Abstract
Chilodonella uncinata, like all ciliates, contains two distinct nuclei in every cell: a germline micronucleus and a somatic macronucleus. The macronucleus develops from the zygotic nucleus through a series of chromosomal rearrangements. Macronuclear development in C. uncinata yields a nucleus with highly amplified gene-sized chromosomes. The macronucleus is transcriptionally active during vegetative growth while there is no expression in the micronucleus except during a brief period following conjugation. Gene family evolution in ciliates occurs through complex processes including gene duplication and an alternative processing of scrambled genes. Here we use quantitative PCR to compare relative expression levels of eight genes (SSU-rDNA, Actin, α-Tubulin and five β-Tubulin sequences) to their abundance as macronuclear chromosomes. We show that three strains of the morphospecies C. uncinata share similar patterns across all loci. For example, we find an inverse correlation among five β-tubulin genes whereby the more abundant macronuclear chromosomes have lower levels of expression compared to less abundant chromosomes. We discuss implication of ours finding, which suggest an epigenetic mechanism maintains chromosome copy number in C. uncinata.
Keywords: Quantitative PCR, Gene expression, Ciliate, Tubulin, Copy Number Variation, Macronucleus
1. Introduction
Over the past decades, considerable insights have been gained into the variation in gene copy number and expression levels in eukaryotic genomes. Gene duplication and the existence of tandem arrays are important sources of this variability (Walsh, 1987). A further phenomenon impacting gene copy number is the widespread presence of extrachromosomal circular DNA in eukaryotes (Cohen and Segal, 2009), which may be involved in genome plasticity. For example, variations in copy number of rDNA are common and found in many organisms (Eickbush and Eickbush, 2007). Moreover, the explosion of molecular data from qPCR (VanGuilder et al., 2008), microarrays (Ranz and Machado, 2006) and more recently transcriptome sequencing (Wang et al., 2009) has allowed for the characterization of considerable variation in expression levels among eukaryotes.
Ciliates such as Chilodonella uncinata (Cl: Phyllopharyngea) are an appropriate model to study variation in gene copy number and expression level because extensive fragmentation yields a somatic macronucleus with gene-sized chromosomes (i.e. individual genes are on unlinked chromosomes; Riley and Katz, 2001). All ciliates are characterized by the presence of two types of nuclei in each cell: a transcriptionally-inactive micronucleus and a macronucleus that is analogous to the somatic nuclei of animals in that it is the site of virtually all gene expression. Following conjugation, the zygotic nucleus divides giving rise to a daughter nucleus, one of which will develop into a new macronucleus through extensive rearrangement that includes fragmentation of the genome, amplification and elimination of micronuclear-limited sequences (Prescott, 1994; Juranek and Lipps, 2007). In addition to gene-sized macronuclear chromosomes, C. uncinata has both gene scrambling, a process whereby coding regions must be ‘unscrambled’ from their non-canonical order in the zygotic genome, and the generation of gene family diversity though alternative processing of germline sequences (Katz and Kovner, 2010). The extensive rearrangements in C. uncinata yield a macronucleus with gene-sized chromosomes that divides through amitosis.
Cytoskeletal genes, the focus of this study are involved in generation of microtubules and actin filaments. Microtubules form major component of the eukaryotic cytoskeleton and are implicated in intracellular function, nuclear and cell division, conjugation, as well as in cilia movement. Microtubules are filamentous structures principally assembled of heterodimers of α- and β-tubulin (Bryan and Wilson, 1971). Most eukaryotic cells possess a multigene tubulin family disperse throughout their genome and thus express several isotypes of tubulin. These forms of tubulin are generally conserved but their complexity, heterogeneity, and diversity vary among species (MacRae and Langdon, 1989). For example, four functional α- and β-tubulin are present in Drosophila (Raff 1984), six in mice (Lewis et al., 1985) and only two of each gene in the green alga Chlamydomonas (Brunke et al., 1982; Montereiro and Cox, 1987). This broad diversity may be explained by a tissue-specific expression or specific life cycle stages (Burland et al., 1988).
Previous work on C. uncinata revealed the presence of one α-tubulin and multiple β-tubulin sequences in the macronuclear genome (Katz et al., 2003; Zufall et al., 2006; Zufall and Katz, 2007). Five sequences from β-tubulin show variability in their degree of conservation. Two sequences, shared P1 (SP1) and shared P2 (SP2) are highly conserved (identical amino acid sequences and < 2 % difference at the nucleotide level) but three macronuclear chromosome P1, P2, and P3 are more divergent with up to 13.5 % nucleotide differences (Robinson and Katz, 2007; Katz et al., 2011). Recent analyses of the micronuclear loci from the β-tubulin gene family reveal that an alternative processing of scrambled genes generates some of the diversity in the macronuclear sequences of this family (Katz and Kovner, 2010). For example, the macronuclear β-tubulin P1 and P2 are assembled by alternative processing of overlapping germline loci: MIC-P1, MIC-P2 and MIC-SP1 (Katz and Kovner, 2010).
To gain a better understanding of macronuclear chromosome number and gene expression of macronuclei, we analyze copy number and expression levels of eight different genes (SSU-rDNA, Actin, α-tubulin and five β-tubulins) by quantitative PCR. We characterize these sequences from three cryptic species of the morphospecies C. uncinata, one isolated from Poland and two from different places in the United States (USA-ATCC and USA-SC2) as described in Katz et al. (2011). We focus on the five β-tubulin chromosomes to assess the relationship between macronuclear copy number and expression patterns of gene family members.
2. Materials and Methods
2.1 Ciliate culture, DNA and RNA extraction
The American line USA-ATCC of C. uncinata was originally obtained from ATCC® 50194 and the POL line was isolated in culture from Stefan Radizowki and then deposited as ATCC® PRA-256 (Robinson and Katz, 2007). The third line, named USA-SC2, was collected on September 2007 at Lyman Pond (Smith College), Northampton, Massachusetts (N42 19’07”; W72 38’24”) as described in Katz et al. (2011) The presence of cryptic species within the morphospecies C. uncinata is supported by the concordant topologies of multiple loci, indicating a lack of recombination among strains (Katz et al. 2011). All strains were cultured at room temperature in the dark with filtered and autoclaved pond water and a rice grain.
Cultures were filtrated through a 5 µm filter to remove bacteria and collected on 0.22 µm filter, then pelletted by centrifugation at 5,000 rpm for 10 minutes. Numbers of cells were estimated by a direct count of 1 µl three times. The total genomic DNA of these three strains was extracted using the DNeasy plant kit from Qiagen (cat. # 69104). RNA was extracted from ciliate cell pellets using the Qiagen RNeasy Mini kit (cat. # 74104) and an additional step of DNase was completed at 37 °C for 30 minutes. The high capacity cDNA Reverse Transcription kit from Applied Biosystems was used to generate cDNA according to the manufacturer’s instructions.
2.2 Gene expression analysis by real-time quantitative PCR
Primers sets were designed with Primer 3 (Rozen and Skaletsky, 2000) for the C. uncinata target genes. Primers for SSU-rDNA, actin, α-tubulin, and four β-tubulin genes (β-tubulin-P1, β-tubulin-P2, β-tubulin-SP1, and β-tubulin-SP2) were designed to work on all three strains whereas strain-specific primers were generated for the more divergent β-tubulin gene P3 (β-tubulin-P3; Table 2). PCR amplifications were performed in a final volume of 25 µl, containing 1 µl (200 ng of cDNA) of a template DNA, 10 pg of each primer, 12.5 µl of Power SYBR Green master mix (Applied Biosystems), and 9.5 µl of water. The PCR thermal cycling protocol included initial steps of polymerase activation at 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles of 15s at 95 °C (denaturing) and annealing/extension at 62 °C for 1 min. Following amplification, melting curve analysis was performed at temperatures in the ranges of 50–95 °C, with the temperature increasing at a rate of 0.5 °C every 10s to determine amplification product specificity (Table 2). Measurements of the SYBR Green fluorescence were recorded during the annealing/extension step. For each experiment three independent cultures were tested in triplicates with negative control. All PCR reactions were performed in an ABI Prism 7300 Real-time PCR (Applied Biosystems).
Table 2.
Specific primers to target different gene of three strains of Chilodonella uncinata for Quantitative PCR.
| Gene | Primer | Primer sequence 5’→ 3 | Length (bp) |
Tm of Amplicon (°C) |
|---|---|---|---|---|
| SSU-rDNA | 985f | GAGTATGGTCGCAAGGCTGAAAC | 162 | 80.1 |
| 1147r | GCACCACCATCCCTCAAATCAAG | |||
| Actin | actf | GACTACGAGACCGAGCTCCAGGAGT | 160 | 82.5 |
| actr | TGGACACCTCCGAAGTCCTTGCCCAA | |||
| α-tubulin | 504f | CGAGGCCATCTACGATGTTT | 212 | 82.3 |
| 716r | GAGATGATCGGGGCATAAGA | |||
| β-tubulin-SP1 | 615f | CGGCAGATACCTCACTGCCT | 154 | 79.8 |
| 769r | TTGGAGGGTATCACAGATGGAGC | |||
| β-tubulin-SP2 | 394f | TCCGGAGTCACCTGCTGCTTG | 153 | 81.3 |
| 547r | GTCAGGGCTCTGTATTGCTGG | |||
| β-tubulin-P3 | 589f | TGCCTCTGCTCTATGAATAACAAG | 112 | 77.3 |
| POL | 701r | CGGGTTTGTAATTGCTGAACTGCA | ||
| β-tubulin-P3 | 589f | TGCCTTTGCTCGATGAACAACAAG | 109 | 78.6 |
| USA-ATCC | 698r | GTTTGAAGTTGTTGAACCGCGAT | ||
| β-tubulin-P3 | 35f | TCTCGCAGTACGAAGCTGAA | 238 | 81.4 |
| USA-SC2 | 273r | GCCAGTGTACGAGTGCAAGA | ||
| β-tubulin-P2 | 394f | TCCGGAGTCACCTGCTGTCTC | 144 | 82.5 |
| 538r | TGTATCCTTGTGAGCCGCGA | |||
| β-tubulin-P1 | 615f | CGGCAGATACCTGACTGCTGCT | 154 | 80.9 |
| POL | 769r | TCGGCGGTATGTCGCAGATACTGG | ||
| β-tubulin-P1 | 615f | CGGCAGATACCTGACTGCTGCT | 154 | 80.6 |
| USA-ATCC | 769r | GCGGTGGGATATCACAGATGCTGC |
2.3 Standard curve and amplification efficiency
Specific standard curves were generated for all genes of the three C. uncinata. We amplified SSU-rDNA, actin, α-tubulin, β-tubulin-SP1, SP2, P1, P2 and P3 from gDNA for each strain using primers described earlier (Robinson and Katz, 2007; Tekle et al., 2008) (Table 1). These PCR products were cleaned using the microClean DNA clean-up reagent (The Gel Company cat. # 2MCL-05), cloned with the Zero Blunt TOPO Cloning kit (Invitrogen cat. # 45-0245) and verified by sequencing. Six tenfold dilution standards were prepared freshly each time from minipreps of these genes, ranging 1 to 1 × 10−6 ng/µl. For each standard, six dilutions in triplicates were obtained and plotted against Ct values. The slope of the standard curves were calculated and the amplification efficiency (E) was estimated as E = (10−1/slope) −1. We obtained efficiency between 95–105 % for all genes.
Table 1.
Genbank accession numbers of macronuclear genes for three strains.
| Gene | POL | USA-ATCC | USA-SC2 |
|---|---|---|---|
| ssu-rDNA | JN111976 | AF300284.1 | JN111977 |
| Actin | EU047825 | DQ665938 | JN111998 |
| α-tubulin | JN111994 | AY041130.1 | JN111996 |
| β-tubulin-SP1 | GQ465330 | EU047814 | JN111984 |
| β-tubulin-SP2 | DQ665918 | EU047815 | JN111985 |
| β-tubulin-P1 | EU047816 | EU047817 | tbd |
| β-tubulin-P2 | EU047818 | EU047819 | JN111991 |
| β-tubulin-P3 | EU47820.1 | EU47821 | tbd |
2.4 Restriction Fragment Length Polymorphism
Because β-tub-SP1 and β-tub-SP2 are very similar, we verified the selectivity of PCR for these products by restriction analysis. First, we amplified sequences by PCR with the specific qPCR primers (Table 2). The program was an initial step of 15s at 98 °C followed by 37 cycles at 98 °C for 15s, 62 °C for 15s, and 72 °C for 90s. A final incubation at 72 °C for 10 min was added to the last cycle. For β-tub-SP1 we tested PCR products with Acl I (restriction enzyme that cuts β-tub-SP2 sequence) and with Sal I (enzyme that cuts β-tub-SP1 sequence). The same experiment was completed for β-tub-SP2 with Ban I (cuts β-tub-SP1 sequence) and with Hinc II (cuts β-tub-SP2 sequence) (New England Biolabs, Ispwich, MA). For each test, a positive control was added. The digestion was left 1hr at 37 °C with 10 µl of DNA, 2 µl of buffer, 1 µl of restriction enzyme, 2 µl BSA (10X) and 5 µl of water for a final volume of 20 µl.
3. Results
3.1 Determination of macronuclear gene copy number and expression level by quantitative PCR
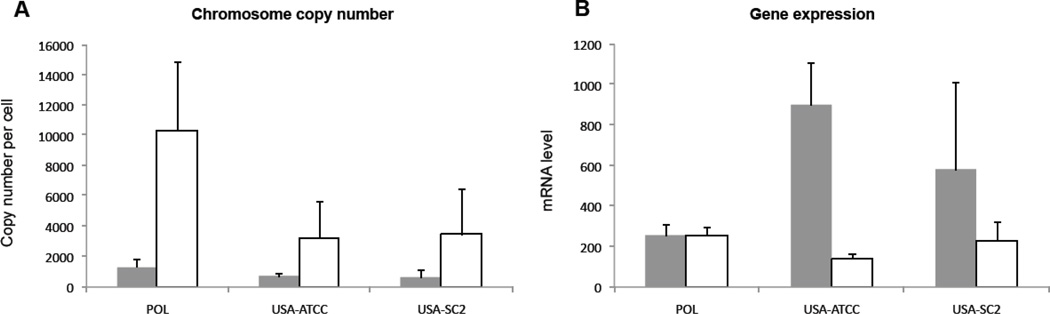
We used specific quantitative PCR primers for all target genes of the three C. uncinata strains, as described in Materials and Methods, to distinguish macronuclear chromosome copy number (quantification of genomic DNA) and gene expression (quantification of mRNA). All three lines of C. uncinata (USA-ATCC, POL and USA-SC2) have the same overall pattern of gene copy number when amplified from total genomic DNA. For example, the α-tubulin gene has a higher level than the actin gene (Fig. 1). For the mRNA expression level, we also observe the same overall pattern for the three lines but with a different relationship compared to DNA as the actin expression level is higher than the α-tubulin gene (Fig. 1). For example, the USA-ATCC line has an estimated 3438 α-tubulin macronculear chromosomes per cell and 644 for actin, while it has expression levels of 228 and 850 per 200 ng, respectively.
Fig. 1.
Macronuclear chromosome copy number and mRNA levels of two genes; Actin (grey), and α-tubulin (white) determined by quantitative PCR: (A) macronuclear chromosome copy number per cell and (B) mRNA expression levels. For each gene, three individual qPCR experiments were performed and triplicates were analyzed (n = 9). Error bars represent the standard deviation. POL, USA-ATCC and USA-SC2 refer to the different strains of C. uncinata used.
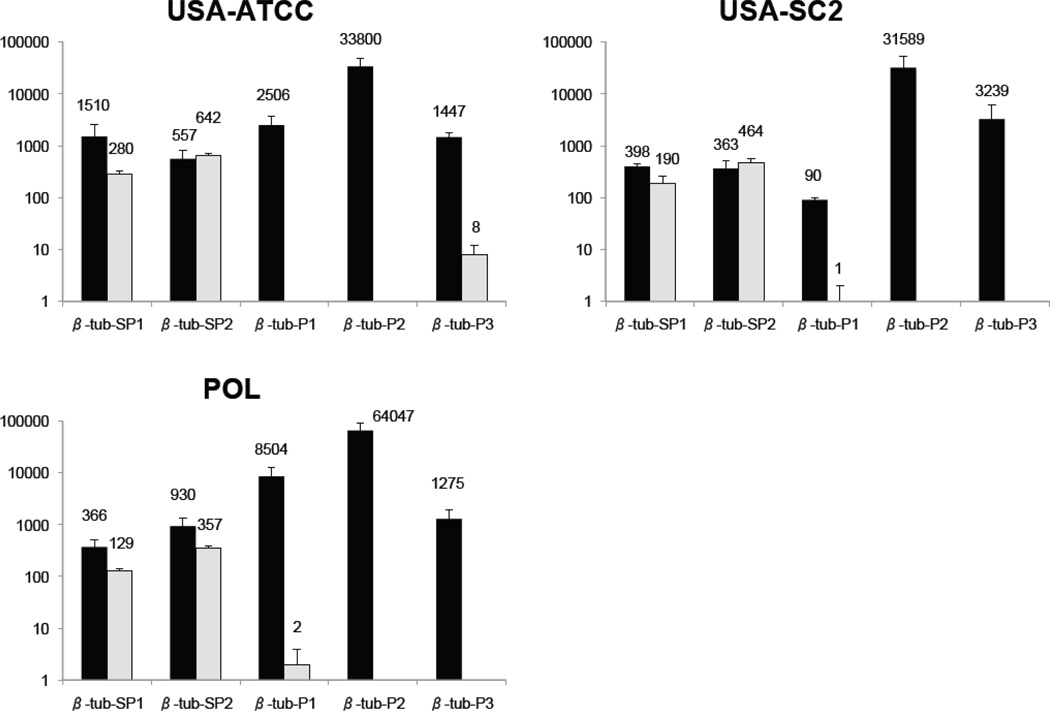
Analyses on the five β-tubulin genes also yield similar estimates for copy number and mRNA expression levels among the three strains. The decreasing order of DNA abundance detected is: β-tub-P2, β-tub-P1, β-tub-P3, β-tub-SP1, and β-tub-SP2 (Fig. 2). The one exception is for β-tub-P1 from USA-SC2 where we observe a low copy number per cell. The three genes with highest macronuclear chromosome copy number (β-tub-P2, β-tub-P1, β-tub-P3) do not have measurable expression at the mRNA level (Fig. 2). In contrast, both β-tub-SP1 and β-tub-SP2 have quantifiable gene expression, though not at the same level as β-tub-SP2 is always higher than β-tub-SP1 (Fig. 2, Table 3).
Fig. 2.
Copy numbers of macronuclear chromosomes (black) and levels of expression of β-tubulin gene as mRNA (grey) determined by quantitative PCR. For each gene sequence, three individual qPCR experiments were performed and triplicates were analyzed (n = 9). Error bars represent the standard deviation. POL, USA-ATCC and USA-SC2 refer to the different strains of C. uncinata used. The Y-axis is a logarithmic scale.
Table 3.
Ratio expression (in %) of target genes with α-Tubulin as reference gene for Quantitative PCR.
| Gene | POL DNA |
USA-ATCC DNA |
USA-SC2 DNA |
POL mRNA |
USA-ATCC mRNA |
USA-SC2 mRNA |
|---|---|---|---|---|---|---|
| SSU-rDNA | 571 | 846 | 1622 | NA | NA | NA |
| Actin | 13 | 22 | 19 | 99 | 658 | 373 |
| β-tub-SP1 | 4 | 47 | 12 | 50 | 204 | 83 |
| β-tub-SP2 | 9 | 17 | 11 | 139 | 469 | 204 |
| β-tub-P1 | 82 | 78 | 3 | 1 | 0 | 0 |
| β-tub-P2 | 619 | 1052 | 919 | 0 | 0 | 0 |
| β-tub-P3 | 12 | 45 | 94 | 0 | 5 | 0 |
NA = Not Applicable because we used an oligodT primer for the reverse transcription step.
The ratios of copy number and mRNA level for each gene, calculated using the α-tubulin gene as reference, allow comparisons across strains (Table 3). For example, the USA-SC2 line has divergent data for β-tub-P1 with a ratio of only 3 % whereas the two other lines have 82 % and 78 % (Table 3). This line also has a higher relative copy number for β-tub-P3 (94 %) compare the other lines (12 % and 45 %). Intriguingly, the USA-ATCC and POL strains share similar ratios and are more closely related to each other than either is to USA-SC2 (Katz et al. 2011).
We estimate the macronuclear copy number for the SSU-rDNA gene, which is the most abundant in the macronucleus for all three lines. The copy number of macronuclear rDNA is 500–1600 fold above the α-tubulin copy number (Table 3). We don’t have the expression level for this locus because we used an oligodT primer for the reverse transcription step in cDNA synthesis.
3.2 Restriction analysis of closely-related β-tubulin gene sequences
To confirm the differential amplification of the closely related β-tub-SP1and β-tub-SP2 sequences, we digested PCR products with different restriction enzymes as described in materials and methods. The enzyme specific to β-tub-SP1 cut PCR products generated with SP1 specific primers, and failed to digest PCR products generated by SP2 specific primers. A reciprocal experiment showed the same results as the enzyme specific for β-tub-SP2 showed a complete digestion of PCR products generated with SP2 specific primers, and failed to cut PCR products performed with SP1 specific primers. Hence, ours SP1 and SP2 β-tubulin primers were highly selective for target loci.
4. Discussion
The present study on macronuclear chromosome abundance and expression levels in multiple lines of the ciliate morphospecies Chilodonella uncinata reveals three main results: (1) macronuclear chromosome copy number does not predict expression pattern for actin and α-tubulin genes (Fig. 1); (2) there is an inverse correlation whereby chromosomes encoding divergent β-tubulin amino acid sequences are more abundant in the macronucleus but these loci are expressed at relatively low levels, while more conserved sequences have lower macronuclear chromosome copy number and higher levels of expression (Fig. 2); (3) the three lines have similar patterns for both copy number of chromosomes and expression levels.
For the first observation, the three lines of C. uncinata have the same pattern of macronuclear chromosomes copy number when amplified from total genomic DNA: the SSU-rDNA gene has the highest level followed by the α-tubulin gene, then the actin gene (Table 3). For the mRNA level of expression, an inverse relationship was observed; actin chromosome copy number is higher than α-tubulin expression (Fig. 1, Table 3). The rDNA chromosome has the highest chromosome copy number in this study and all ciliates species studied to date (Prescott, 1994). For example, in Tetrahymena species, the copy number of macronuclear rDNA is 9,000 or 200-fold above the copy number for other DNA genes (Larson et al., 1991). Chromosome copy number variation in cytoskeletal genes is also seen in other ciliates such as Stylonychia lemnae (Cl: Spirotrichea) where α- and β-tubulin chromosomes occur at a ratio of 1: 5 (Conzelmann and Helftenbein, 1987).
The pattern of copy number of β-tubulin macronuclear chromosomes is generally consistent with previous work on two of the lines of C. uncinata (Robinson and Katz, 2007). This earlier study, based on standard PCR and abundance in clone libraries, established the presence of five macronuclear chromosomes for the β-tubulin gene family. We confirm the presence of the five β-tubulin macronuclear chromosomes and assess copy number by quantitative PCR. We validate observations from clone library such as the relative numbers of macronuclear chromosomes. For example, the POL line shows lowest number for β-tub-SP1 and β-tub-SP2 in both clone libraries (Robinson and Katz, 2007) and qPCR (this study). Here, we further demonstrate that the β-tub-SP1 and β-tub-SP2 are more highly expressed. One notable dissimilarity between these two studies is the amount of β-tub-P2 macronuclear chromosome, which is rare in clone libraries but abundant as measured by qPCR. We hypothesize that this difference may be due to PCR bias as universal β-tubulin primers were used to generate the clone library results as opposed to the specific primers used for this study.
The clone libraries (Robinson and Katz, 2007) and qPCR (this study) studies were carried out almost five years apart on clonal lines that have been maintained in culture, and surprisingly, reveal similar patterns of macronuclear chromosome copy number for the five β-tubulin sequences. These results suggest that C. uncinata possess the ability to maintain a constant copy number of some genes within the macronucleus for a very long period of both asexual division and conjugation (which we observe consistently though at low frequencies (<5%) in our cultures). This pattern of conserved macronuclear chromosome copy number occurs despite the fact that amitotic division of ciliate macronuclei can potentially generate an unequal distribution of DNA content. The consistent pattern of macronuclear chromosomes copy number in this study is similar to patterns found in other ciliates with extensively-processed genomes including two ciliates in the class Spirotrichea, Euplotes crassus and Oxytricha fallax (Herrick et al., 1987; Baird and Klobutcher, 1991). Together, these data strengthen the hypothesis of an epigenetic regulation mechanism of DNA molecules in the macronucleus of ciliates (Prescott, 1994).
Based on the qPCR data and sequence analysis, we see a link between patterns of gene expression and both levels of amino acid conservation and codon bias. A comparison between the five macronuclear chromosomes of β-tubulin shows that β-tub-SP1 and β-tub-SP2 are closely related (around 95% of nucleotide identity) while the three other sequences, β-tub-P2, β-tub-P1, β-tub-P3, are up to 54 % divergence from one another. Expression levels of the three more divergent chromosomes are undetectable by qPCR, yet these genes are not pseudogenes (no indels or codon stops detected, despite divergence). Previous analysis of C. uncinata sequences also indicates that the effective numbers of codons used among β-tubulin family differ with lower numbers (i.e. more biased codon usage) for the conserved sequences (β-tub-SP1 and β-tub-SP2) compared to the more divergent macronuclear chromosomes (Zufall and Katz, 2007; Katz et al., 2011). Our quantitative PCR analyses demonstrate a further correlation between codon usage bias and expression levels: genes that are more biased sequences β-tub-SP1 and β-tub-SP2 are more highly expressed. This hypothesis is consistent with a previous study in Tetrahymena pyriformis of β-tubulin genes expression investigated by Northern blot hybridization (Soares et al., 1991).
The low level of expression for three divergent β-tubulin macronuclear chromosomes (β-tub-P1, β-tub-P2, and β-tub-P3) may be due to varying expression patterns within different life cycle stages. In Tetrahymena pyriformis, the expression pattern of two different β-tubulin seems to be different during reciliation (Soares et al., 1991). In the ciliate Euplotes focardii, transcription of two γ-tubulin are different with one gene cell-cycle dependant whereas the other one is constant during life cycle but at very low levels of expression (Maziale et al., 2008). The DNA and RNA extractions for our study were made on predominantly vegetative cells (90–95% of cells were not in conjugation or division states). Our results are also corroborated by transcriptome analysis of predominantly vegetative cells in which we see only β-tub-SP1 and β-tub-SP2 present (Grant et al., submitted).
A further explanation for the complex pattern of macronuclear chromosome copy number is related to the heteromeric nature of the macronucleus of C. uncinata. One unique characteristic within the class Phyllopharyngea is that the heteromeric macronucleus is made up of both DNA rich (dense chromatin granules) and DNA poor regions (Raikov, 1996). Perhaps the DNA rich region contains genes at high copy number that are rarely expressed. Under this scenario, requirements for nuclear architecture underlying heteromeric nuclei may drive chromosome copy number and expression patterns.
The data presented here indicate that macronuclear chromosome copy number and gene expression level are related as conserved genes are rare in macronuclei but highly expressed while divergent genes are abundant but less expressed. Moreover, this pattern is shared among three lines of C. uncinata and across time. Together, these data support the hypothesis that epigenetics plays an important role in patterns somatic DNA content across the eukaryotic tree of life (Parfrey and Katz, 2010).
Highlights.
-
□
Copy number is inversely correlated with expression in some gene families
-
□
Copy number does not predict expression level for protein-coding genes
-
□
Epigenetics drives chromosome copy number in the ciliate Chilodonella uncinata
-
□
Somatic genome content is regulated by epigenetic mechanisms in eukaryotes
Acknowledgements
We would like to thank Jessica R. Grant and Laura W. Parfrey for technical help. This work was supported by awards from the National Institutes of Health (AREA award 1R15GM081865-01) and the National Science Foundation (DEB 0919152 and DEB 0816828) to L.A.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baird SE, Klobutcher LA. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J Protozool. 1991;38:136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- Brunke KJ, Young EE, Buchbinder BU, Weeks DP. Coordinate regulation of the four tubulin genes of Chlamydomonas reinhardi. Nucl Acids Res. 1982;10:1295–1310. doi: 10.1093/nar/10.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J, Wilson L. Are Cytoplasmic Microtubules Heteropolymers? Proc Nat Acad Sci U.S.A. 1971;68:1762–1766. doi: 10.1073/pnas.68.8.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland TG, Paul ECA, Oetliker M, Dove WF. A gene encoding the major beta tubulin of the mitotic spindle in Physarum polycephalum plasmodia. Mol Cell Biol. 1988;8:1275–1281. doi: 10.1128/mcb.8.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Segal D. Extrachromosomal Cirucular DNA in Eukaryotes: Possible Involvement in the Plasticity of Tandem Repeats. Cytogenet Genome Res. 2009;124:327–228. doi: 10.1159/000218136. [DOI] [PubMed] [Google Scholar]
- Conzelmann KK, Helftenbein E. Nucleotide sequence and expression of two β-tubulin genes in Stylonychia lemnae. J Mol Biol. 1987;198:643–653. doi: 10.1016/0022-2836(87)90207-5. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. Finely Orchestrated Movements: Evolution of the Ribosomal RNA Genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JR, Lahr D, Rey FE, Gordon JI, Knight R, Molestina RE, Katz LA. Gene discovery in the transcriptomes of three diverse microbial eukaryotes: Corallomyxa tenera, Chilodonella uncinata, and Subulatomonas tetraspora. 2011 [Google Scholar]
- Herrick G, Hunter D, Williams K, Kotter K. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev. 1987;1:1047–1058. doi: 10.1101/gad.1.10.1047. [DOI] [PubMed] [Google Scholar]
- Juranek SA, Lipps HJ. New Insights into the Macronuclear Development in Ciliates. Int. Rev. Cytol. 2007;262:219–251. doi: 10.1016/S0074-7696(07)62005-1. [DOI] [PubMed] [Google Scholar]
- Katz LA, Lasek-Nesselquist E, Snoeyenbos-West OLO. Structure of the micronuclear a-tubulin gene in the phyllopharyngean ciliate Chilodonella uncinata: implications for the evolution of chromosomal processing. Gene. 2003;315:15–19. doi: 10.1016/j.gene.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Katz LA, Kovner AM. Alternative Processing of Scrambled Genes Generates Protein Diversity in the Ciliate Chilodonella uncinata. J Exp Zool. 2010;314B:480–488. doi: 10.1002/jez.b.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, DeBerardinis J, Hall MS, Kovner AM, Dunthorn M, Muse SV. Heterogeneous rates of molecular evolution among cryptic species of the ciliate morphospecies Chilodonella uncinata. J Mol Evol. 2011;73:266–272. doi: 10.1007/s00239-011-9468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DD, Umthun AR, Shaiu W-L. Copy number control in the Tetrahymena macronuclear genome. J Protozool. 1991;38:258–263. doi: 10.1111/j.1550-7408.1991.tb04439.x. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Gwo-Shu LM, Cowan NJ. Five mouse tubulin isotypes and their regulated expression during development. J Cell Biol. 1985;101:852–861. doi: 10.1083/jcb.101.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae TH, Longdon CM. Tubulin synthesis, structure, and fonction: what are the relionships? Biochem Cell Boil. 1989;67:770–790. doi: 10.1139/o89-116. [DOI] [PubMed] [Google Scholar]
- Marziale F, Pucciarelli S, Ballarini P, Melki R, Uzun A, Llyin A, Detrich HW, III, Miceli C. Different roles of two γ-tubulin isotypes in the cytoskeletons of the Antartic ciliate Euplotes focardii. FEBS Journal. 2008;275:5367–5382. doi: 10.1111/j.1742-4658.2008.06666.x. [DOI] [PubMed] [Google Scholar]
- Monteiro MJ, Cox RA. Primary structure of an α-tubulin gene of Physarum polycephalum. J Mol Biol. 1987;193:427–438. doi: 10.1016/0022-2836(87)90257-9. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Katz LA. Dynamic genomes of eukaryotes and the maintenance of genomic integrity. Microbe. 2010;5:156–164. [Google Scholar]
- Prescott DM. The DNA of Ciliated Protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff EC. The genetics of microtubule systems. J Cell Biol. 1984;99:1–10. doi: 10.1083/jcb.99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikov IB. Nuclei of Ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates Cells as Organisms. Gustav Fischer: 1996. pp. 221–243. [Google Scholar]
- Ranz JM, Machado CA. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol Evol. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Riley JL, Katz LA. Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol. Biol. Evol. 2001;18:1372–1377. doi: 10.1093/oxfordjournals.molbev.a003921. [DOI] [PubMed] [Google Scholar]
- Robinson T, Katz LA. Non-Mendelian inheritance of two cytoskeletal genes in the ciliate Chilodonella uncinata. Mol Biol Evol. 2007;24:2495–2503. doi: 10.1093/molbev/msm203. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Soares H, Cyrne L, Barahona I, Rodrigues-Pousada C. Different patterns of expression of β-tubulin genes in Tetrahymena pyriformis during reciliation. Eur J Bioche. 1991;197:291–299. doi: 10.1111/j.1432-1033.1991.tb15910.x. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Grant JR, Anderson LOR, Cole JC, Nerad T, Katz LA. Multigene phylogenetic analyses of diverse amoebae and assessment of stability of clades within ‘Amoebozoa’ based on rate corrected SSU-rDNA analysis. Mol Phylogenet Evol. 2008;47:339–352. doi: 10.1016/j.ympev.2007.11.015. [DOI] [PubMed] [Google Scholar]
- VanGuilder HD, Vrana KE, Freeman WN. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- Walsh JB. Persistance of Tandem Arrays: Implications for Satellite and Simple-Sequence DNAs. Genetics. 1987;115:555–567. doi: 10.1093/genetics/115.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall RA, McGrath C, Muse S, Katz LA. Genome architecture drives protein evolution in ciliates. Mol Biol Evol. 2006;23:1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]
- Zufall RA, Katz LA. Micronuclear and macronuclear forms of β-tubulin genes in the ciliate Chilodonella uncinata reveal insights into genome processing and protein evolution. J Eukaryot Microbiol. 2007;54:275–282. doi: 10.1111/j.1550-7408.2007.00267.x. [DOI] [PubMed] [Google Scholar]