Torpedo electroplax proteins would be candidates for human ESTs expressed at the human NMJ. A total of 9,719 primary electroplax cDNA clones were sequenced. We identified 44 human ORFs showing high (>63%) amino acid identity to Torpedo electroplax transcripts with enrichment for mRNA splicing motifs (SH2 and pre-mRNA splicing domains), an observation potentially important for the strict nuclear domains maintained by myonuclei underlying the NMJ. We generated antibodies against two uncharacterized human genes (C19orf29 [Drosophila cactin] and C15orf24) and showed that these were indeed expressed at the murine NMJ. Cactin, a member of the Rel transcription factor family in Drosophila, localized to the postsynaptic cytosol of the NMJ and nuclear membrane. C15orf24 protein localized to the murine postsynaptic sarcolemma. We show a novel approach towards identifying proteins expressed at a subcellular specializations using evolutionary diversity of organ function and cross-species mapping.
Keywords: Electric organ, Neuromuscular Junction (NMJ), Proteome, Torpedo californica, Novel Gene Discovery, Open Reading Frame (ORF)
Introduction
The availability of complete genomic sequences for a wide array of species has facilitated cross-species comparisons of sequence data, and identification of highly conserved transcript units. For many of the conserved transcript units, EST data supports the expression of the transcript, and a protein has been identified with some function assigned to that protein. However, there remain many highly conserved transcript units where EST support is limited, and there has been no characterization of the predicted protein product. Genomics methods of identifying transcripts have typically used cDNA libraries constructed from whole tissues, whole embryos (during development), or immortalized cell lines. With the emergence of next-generation sequencing technologies, the possibility of identifying rare sequences is becoming a possibility [1, 2]. There are many highly specialized subcellular elements of cells, and both the cells harboring the specializations and the specializations may show very low abundance in any tissue or organ, or be expressed at a narrow time window during development. Such cells and subcellular specializations may not be expressed in immortalized cell culture models and/or difficult to detect in whole tissue extracts unless subcellular profiling is employed such as laser microdissection coupled to molecular profiling [3].
We hypothesized that it would be possible to take advantage of evolutionary diversity of cell and organ function, where a small specialization in humans has been grown to very large size in specific species. Examples of such species-specific augmentation of cells and subcellular specializations include the giant axon of squid [4, 5], and the electric organ of certain rays and eels [6-9]. A cross-species genomics approach might enable the identification of components of the species-specific organ, and then the human orthologues identified via cross-species homology comparison.
Myofibers are large syncytial cells. Typically, thousands of myonuclei, each derived from distinct myogenic precursor cells (myoblasts) fuse into the large myofiber that is visible to the naked eye. A very small membrane specialization of the myofiber is the NMJ, the point at which the motor neuron innervates the myofiber. The myofiber nuclei immediately underlying the NMJ express a distinct transcriptional program (nuclear domain), including localized expression of the acetylcholine receptor subunit genes and other NMJ-specific proteins. The NMJ is a 30-50 micron localized region of the myofiber plasma membrane, and comprises only a very small fraction (0.1%) of myofiber membrane surface area[10]. Due to the limited size of the NMJ, biochemists turned to the Torpedo electroplax, where this species has hypertrophied the NMJ to the size of a kilogram organ. The electroplax organ consists of stacks of hexagonally shaped modified and specialized muscle fibers (each called an electroplaque) which have lost the ability to contract, composed largely of NMJ-like structures [9, 11]. Instead of functioning as contractile muscle, the electroplax of electric fish has the primary function of generating electrical shock, up to 600V through water, in response to outside stimuli such as predator or prey [12]. During the 1970s and 1980s, initial studies using the Pacific electric ray (Torpedo californica) electric organ led to the first biochemical identification, purification and visualization of the key transmembrane ion channel, the acetylcholine receptor (AChR) [13-15]. Further molecular components of nerve synapses and neuromuscular junctions such as agrin and acetyl cholinesterase were consequently discovered using Torpedo electric organ [6, 8, 9, 16].
Despite many years of research, only about a dozen NMJ-associated proteins have been identified. The relative small size of the NMJ coupled with the lack of genome/proteome databases for Torpedo californica have hindered attempts to identify a more complete picture of the NMJ. For example, based on observed physiological or biochemical cascades, the existence of two proteins/complexes (MASC, RATL) have been hypothesized, yet the identity of these proteins remains unknown [17-19]. Also, the NMJ is a model for nuclear domains and plasma membrane specializations, with distinct mRNA and protein regulation required to maintain a single NMJ per cell, yet the molecular mechanics of establishing and maintaining the nuclear domains are not well understood. Identification of novel molecular constituents of the NMJ should provide insights into synapses, nuclear domains, regulation of membrane specializations, and disorders of these processes in poorly understood neuromuscular conditions such as spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS).
We have recently reported in a pilot study, a partial proteome and transcriptome profile of Torpedo electric organ [7]. In this previous study, we constructed a cDNA library of Torpedo electroplax, sequenced 607 cDNA clones, conducted cross-species homology searches, and generated a custom MS/MS spectral matching database for proteomics identification of proteins (cytoplasmic TOF/TOF MS/MS, and membrane LC-LTQ MS/MS). In this current report, we extend this analysis by sequencing an additional 9,719 primary cDNA clones. Our ability to identify novel components of the mammalian NMJ was validated by showing that the proteins encoded by two previously uncharacterized human open reading frames (C19orf29 [Drosophila cactin], C15orf24) both localize to murine neuromuscular junctions.
Materials and Methods
Mouse and Torpedo Specimens
Two (4-mo-old C57Bl/10) mice were euthanized by cervical dislocation, and tibialis anterior (TA), intercostals (IC) muscles and spinal cords (SC) were collected. The tissue was immediately frozen in isopentane cooled in liquid nitrogen and stored at _80°C. Specimens were used for, immunostaining, and western blott assays. All animal procedures and experiments complied fully with the principles set forth in the “Guide for the Care and Use of Laboratory Animals” prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Resources, National Research Council, and were approved by the Children’s National Medical Center’s Institutional Animal Use and Care Committee. Torpedo californica specimens were obtained from our collaborator Dr. Khurana (University of Pennsylvania).
cDNA library construction and sequencing, and custom proteome database
5′-biased cDNA library construction was done using polyA+ mRNA from flash frozen Torpedo electroplax organ, as previously described [7]. Torpedo clones were sequenced from the presumed 5′ end of the transcript/clone using the T3 primer (5′ATTAACCCTCACTAAAGGGA 3′). Clones were sequenced by High-Throughput Sequencing Solutions (University of Washington High-Throughput Genomics Unit, Seattle, WA). Templates were generated by Rolling Circle Amplification (Templi-Phi --GE Healthcare) and sequenced using Big-Dye chemistry (ABI), with products resolved and read using a ABI 3730xl (50cm Capillary). Sequencher software (Gene Codes, Ann Arbor, MI) was used to construct contigs of all sequences.
cDNA sequences (in FASTA format) were imported into GPS v3.5 Explorer software, translated into six reading frames, and in silico trypsin digested. This database was then searched using the same parameters as above [alkylation of Cys residues (+ 57 Da) and possible Met residues oxidation (+ 16Da)]. The resulting file was indexed as a local library searchable by peptide mass spectra.
Antibody production
Affinity-purified polyclonal antibodies were produced against two ORFs, chromosome 15 ORF 24 [(C15orf24) human gene ID: 56851, epitope: LPKVVNTSDPDMRRE; Torpedo accession number: EH115594), and chromosome 19 ORF 29 (C19orf29, human gene ID: 58509, epitope: GTSVDPVEPVEPEE; Torpedo accession number EH115553) the putative human orthologue of Drosophila cactin. The peptides were conjugated to KLH and coimmunized into rabbits, with resulting sera affinity purified using each immobilized peptide separately (Bethyl Laboratories).
Immunostaining
Mouse tibialis anterior muscles were dissected, snap frozen in isopentane cooled in liquid nitrogen, and sectioned at 8 μm. Sections were blocked with horse serum, and probed with C19orf24 or cactin primary antibodies (1:1000) that were detected by anti-rabbit Alexa Fluor 594 secondary antibodies (Molecular Probes). Sections were counterstained with anti-bungarotoxin Alexa Fluor 488 (Molecular Probes) to label the neuromuscular junction and with DAPI to label nuclei and imaged on a Zeiss ApoTome (Thornwood, NY) microscope. Controls for specificity of the immunostaining included slides incubated with secondary antibodies alone, primary antibodies mixed with purified antigenic peptide (epitope blocking), and preimmune sera from the same animal (data not shown).
Immunoblotting
Frozen torpedo electroplax tissue was obtained from Aquatic Research Consultants (San Pedro, CA). Mouse tibialis anterior muscles, gastrocnemius muscles, and spinal cord tissues were dissected and snap frozen on dry ice. All tissues were pulverized in liquid nitrogen, homogenized in PBS with 1% Tween, sonicated, and spun at 800 x g for 15 minutes to separate the cytosolic and membrane fractions. Protein concentration was determined by BioRad DC Protein Assay and 10 μg of extracts were analyzed by SDS-PAGE. Proteins were transferred to hybond nitrocellulose (Amersham) and the blots were probed with primary antibodies to cactin and secondary anti-rabbit-HRP antibodies. Signal was detected using the ECL system (Amersham).
Phylogenic Analyses
Alignments were done in Mesquite2.74 using ClustalW [20] with gap opening and extension costs set to 8/6.Bayesian analysis for each gene was performed using MrBayes V3.1.2 [21]. The Markov chain Monte Carlo was run with four chains for 1 million generations, sampling the Markov chain every 1000 generations, and the sample points of the first 250,000 generations were discarded as “burnin”.Other settings in MrBayes were lsetnst=6 rates=invgamma;unlink statefreq=(all) revmat=(all) shape=(all) pinvar=(all); prsetapplyto=(all) ratepr=variable. The taxon set, with the first one listed interpreted as primary outgroup, in analysis one was Homo sapiens, Torpedo californica, Mus musculus, Rattus norvegicus, Danio rerio. The taxon set in the second analysis was as above but with an addition of the primary outgroup Drosophila melanogaster.
Results
Sequencing of a cDNA library from Torpedo californica electroplax
We previously described the construction of a Torpedo electroplax cDNA library containing 5.16 × 106 primary colony forming units (cfu/ml) in pBluescript II KS+, enriched for 5′ ends of transcripts. In order to expand our transcriptome and proteome database of Torpedo electroplax from the 607 clones previously reported [7], we sequenced an additional 9,719 primary non-amplified colonies from this cDNA library. The average sequence length was 710 bp (uni-directional sequencing, average insert size 1.3 ± 0.86 kbp). All sequences were deposited in NCBI GenBank (GenBank sequence IDs EW688567-EW698285). We compiled the new cDNA sequences with the 607 clones sequenced previously [7], for total cDNA clone number of 10,326. 28% of clone sequences showed no overlap with other sequences (singletons), although non-overlapping clones of the same subsequence could not be ruled out.
The 10,326 clones assembled into 4,243 non-overlapping contigs. However, amongst singletons we detected non-overlapping clones (e.g. AChE) that represented the same gene. These 4,243 sequences were searched against non-redundant (NCBInr) database, using first human sequences to define the highest homologies (>75% amino acid identity), then zebrafish, mice and rat for those not showing strong alignment to human. Of these, 1,245 sequences showed homology to human.
Characterization of novel NMJ proteins
Within the 1,245 Torpedo cDNAs that we found strong support for mapping of a human orthologue, 44 were uncharacterized human ORFs (Table 1). These were candidates for novel protein components of the mammalian NMJ. To test this, we produced affinity-purified polyclonal antisera against two of the ORFs (C19orf29; C15orf24) that we had previously identified [7]. These two ORFs were selected based upon their particularly high conservation (C19orf29 96% identity between human and Torpedo over 55 amino acids; C15orf24 91% identity over 152 amino acids), and the suitability for antibody production (hydrophilic peptides). These two ORFs were detected by singleton ESTs from Torpedo.
Table 1.
Torpedo electroplax cDNA sequences that show homology to human open reading frames (ORFs).
| Torpedo | Human | |||
|---|---|---|---|---|
|
Torpedo cDNA GenBank Accession Number |
No. of cDNA Clones |
Gene/Protei ID Official Symbol |
Amino Acid | Function/Similar to |
| % Identity | ||||
| RNA Processing | ||||
| EH115702 | 1 | EAW75176.1; C20orf14 | 98% | PRP6 pre-mRNA processing factor |
| Hypothetical Function | ||||
| EW689494 | 1 | CAI14998.1 | 77% | Novel SH2 domain-containing protein |
| Contig of (EW692562; EW692829) | 2 | XP_935194 | 95% | Endosomal Targeting |
| EW691177 | 1 | EAW60225.1; C22orf5 | 90% | Transmembrane protein 184B |
| EW690431 | 1 | NP_057039.1; C2orf4 | 94% | Mediator of ErbB2-driven cell motility |
| EW689749 | 1 | NP_060387.2; C14orf10 | 92% | Serine/threonine-PP2A |
| Contig of (EW691292; EW692097) | 2 | EAW61630.1 | 75% | Isoform CRA_b |
| Unknown Function | ||||
| Contig of (EW697079; EW695860; EW697008; EW696100; EW696827; EW696779; EW695774; EW695604; EW695604; EW696506; EW695652; EW695415; EW696081; EW695704; EW695756; EW695676; EW695943; EW695655; EW697364; EW696424; EH115873; EH115813) |
22 | AAH02531.1, C20orf149 | 63% | Unknown |
| Contig of (EW694144; EW694078; EW694237; EW694146;EW694206; EW694024) |
6 | EAW63210.1; C8orf40 | 82% | Unknown |
| Contig of (EW693112; EW693254; EW693537; EW693542) |
4 | EAW77312.1; C15orf48 | 72% | Unknown |
| Contig of (EW693253; EW693111; EW693495; EW693493) |
4 | AAI09304.1; C10orf38 | 76% | |
| Contig of (EW692974; EW692531; EW692798) |
3 | NP_077291.1; C7orf23 | 88% | Unknown |
| Contig of (EW692796; EW692530; EW693070) |
3 | NP_060932.2; C3orf10 | 100% | Unknown |
| Contig of (EW691293; EW692020) | 2 | NP_077270.1; C6orf106 | 92% | Unknown |
| Contig of (EW691291; EW691704) | 2 | AAH19351.2; C4orf31 | 89% | Unknown |
| Contig of (EW692064; EW691290) | 2 | NP_060366.1; C20orf11 | 97% | Unknown |
| Contig of (EW691287; EW692192) | 2 | EAW69978.1; C1orf57 | 71% | Unknown |
| Contig of (EW691286; EW691724) | 2 | AAH10908.1; C1orf123 | 90% | Unknown |
| Contig of (EW691289; EW691888) | 2 | AAH04818.1; C14orf129 | 81% | Unknown |
| Contig of (EW691288; EW691858) | 2 | AAH02750.1; C11orf10 | 100% | Unknown |
| EW689747 | 1 | AAH02863.1; C9orf156 | 86% | Unknown |
| EW688678 | 1 | EAW88338.1; C9orf140 | 64% | Unknown |
| EH115833 | 1 | EAW94148.1; C7orf25 | 91% | Unknown |
| EH115528 | 1 | AAH11709.1; C6orf130 | 80% | Unknown |
| EW688651 | 1 | EAW62287.1; C5orf15 | 85% | Unknown |
| EW690380 | 1 | EAW70869.1; C2orf33 | 73% | Unknown |
| EW689205 | 1 | EAW59669.1; C22orf13 | 90% | Unknown |
| EW689145 | 1 | CAM14447.1; C20orf4 | 78% | Unknown |
| EW689145 | 1 | CAM14447.1; C20orf4 | 78% | Unknown |
| EW691211 | 1 | CAH73931.1; C1orf58 | 89% | Unknown |
| EW690281 | 1 | NP_110433.1; C1orf21 | 80% | Unknown |
| EW689261 | 1 | EAW94893.1; C1orf151 | 90% | Unknown |
| EH115553 | 1 |
NP_001074012.1; C19orf29 |
96% | Unknown |
| EW689526 | 1 | EAW94413.1; C17orf71 | 73% | Unknown |
| EH115594 | 1 | NP_064539.1; C15orf24 | 91% | Unknown |
| EW688992 | 1 | AAH30119.2; C15orf109 | 88% | Unknown |
| EW690655 | 1 | AAH21701.1; C14orf147 | 95% | Unknown |
| EW690632 | 1 | EAW80691.1; C14orf101 | 77% | Unknown |
| EW689791 | 1 | EAX08708.1; C13orf21 | 89% | Unknown |
| EW691179 | 1 | EAW58230.1; C12orf44 | 94% | Unknown |
| EW690914 | 1 | AAH02750.1; C11orf10 | 100% | Unknown |
| EW689274 | 1 | XP_946758.2 | 76% | Unknown |
| EW689251 | 1 | XP_001129158.1 | 100% | Unknown |
| EW689442 | 1 | NP_067037 | 94% | Unknown |
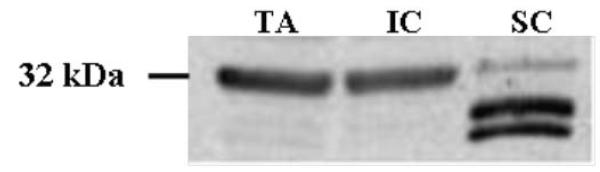
C15orf24 is predicted to transcribe a 726 bp mRNA corresponding to a 242 amino acid protein according to human genomic DNA data. Based on the predicted protein sequence, Gene Ontology (GO) annotation suggested that C15orf24 codes for an extracellular or membrane protein. Antibodies to C15orf24 were used for immunoblotting assays using mouse tibialis anterior (TA), intercostal (IC) muscle and spinal cord (SC) protein extracts (Figure 1). A 32 kDa band was identified in TA and IC muscle extracts whereas a less prominent band was present in the spinal cord protein extract, with smaller molecular weight species that represented isoforms, degraded products, or cross-reactive species. The apparent discrepancy in the calculated (26 kDa) and detected (32 kDa) molecular weight could be attributed to post-translational modifications. C15orf24 is predicted to code for a membrane or secreted protein. Such proteins are often highly post-translationally modified. For example, each glycosylated site could contribute up to 2kDa to protein mass. As such more investigation is necessary to identify potential post-translational modification.
Figure 1. Immunoblot of the protein product of C15orf24.
Shown is immunoblot analysis of the C15orf24, with detection of a band of approximately 32 kDa in mouse tibialis anterior (TA), intercostal muscles (IC) and Spinal cord (SC).
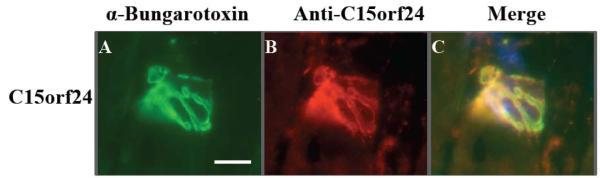
Alpha-bungarotoxin (α-BTX) binds with high affinity to α-acetylcholine receptor (α-AChR) in the myofiber membrane of the NMJ and was therefore used for co-staining of NMJs in mouse TA muscle. Co-localizations of α-BTX and the ORF proteins provide sub-structure information (plasma membrane vs. other subcellular localization; myofiber vs. motorneuron localization). Immunostaining assays using the C15orf24 antibody showed specific staining of the murine NMJ (Figure 2). Co-staining with α-bungarotoxin showed complete co-localization of the C15orf24 protein product and bungarotoxin (Figure 2), suggesting that the protein encoded by C15orf24 localizes to the NMJ at the post-synaptic membrane, in the myofiber plasma membrane. Thus, a singleton electroplax EST with high sequence similarity between Torpedo and human is shown to be a novel component of the myofiber membrane specialization of the NMJ.
Figure 2.
C15orf24 antibody detects a protein localized to the murine neuromuscular junction. Affinity purified antibodies against C14orf24 peptide were tested on longitudinal cryosections from murine tibialis anterior muscle. Fluorescently labeled bungarotoxin labels the post-synaptic (myofiber) membrane (Panel A). Antibodies to C15org24 protein showed a pattern very similar to bungarotoxin (Panel B), and double labeling of the same section showed precise co-localization (Panel C). Sections were also stained with Hoechst (blue) to localize subsynaptic myonuclei (arrow in Panel C). Scale bar = 20 μm.
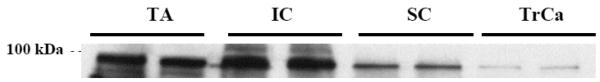
For C19orf29 (NCBI Accession number AY917150), sequence analysis predicts that this gene is the human orthologue of the Drosophila cactin gene, an inhibitor of kappa B (IκB) interacting protein [22]. The human cactin is predicted to code for a protein with 758 amino acids, and consistent with this, we detected as a band of around 100 kDa on a Western blot of mouse muscle (Figure 3). Immunoblot analysis of Torpedo electroplax tissue, murine muscle (gastrocnemius, tibialis anterior), and spinal cord showed that cactin is detected most strongly in muscle tissues (Figure 3).
Figure 3. Immunoblot of the C19orf29 ORF (cactin).
Shown is immunoblot analysis of murine tibialis anterior (TA), intercostal (IC), spinal cord (SC) muscle extracts and Torpedo electroplax (TrCa) using polyclonal antibody against protein encoded by C19orf19. The protein product of C19orf29 is detected at the predicted molecular weight (100kDa), and is seen enriched in membrane fractions of both Torpedo electroplax, and murine TA and gastrocnemius.
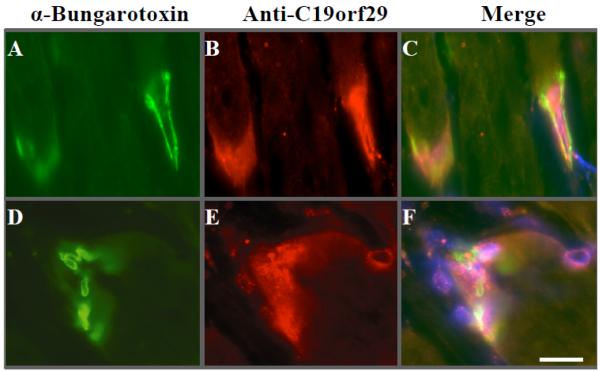
Co-immunostaining of cactin and bungarotoxin in frozen sections of murine muscle showed NMJ-specific localization of cactin (Figure 4). Control sections tested negative, including secondary antibodies alone, primary antibodies mixed with purified antigenic peptide (epitope blocking), and preimmune sera from the same animal (data not shown). Within the NMJ substructures, cactin was localized to the myofiber cytoplasm immediately underlying the NMJ myofiber membrane, and also to myonuclei underlying the NMJ (subsynaptic nuclei) (Figure 4). The nuclear staining appeared to be at the periphery of the nuclei, likely at the myonuclear envelope. This data shows that the human C19orf29 (presumed orthologue of Drosophila cactin) is expressed at the NMJ, and shows highly restricted expression to cytoplasm and nuclear envelop only within the NMJ nuclear domain (subcellular specialization).
Figure 4. Immunostaining of frozen sections of murine tibialis anterior with antibodies to the protein encoded by C19orf29 showed localization to the NMJ.
Co-localization with bungarotoxin (Panels A, C) showed different localization for C19orf29 and bugarotoxin (Panel C). Hoechst staining (blue) was also done to localize subsynaptic myonuclei (Panel F, arrow). C19orf29 was localized to the myofiber cytoplasm adjacent the NMJ (Panel B), and also the myonuclei immediately underlying the NMJ (Panel E, F), with apparent nuclear envelope localization (arrowhead Panel E, F). Scale bar = 20 μm.
Discussion
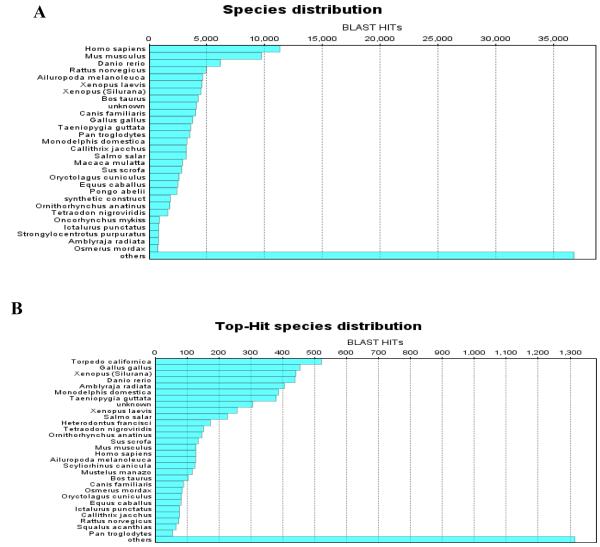
We present a cross-species genomics approach where a highly specialized organ, the electric organ of Torpedo californica, was used as a means to identify novel components of the mammalian neuromuscular junction. We sequenced a total of 10,326 cDNA clones from the electric organ. We used Blast2Go suit software [23] to analyze generated Torpedo californica sequences. Blast2Go integrates functional annotation of DNA or protein sequences based on the Gene Ontology (GO). Our analyzing using Blast2Go showed that the majority of sequences (~5,000) of sequences match wither to UniProt/TrEMBL or UniProtKB/Swiss-Prot databases. Species distribution showed that identified sequences have the highest similarity to Homo sapiens, Mus musculus, and danio rerio respectively (Figure 5A). Among these sequences, top hits belonged to Torpedo californica, Gallus gallus, Xenopus and Danio rerio respectively (Figure 5B). Since our sequences are already deposited in GenBank we expected to identify Torpedo californica as the top hit. However, identifying Gallus gallus as the second best hit was not expected and may indicate a more comprehensive existing Gallus gallus genome database compared to Xenopus and Danio rerio. We also used Blast2Go to determine the distribution of Torpedo californica sequences based on their role in biological processes (Supplemental Table 1), molecular function (Supplemental Table 2), and subcellular association (Supplemental Table 3). These analyses showed a high association with neuromuscular process, neurological system process, protein binding, and membrane fractions (Supplemental Tables 1, 2, 3). We inspected the 10,326 cDNA sequences by homology search for human ORFs of unknown function. We found that at least 44 transcripts showed a significant (>63%) degree of amino acid identity to uncharacterized human ORFs (Table 1). Although the main function of most of these proteins is unknown (i.e. contain conserved domains of unknown function), some of these transcripts contain domains involved in RNA processing, chromatin binding, or are homologous to Src homology 2 (SH2) domain (Table 1). Detection of these proteins emphasizes the importance of molecular machinery that is important for the establishment and maintenance of myonuclear domains. Furthermore bi-directional exchange of molecular information between muscle and nerve is important for stability of the NMJ [24]. Muscle derived growth factors for example influence motorneuron survival where as trophic factors such as agrin influence specific gene expression in subsynaptic myonuclei. Agrin, initially identified in Torpedo electric organ, is one of the main factors involved in initiating the clustering of postsynaptic scaffolding via MuSK signaling [8, 25]. Despite the fact that agrin is present in the adult synaptic regions in both nerve and muscle, it is expressed throughout the basal lamina during myofiber development [26]. However, it was later discovered that adult muscle fibers (and not motorneurons), express alternatively spliced agrin which lacks the C-terminal domain important for AChR clustering [27-30] More importantly, tyrosin phosphatases such as SH2 are important for facilitating post-synaptic agrin/MuSK signaling and more importantly AChR clustering at the NMJ [31-33].
Figure 5. Distribution of Torpedo californica sequences based on their percent identity when compared to existing databases.
A) Torpedo californica sequences show a high similarity to human sequences, however, when accounted for percent identity B) Torpedo sequences are more similar to other species including the puffer fish (Danio rario). Torpedo californica appears in on the top-hit species because all of our sequences are already deposited in GenBank.
Alternative splicing of α-dystrobrevin (DB) is a valid example of subsynaptic myonuclear domain specialization. DB exists in multiple isoforms including DB1 and DB2 [34, 35]. DB1, strictly expressed at NMJs and myotendinous junctions (MTJs), has an extended C-terminal domain, which is tyrosine phosphorylated, where as DB2 lacks this domain and is therefore expressed throughout the muscle fiber including NMJ and MTJ. Together, our findings provides suitable candidate human ORFs for future analysis towards identification of novel components of the NMJ.
One of the most interesting contigs of cDNA sequences is a one comprised of 22 clones (Table 1). The translated contig codes for 111 amino acids which has a 63% sequence identity (total of 71 amino acids) to translated human chromosome 20 ORF 149 (C20orf149). The high cDNA clone number may correlate with the degree of importance of this transcript in function and maintenance of the electric organ suggesting it as an ideal potential NMJ candidate.
Two ORFs were selected for antibody production based on their cDNA library source, computated subcellular localization and presence of functional domains. C15orf24 contains a domain of unknown function (DUF 2012) which belongs to the eukaryotic family of uncharacterized proteins. Our immunohistochemistry experiments showed that the protein coded by C15orf24 (C15orf24_P) localized to mouse NMJs (Figure 2). C15orf24_P localized to the postsynaptic membrane following a close proximity to acetylcholine receptors (AChRs). Further investigations are important to define the molecular functions of this protein at the NMJ and other electrical synapses.
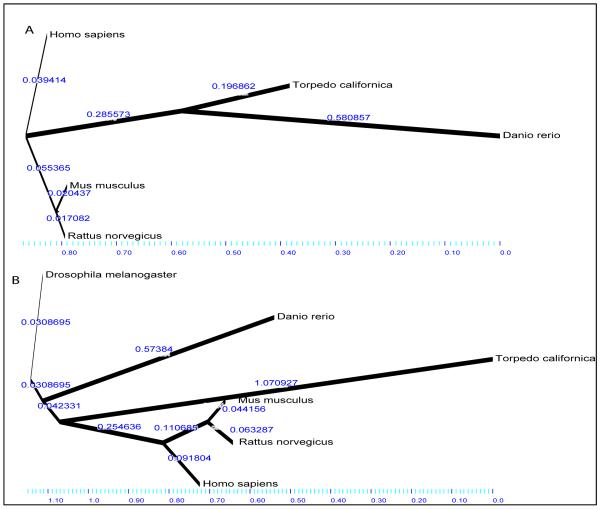
We have previously reported sequencing a Torpedo californica cDNA clone homologs to a human open reading frame on chromosome 19 (C19orf29) [7], which is homologous to the Drosophila cactin protein. Fly cactin was originally identified by yeast two hybrid system as a coil-coil protein with N-terminal Arg-Asp (RD)-like domain that directly interacts with the fly cactus protein [22]. In flies, cactin functions with the Rel family of transcription factors which are involved in vertebrate and invertebrate embryonic development and dorsal-ventral embryonic polarity [22, 36, 37]. Nuclear factor kappaB (NF-κB) is a member of Rel family which is important for neuronal synaptic plasticity [38]. In Drosophila, three genes, dorsal (dl), dorsal related immunity factor (Dif) and relish (Rel), encode NF-kB/Rel proteins [39-41]. Dif and dl are sequestered in the cytoplasm by dimerization with cactus. Upon phosphorylation and degradation of cactus, Dif and dl are released and translocated to the nucleus [42]. To test subcellular localization of C19orf29 (human cactin), we developed polyclonal antibodies against this protein and detected its localization to postsynaptic scaffolding as well as nuclear membrane of both subsynaptic and non-subsynaptic myonuclei (Figure 4). Our results, while adding cactin to the list, reinstate previous studies describing synaptic activities of Rel family proteins [43, 44]. Our observation that cactin is localized to the subsynaptic cytoplasm suggests that it is regulated by synaptic electrical activity. Indeed, the role of electrical impulses and their role in differential gene expression program of subsynaptic myonuclei is well studied [26, 45]. We conducted phylogenic Bayesian study of both C15ORF24 and C19ORF24 with branch lengths proportional to inferred divergence times (Figure 6). C15ORF24 sequence was compared to Homo sapiens, danio rerio, Mus musculus, Rattus norvegicus and Drosophila melanogaster orthologues. We found that C15ORF24 showed a substantial divergence that is shared between Torpedo californica and Danio rario (puffer fish), with additional evolutionary burst in the latter (Figure 5A). We analyzed C19ORF29 with sequences available from Homo sapiens, danio rerio, Mus musculus, Rattus norvegicus and Drosophila melanogaster. C19ORF29 showed extremely long and isolated divergence in Torpedo califirnica, which is not shared with any other taxon in the analysis (Figure 5B). More studies are required to reveal the specific role of these two ORFs at the NMJ and neuronal synapses.
Figure 6. The Bayesian tree for each gene with branch lengths proportional to inferred divergence times.
A) C15ORF24 exhibits substantial divergence that is shared between Torpedo californica and Danio rario, with an additional evolutionary burst in the latter. B) C19ORF29 on the other hand, shows extremely long and islolated divergence in Torpedo californica, which is not shared with any other taxon in this analysis.
This study, to our best knowledge, provides the largest number of molecules potential for synapse formation and stability. Furthermore, we show the NMJ association of two ORF-coded proteins to murine neuromuscular junctions. Future studies should focus on first localizing potential targets introduced in this study and further characterizing their specific function at the neuromuscular junction.
Supplementary Material
Research Highlights.
The amount of whole genome sequencing of many species is expanding rapidly. Whole genome sequencing may provide information on general aspects of evolution, but often does not impart more specific knowledge of adaptation of specific cells or organs. We chose to study an example of relatively extreme evolution, the Torpedo electric organ, where embryonic muscle tissue differentiates into a large organ capable of generating electric shock through water. We describe the sequencing of 9,719 cDNA clones (ESTs) from the Torpedo electric organ. This EST sequence resource was then utilized to identify two potential human/mouse undefined transcript units that were candidates for components of the mammalian neuromuscular junction. Antibodies to the human proteins encoded by these transcript units labeled the neuromuscular junction in murine myofibers. Our approach presents the more generalizable model where examples of extreme evolution can be leveraged to increase understanding of features of mammalian molecular physiology.
Acknowledgements
Supported by Children’s National Medical Center (CNMC) Intellectual and Developmental Disabilities Research Center (IDDRC) grant (P30HD40677), the W.M. Keck Foundation, and the Erynn Godla Family via the Juvenile ALS Foundation (www.jals.org). We would like to thank Dr. Matjaz Kuntner the head of Institute of BiologyScientific Research Centre of the Slovenian Academy of Sciences and Arts and Dr. Ingi Agnarsson, Assistant Professor and director of Museum of Zoology University of Puerto Rico for their help in generating and processing the phylogenic analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nazarian J, Bouri K, Hoffman EP. Intracellular expression profiling by laser capture microdissection: three novel components of the neuromuscular junction. Physiol Genomics. 2005;21(1):70–80. doi: 10.1152/physiolgenomics.00227.2004. [DOI] [PubMed] [Google Scholar]
- 4.Atchison WD, Luke VS, Narahashi T, Vogel SM. Nerve membrane sodium channels as the target site of brevetoxins at neuromuscular junctions. Br J Pharmacol. 1986;89(4):731–8. doi: 10.1111/j.1476-5381.1986.tb11177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clay JR. Axonal excitability revisited. Prog Biophys Mol Biol. 2005;88(1):59–90. doi: 10.1016/j.pbiomolbio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Miller C. Open-state substructure of single chloride channels from Torpedo electroplax. Philos Trans R Soc Lond B Biol Sci. 1982;299(1097):401–11. doi: 10.1098/rstb.1982.0140. [DOI] [PubMed] [Google Scholar]
- 7.Nazarian J, Hathout Y, Vertes A, Hoffman EP. The proteome survey of an electricity-generating organ (Torpedo californica electric organ) Proteomics. 2007;7(4):617–27. doi: 10.1002/pmic.200600686. [DOI] [PubMed] [Google Scholar]
- 8.Nitkin RM, Smith MA, Magill C, Fallon JR, Yao YM, Wallace BG, McMahan UJ. Identification of agrin, a synaptic organizing protein from Torpedo electric organ. J Cell Biol. 1987;105(6 Pt 1):2471–8. doi: 10.1083/jcb.105.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sealock R. Visualization at the mouse neuromuscular junction of a submembrane structure in common with Torpedo postsynaptic membranes. J Neurosci. 1982;2(7):918–23. doi: 10.1523/JNEUROSCI.02-07-00918.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsim KW, Barnard EA. The signaling pathways mediated by P2Y nucleotide receptors in the formation and maintenance of the skeletal neuromuscular junction. Neurosignals. 2002;11(1):58–64. doi: 10.1159/000057322. [DOI] [PubMed] [Google Scholar]
- 11.Sheridan MN. The Fine Structure of the Electric Organ of Torpedo Marmorata. J Cell Biol. 1965;24:129–41. doi: 10.1083/jcb.24.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent GC. Comparative anatomy of the vertebrates. Mosby-Year Book; St. Louis, MO: 1992. [Google Scholar]
- 13.Kistler J, Stroud RM. Crystalline arrays of membrane-bound acetylcholine receptor. Proc Natl Acad Sci U S A. 1981;78(6):3678–82. doi: 10.1073/pnas.78.6.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kistler J, Stroud RM, Klymkowsky MW, Lalancette RA, Fairclough RH. Structure and function of an acetylcholine receptor. Biophys J. 1982;37(1):371–83. doi: 10.1016/S0006-3495(82)84685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra AK, McCarthy MP, Stroud RM. Three-dimensional structure of the nicotinic acetylcholine receptor and location of the major associated 43-kD cytoskeletal protein, determined at 22 A by low dose electron microscopy and x-ray diffraction to 12.5 A. J Cell Biol. 1989;109(2):755–74. doi: 10.1083/jcb.109.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr C, Fischbach GD, Cohen JB. A novel 87,000-Mr protein associated with acetylcholine receptors in Torpedo electric organ and vertebrate skeletal muscle. J Cell Biol. 1989;109(4 Pt 1):1753–64. doi: 10.1083/jcb.109.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apel ED, Glass DJ, Moscoso LM, Yancopoulos GD, Sanes JR. Rapsyn is required for MuSK signaling and recruits synaptic components to a MuSK-containing scaffold. Neuron. 1997;18(4):623–35. doi: 10.1016/s0896-6273(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 18.Glass DJ, Bowen DC, Stitt TN, Radziejewski C, Bruno J, Ryan TE, Gies DR, Shah S, Mattsson K, Burden SJ, DiStefano PS, Valenzuela DM, DeChiara TM, Yancopoulos GD. Agrin acts via a MuSK receptor complex. Cell. 1996;85(4):513–23. doi: 10.1016/s0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, Glass DJ, Yancopoulos GD, Sanes JR. Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J Cell Biol. 1999;146(5):1133–46. doi: 10.1083/jcb.146.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294(5550):2310–4. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin P, Huang LH, Steward R. Cactin, a conserved protein that interacts with the Drosophila IkappaB protein cactus and modulates its function. Mech Dev. 2000;94(1-2):57–65. doi: 10.1016/s0925-4773(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 23.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rezaee M, Isokawa K, Halligan N, Markwald RR, Krug EL. Identification of an extracellular 130-kDa protein involved in early cardiac morphogenesis. J Biol Chem. 1993;268(19):14404–11. [PubMed] [Google Scholar]
- 25.Godfrey EW, Nitkin RM, Wallace BG, Rubin LL, McMahan UJ. Components of Torpedo electric organ and muscle that cause aggregation of acetylcholine receptors on cultured muscle cells. J Cell Biol. 1984;99(2):615–27. doi: 10.1083/jcb.99.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 27.Ferns MJ, Hall ZW. How many agrins does it take to make a synapse? Cell. 1992;70(1):1–3. doi: 10.1016/0092-8674(92)90525-h. [DOI] [PubMed] [Google Scholar]
- 28.Ferns M, Hoch W, Campanelli JT, Rupp F, Hall ZW, Scheller RH. RNA splicing regulates agrin-mediated acetylcholine receptor clustering activity on cultured myotubes. Neuron. 1992;8(6):1079–86. doi: 10.1016/0896-6273(92)90129-2. [DOI] [PubMed] [Google Scholar]
- 29.Ruegg MA, Tsim KW, Horton SE, Kroger S, Escher G, Gensch EM, McMahan UJ. The agrin gene codes for a family of basal lamina proteins that differ in function and distribution. Neuron. 1992;8(4):691–9. doi: 10.1016/0896-6273(92)90090-z. [DOI] [PubMed] [Google Scholar]
- 30.Tsim KW, Ruegg MA, Escher G, Kroger S, McMahan UJ. cDNA that encodes active agrin. Neuron. 1992;8(4):677–89. doi: 10.1016/0896-6273(92)90089-v. [DOI] [PubMed] [Google Scholar]
- 31.Madhavan R, Zhao XT, Ruegg MA, Peng HB. Tyrosine phosphatase regulation of MuSK-dependent acetylcholine receptor clustering. Mol Cell Neurosci. 2005;28(3):403–16. doi: 10.1016/j.mcn.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Zhao XT, Qian YK, Chan AW, Madhavan R, Peng HB. Regulation of ACh receptor clustering by the tyrosine phosphatase Shp2. Dev Neurobiol. 2007;67(13):1789–801. doi: 10.1002/dneu.20556. [DOI] [PubMed] [Google Scholar]
- 33.Zhao XT, Zhang Z. The role of protein tyrosine phosphatases Shp-2 involved in the formation of the neuromuscular junction. Zhonghua Yi Xue Za Zhi. 2006;86(15):1052–6. [PubMed] [Google Scholar]
- 34.Balasubramanian S, Fung ET, Huganir RL. Characterization of the tyrosine phosphorylation and distribution of dystrobrevin isoforms. FEBS Lett. 1998;432(3):133–40. doi: 10.1016/s0014-5793(98)00804-7. [DOI] [PubMed] [Google Scholar]
- 35.Wagner KR, Cohen JB, Huganir RL. The 87K postsynaptic membrane protein from Torpedo is a protein-tyrosine kinase substrate homologous to dystrophin. Neuron. 1993;10(3):511–22. doi: 10.1016/0896-6273(93)90338-r. [DOI] [PubMed] [Google Scholar]
- 36.Belvin MP, Anderson KV. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 37.Rusch J, Levine M. Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Curr Opin Genet Dev. 1996;6(4):416–23. doi: 10.1016/s0959-437x(96)80062-1. [DOI] [PubMed] [Google Scholar]
- 38.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6(10):1072–8. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 39.Ayyar S, Pistillo D, Calleja M, Brookfield A, Gittins K, Goldstone C, Simpson P. NF-kappaB/Rel-Mediated Regulation of the Neural Fate in Drosophila. PLoS ONE. 2007;2(11):e1178. doi: 10.1371/journal.pone.0001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo--shaping and transducing a morphogen gradient. Curr Biol. 2005;15(21):R887–99. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 41.Royet J, Reichhart JM, Hoffmann JA. Sensing and signaling during infection in Drosophila. Curr Opin Immunol. 2005;17(1):11–7. doi: 10.1016/j.coi.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110(3):791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- 43.Beramendi A, Peron S, Megighian A, Reggiani C, Cantera R. The inhibitor kappaB-ortholog Cactus is necessary for normal neuromuscular function in Drosophila melanogaster. Neuroscience. 2005;134(2):397–406. doi: 10.1016/j.neuroscience.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 44.Heckscher ES, Fetter RD, Marek KW, Albin SD, Davis GW. NF-kappaB, IkappaB, and IRAK control glutamate receptor density at the Drosophila NMJ. Neuron. 2007;55(6):859–73. doi: 10.1016/j.neuron.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2(11):791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.