Abstract
Sirtuin proteins regulate diverse cellular pathways that influence genomic stability, metabolism, and ageing1,2. SIRT7 is a mammalian sirtuin whose biochemical activity, molecular targets, and physiologic functions have been unclear. Here we show that SIRT7 is an NAD+-dependent H3K18Ac (acetylated lysine 18 of histone H3) deacetylase that stabilizes the transformed state of cancer cells. Genome-wide binding studies reveal that SIRT7 binds to promoters of a specific set of gene targets, where it deacetylates H3K18Ac and promotes transcriptional repression. The spectrum of SIRT7 target genes is defined in part by its interaction with the cancer-associated ETS transcription factor ELK4, and comprises numerous genes with links to tumour suppression. Notably, selective hypoacetylation of H3K18Ac has been linked to oncogenic transformation, and in patients is associated with aggressive tumour phenotypes and poor prognosis3–6. We find that deacetylation of H3K18Ac by SIRT7 is necessary for maintaining essential features of human cancer cells, including anchorage-independent growth and escape from contact inhibition. Moreover, SIRT7 is necessary for a global hypoacetylation of H3K18Ac associated with cellular transformation by the viral oncoprotein E1A. Finally, SIRT7 depletion markedly reduces the tumourigenicity of human cancer cell xenografts in mice. Together, our work establishes SIRT7 as a highly selective H3K18Ac deacetylase and demonstrates a pivotal role for SIRT7 in chromatin regulation, cellular transformation programs, and tumour formation in vivo.
The chromatin silencing factor Sir2 (Silent Information Regulator-2) catalyzes NAD+-dependent histone deacetylation to regulate genomic stability and cellular senescence in budding yeast1,2. In mammals, SIRT7 is the only sirtuin (Sir2 family member) for which a clear enzymatic activity has remained elusive. Indirect evidence has led to the suggestion that SIRT7 deacetylates the tumour suppressor p537, although in vitro and cellular data do not support this model (Supplementary Fig. 1, reference 8). In addition, multiple studies have failed to detect direct deacetylase activity of SIRT7 on histones or other substrates1.
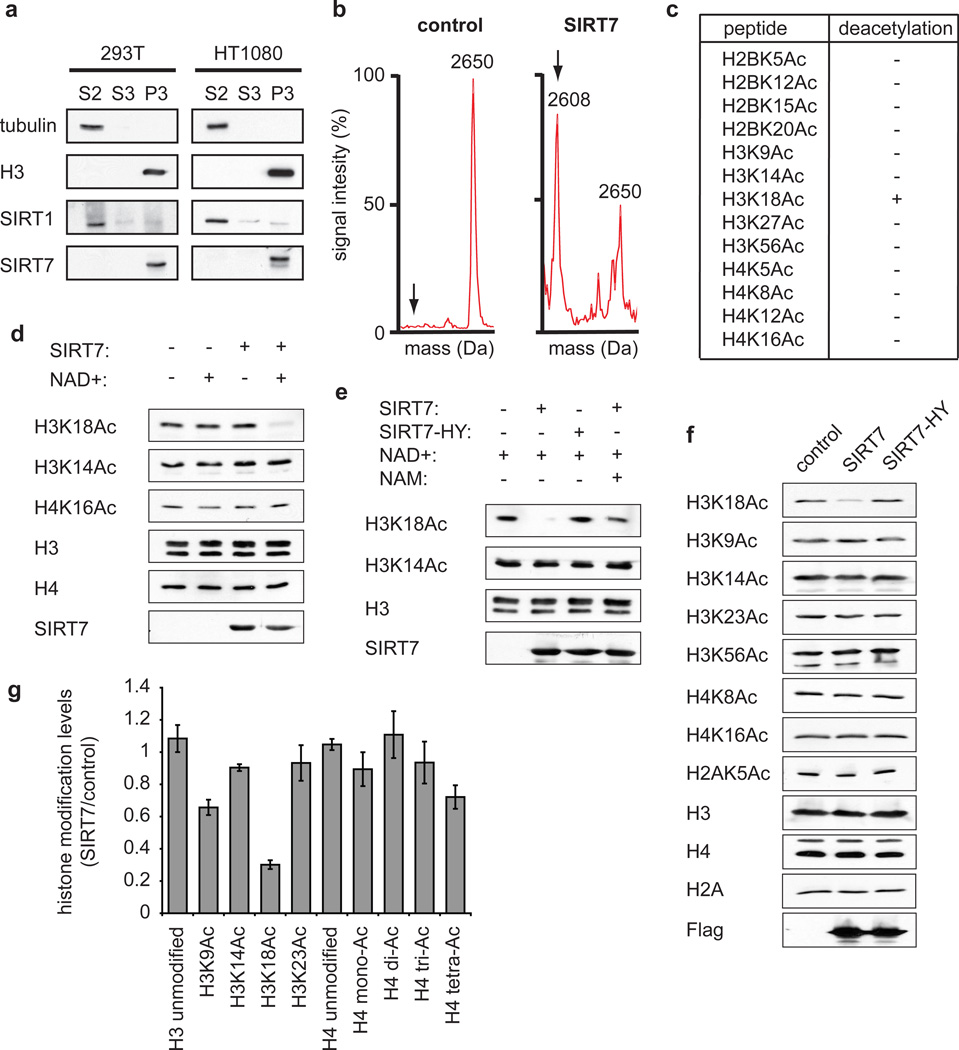
In biochemical fractionation studies, we detected SIRT7 almost exclusively in a chromatin-enriched fraction, suggesting that SIRT7 might function at chromatin (Fig. 1a). We therefore used mass spectrometry to screen for potential NAD+-dependent histone deacetylase activity of SIRT7 in vitro. Notably, SIRT7 exhibited highly specific deacetylase activity on peptides containing H3K18Ac, but had no activity on 12 other histone acetylation sites tested (Fig. 1b, c). This striking selectivity of SIRT7 for H3K18Ac contrasts with the broader substrate spectrum of other deacetylases such as SIRT1 (Supplementary Table 1) or HDAC19. SIRT7 also exhibited robust and specific NAD+-dependent H3K18Ac-deacetylase activity on full-length histone H3 in purified poly-nucleosomes (Fig. 1d). This activity was abolished by substitution of a conserved histidine residue (H187→Y) in the predicted catalytic domain of SIRT7 and by the general sirtuin inhibitor nicotinamide (Fig. 1e). Selective H3K18Ac deacetylation was also observed in cells following overexpression of the wild-type, but not mutant, SIRT7 protein (Fig. 1f). Moreover, an unbiased proteomic approach using quantitative mass spectrometry independently demonstrated that SIRT7 overexpression induces a dramatic depletion of H3K18Ac in cells (Fig. 1g), whereas changes in other acetylation marks, presumably due to downstream effects on chromatin structure, were more modest or negligible. Together, our data demonstrate that SIRT7 is an NAD+-dependent H3K18Ac deacetylase and the first known deacetylase with high selectivity for the H3K18Ac chromatin mark.
Figure 1. SIRT7 is a chromatin-associated H3K18Ac-specific deacetylase.
a, Western analysis showing chromatin association of SIRT7 in 293T and HT1080 cells. Biochemical fractions S2, S3, and P3 are enriched for cytoplasm, nucleoplasm, or chromatin, respectively. b, Mass spectra showing deacetylation of H3K18Ac peptide by SIRT7 compared to negative control reaction lacking enzyme. Molecular weights of acetylated and deacetylated (arrows) peptides are 2650 and 2608 Daltons, respectively. c, Results of SIRT7 deacetylation reactions using acetylated histone peptides, determined by mass spectrometry as in (b). d, e, Western analysis of H3K18Ac deacetylation activity of wild-type (SIRT7) or mutant (SIRT7-HY) proteins on poly-nucleosomes in vitro, and inhibition by nicotinamide (NAM). f, Western analysis showing H3K18Ac levels in 293T cells transfected with Flag-tagged SIRT7, SIRT7-HY, or control empty vector. g, Changes in global histone acetylation levels in SIRT7 overexpressing versus control 293T cells, determined by quantitative mass spectrometry. Error bars represent standard error of the mean (S.E.M.) of three independent experiments.
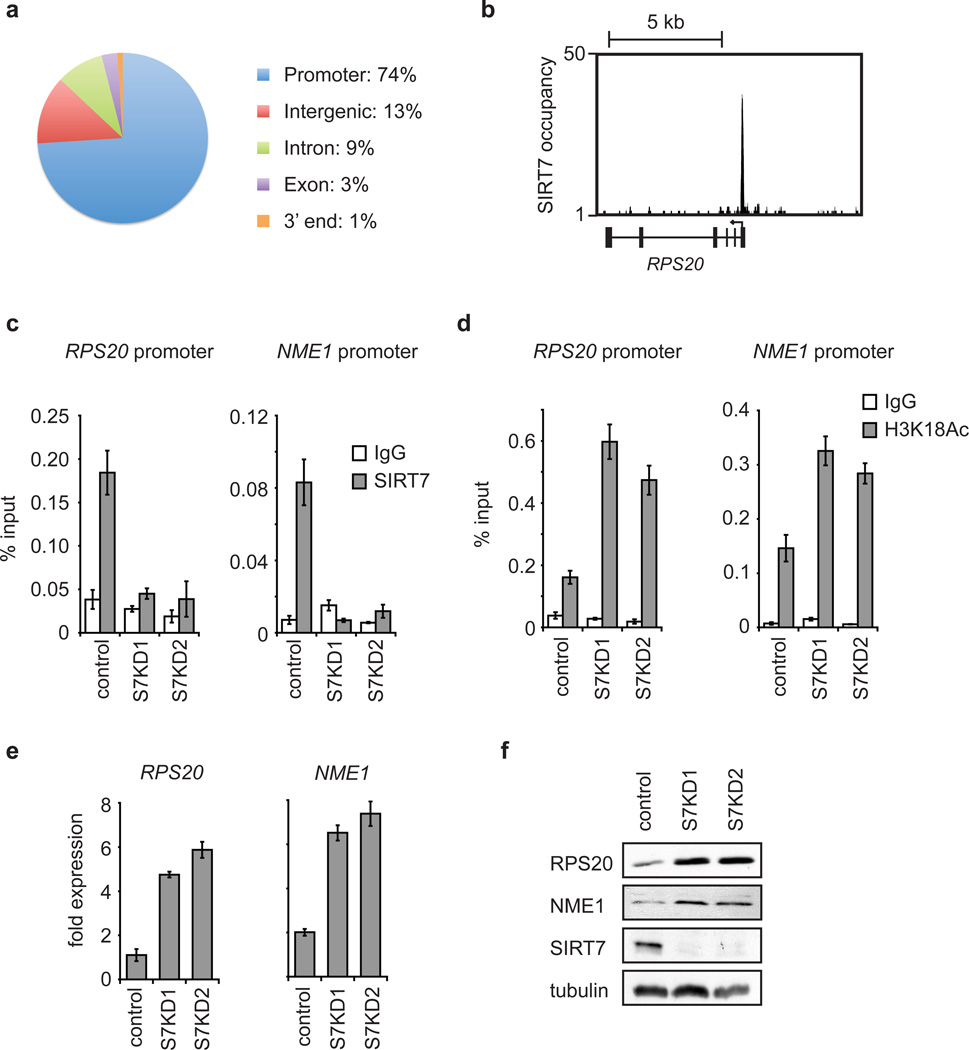
Depletion of H3K18Ac has been associated with aggressive cancer phenotypes and poor patient prognosis5,6, and in cellular studies, has been linked to epigenetic reprogramming during transformation of primary human cells by viral oncoproteins3,4. In addition, H3K18Ac is enriched at gene promoters and correlates with transcriptional activation10. We therefore hypothesized that SIRT7 might deacetylate H3K18Ac at promoters to modulate cancer-related gene expression programs. We first determined the genome-wide occupancy of SIRT7 by ChIP-sequencing. Results from multiple independent ChIP-sequencing experiments identified 276 SIRT7 binding sites (Supplementary Tables 2–4), which were dramatically enriched for proximal promoter regions (Fig. 2a, b, Supplementary Fig. 2a, b). Notably, SIRT7 binding sites also overlapped significantly with previously mapped regions of H3K18Ac enrichment (p-value 1.4e-80)10. Together, these data suggest that SIRT7 is a locus-specific enzyme that is positioned to deacetylate H3K18Ac at promoters of a select set of gene targets.
Figure 2. SIRT7 binds to gene promoters and couples H3K18 deacetylation to transcriptional repression.
a, Enrichment of SIRT7 in promoter proximal regions, determined by ChIP-sequencing. b, Representative SIRT7 ChIP-sequencing peak at the RPS20 gene TSS (arrow). c, ChIP-qPCR (mean +/− S.E.M.) showing SIRT7 occupancy in control or SIRT7 knockdown (S7KD1, S7KD2) HT1080 cells, compared to IgG negative control samples. d, ChIP-qPCR (mean +/− S.E.M.) showing H3K18Ac hyperacetylation in S7KD HT1080 cells. e, Increased expression of SIRT7 target genes in S7KD HT1080 cells determined by qPCR (mean +/− S.E.M.). Signals were normalized to GAPDH expression. f, Western blots of cell extracts corresponding to samples in e.
The identified SIRT7 ChIP-sequencing peaks correspond to 241 protein-coding genes (See Methods for details). Using ChIP-qPCR, we confirmed the binding of SIRT7 at several of the identified promoters and validated the specificity of the ChIP signals by siRNA-mediated depletion of SIRT7 (Fig. 2c, Supplementary Fig. 3). Functional categorization of the SIRT7-bound genes revealed strong enrichment for factors involved in RNA processing, protein translation and cellular macromolecule metabolism, with diverse links to tumour suppressive activities (Supplementary Fig. 2). Interestingly, SIRT7 bound upstream of several ribosomal protein (RP) genes, whose mis-regulation has been linked to cancer in multiple settings (see below), as well as genes found repressed in aggressive cancers or identified in screens for tumour suppressor genes (e.g., NME1 and COPS2)11,12.
We next asked whether SIRT7 deacetylates H3K18Ac at the promoters of specific candidate target genes. SIRT7-depletion led to hyperacetylation of H3K18 at the promoters of the RPS20, RPS7, RPS14, NME1, and COPS2 genes, but not multiple negative control promoters (Fig. 2d, Supplementary Fig. 4). Consistent with this locus specificity, global H3K18Ac levels were not affected by SIRT7 depletion (Supplementary Fig. 5). Importantly, SIRT7 knockdown (S7KD) also led to specific increases in expression of multiple target genes (Fig. 2e, f, Supplementary Fig. 6, 7), whereas depletion of HDAC1 (which can also deacetylate H3K18Ac9) did not (Supplementary Fig. 8). Together, our findings demonstrate that SIRT7 functions in gene-specific transcriptional repression at a select subset of H3K18Ac-containing promoters.
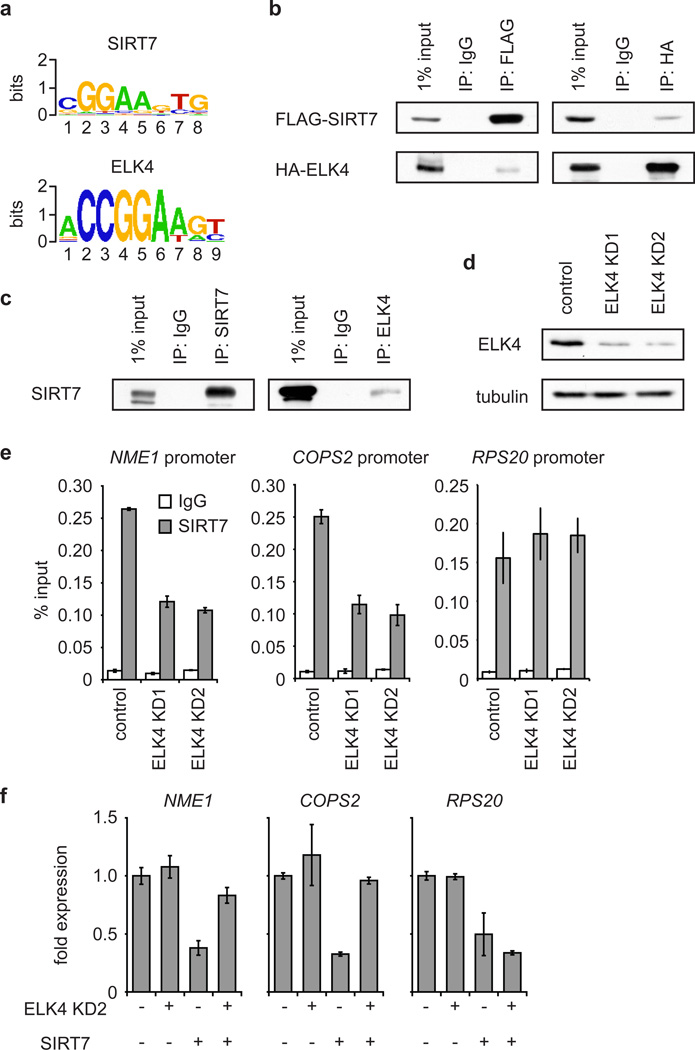
We next asked how selective recruitment of SIRT7 to its target promoters is achieved. SIRT7 lacks known sequence-specific DNA binding domains, leading us to hypothesize that it might interact with other proteins that contain such domains. We therefore identified de novo DNA motifs that are enriched in SIRT7-bound promoter sequences, and compared these motifs to curated transcription factor binding motifs in the JASPAR CORE database13. Of the 50 most significant SIRT7-associated motifs, 25 corresponded to consensus binding sites for the ETS (E26 Transformed Specific) family of transcription factors, many of which are important modulators of cellular transformation and cancer progression14. The SIRT7 consensus motif was most similar to the DNA sequence recognized by the ETS protein ELK4 (Fig. 3a).
Figure 3. SIRT7 is stabilized at target promoters by interaction with the ETS family transcription factor ELK4.
a, Comparison of the SIRT7 consensus motif to the ELK4 consensus motif (e-value: 9.66e-9). b, Western analysis showing co-immunoprecipitation (co-IP) of FLAG-tagged SIRT7 and HA-tagged ELK4 expressed in 293T cells. c, Western blots showing co-IP of endogenous SIRT7 and ELK4 proteins. d, Westerns blots showing knockdown of ELK4 from HT1080 cells with two independent siRNAs. e, Partial reduction of SIRT7 occupancy at target promoters in ELK4 KD HT1080 cells determined by ChIP (mean +/− S.E.M.). f, ELK4 depletion attenuates SIRT7-mediated transcriptional repression in HT1080 cells, as determined by qPCR. Error bars represent S.E.M. of three independent experiments.
Although its molecular function has not been extensively studied, ELK4 has been implicated in both transcriptional activation and repression15. Of the 276 SIRT7 binding sites that we identified by ChIP-sequencing, 57.6% contain at least one ELK4 consensus motif, a significant enrichment over total RefSeq promoters (p-value 3.1e-89) (Supplementary Tables 5, 6). In addition, ~70% of SIRT7 binding sites overlap with ELK4 peaks previously identified by ChIP-sequencing (p-value < 1e-300)16. To examine the potential interplay between SIRT7 and ELK4, we first confirmed that ELK4 binds several of the SIRT7 target promoters that contain the ETS consensus motif (NME1 and COPS2), but not promoters lacking the motif (RPS20 or GAPDH; Supplementary Fig. 9a). Next, in co-immunoprecipitation experiments, we found that SIRT7 interacts physically with ELK4 (Fig. 3b, c; Supplementary Fig. 10a), but not with two other ETS proteins, ELK1 and GABPα (Supplementary Fig. 10b). To assess the functional importance of this interaction, we examined the effects of ELK4 knockdown on SIRT7 ChIP occupancy at specific target promoters. Depletion of ELK4 led to a partial but significant decrease in SIRT7 occupancy at the NME1 and COPS2 promoters but not the RPS20 promoter (Fig. 3d, e; Supplementary Fig. 11a, b), and did not alter global levels of SIRT7 at chromatin (Supplementary Fig. 12). Moreover, ELK4 knockdown led to elevated H3K18Ac levels at the NME1 and COPS2 promoters, but not at promoters lacking the ETS motif (Supplementary Fig. 9b). Together, these findings suggest that ELK4 functions to target SIRT7 to specific promoters for H3K18 deacetylation.
We next examined the effects of ELK4 knockdown on gene repression by SIRT7. ELK4 knockdown did not appreciably alter expression of NME1 and COPS2 under baseline conditions (Fig. 3f), likely because considerable SIRT7 protein remained bound at these promoters (Fig. 3e). This incomplete loss of promoter-bound SIRT7 could reflect the incomplete depletion of ELK4 (Fig. 3d) as well as compensatory activity of other ETS factors in SIRT7-targeting. Indeed, the SIRT7 ChIP-sequencing peaks displayed some overlap (25%; p-value < 1e-300) with binding sites for ELK1. Importantly, however, knockdown of ELK4, but not ELK1 or GABPα, was sufficient to impair gene repression induced by overexpression of SIRT7 (Fig. 3f; Supplementary Figs. 11c, 13). Thus, any compensatory capacity of other ETS factors is exceeded under conditions of elevated SIRT7 expression, and in this setting, ELK4 is the main ETS factor responsible for SIRT7 targeting. Thus, we conclude that the promoter stabilization of SIRT7 by ELK4 is important for SIRT7-mediated gene repression. Moreover, this functional interplay between ELK4 and SIRT7 might be particularly important in settings of elevated SIRT7 expression, which occurs in certain cancers (Supplementary Fig. 14)17–19.
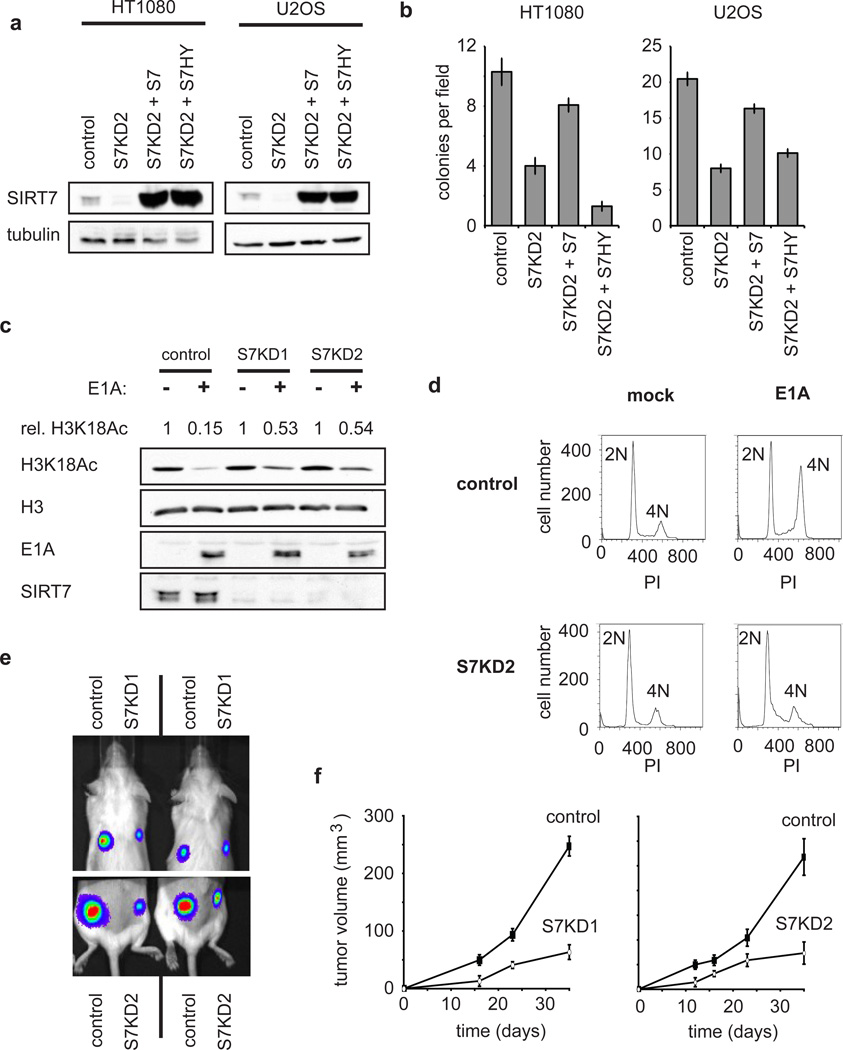
Analysis of SIRT7-occupied genes revealed a clear correlation with factors whose expression is altered in various cancers (Supplementary Fig. 2d). This observation, together with previous reports linking both H3K18Ac3–6 and ELK420 to cancer, suggested that SIRT7 might regulate transformed features of cancer cells. Indeed, SIRT7 depletion in HT1080 and U2OS cells severely impaired both anchorage-independent cellular growth in soft agar and proliferation in low serum, two important hallmarks of transformed cells (Fig. 4a, b; Supplementary Figs. 15, 16). These effects of SIRT7-depletion were associated with both increased cell death and altered cell-cycle progression (Supplementary Fig. 17), and were also observed in prostate cancer cells, a setting where overexpression of both ELK4 and SIRT7 has been observed (Supplementary Fig. 14)20. Importantly, functional reconstitution assays revealed that the catalytic activity of SIRT7 is necessary for maintenance of the cancer cell-specific growth properties (Fig. 4a, b; Supplementary Fig. 16), linking the biochemical activity and cancer-related functions of SIRT7. In addition, simultaneous expression of SIRT7 and ELK4 had a synergistic effect on maintenance of the transformed phenotype of these cells (Supplementary Fig. 18), further highlighting the importance of the molecular interplay between SIRT7 and ELK4.
Figure 4. SIRT7 depletion reverses cancer cell phenotypes and inhibits tumour growth in vivo.
a Western blots showing SIRT7 levels from stable cell lines used in (b). b, Reduced anchorage-independent growth of SIRT7 knockdown cells when plated in soft agar, and reconstitution with wild-type but not mutant SIRT7. Data represent mean +/− S.E.M. of three independent experiments. c, Western analysis showing impaired H3K18 deacetylation induced by E1A in S7KD HT1080 cells. Rel. H3K18Ac: relative levels of H3K18Ac in mock-treated versus E1A expressing cells, normalized to total H3 levels. d, SIRT7 depletion impairs E1A-mediated loss of contact inhibition in primary IMR90 fibroblasts determined by flow cytometry. DNA content (2N or 4N, as determined by propidium iodide (PI) staining) is indicated. e, Representative imaging of tumours derived from SIRT7-knockdown or control cells, following subcutaneous xenograft transplants in immunodeficient mice, 16 days post injection. f, Tumour volume (mean +/− S.E.M.; n=5) as in (e), measured over 35 days.
The adenoviral E1A oncoprotein induces a specific decrease in H3K18 acetylation that is important for its transforming activity3,4. Strikingly, SIRT7 depletion in HT1080 cells severely inhibited this E1A-dependent reduction of H3K18Ac (Fig. 4c). Moreover, expression of E1A in non-dividing, contact-inhibited primary human fibroblasts triggers cell-cycle re-entry and escape from contact inhibition, another hallmark of oncogenic transformation21, and SIRT7 depletion abolished this effect (Fig. 4d). Thus, SIRT7 is required for both the global H3K18Ac deacetylation and escape from contact inhibition that are induced by the E1A oncoprotein. Finally, we examined the effect of SIRT7-knockdown on tumor growth using subcutaneous xenografts of U251 cancer cells in mice, and found that tumour formation was severely impaired by SIRT7 depletion (Fig. 4e, f; Supplementary Fig. 19). Together, our data suggest that H3K18Ac-specific deacetylation by SIRT7 is important for maintaining fundamental properties of the cancer cell phenotype and stabilizing the tumourigenicity of human cancer cells in vivo.
In summary, we have demonstrated that SIRT7 is a promoter-associated, highly selective H3K18Ac deacetylase that mediates transcriptional repression and stabilizes cancer cell phenotypes. These findings suggest that pathological up-regulation of SIRT7 in cancer cells may contribute to the malignant phenotype of certain tumours. Indeed, SIRT7 overexpression is observed in multiple cancer tissues (Supplementary Fig. 14)17–19, and the cBio Cancer Genomics Portal has reported 55 separate instances of SIRT7 gene amplification in patient tumors to date (http://www.cbioportal.org). We note that while SIRT7 is important for maintaining the transformed state of cancer cells, we have not observed a role for SIRT7 in initiating the process of cellular transformation itself. For example, overexpression of SIRT7 in immortalized mouse embryonic fibroblasts or primary human fibroblasts did not lead to oncogenic transformation (data not shown, and reference 8). Thus, our data suggest models in which H3K18Ac deacetylation by SIRT7 modulates the epigenetic stability and tumourigenicity of cancer cells, but how SIRT7 deficiency impacts on tumor initiation and the overall incidence of cancer is likely more complex.
Our observation that SIRT7 represses several RP genes is intriguing, as mutations in RP genes have been linked to cancer progression22,23. For example, the SIRT7 target gene RPS14 is a disease gene of the human 5q− syndrome, a myelodysplastic disorder that frequently progresses to acute myeloid leukemia23, and multiple RPs have been identified as haploinsufficient tumor suppressors in zebrafish22. The molecular mechanisms underlying the links between RP protein insufficiency and cancer are unclear, but have been hypothesized to involve imbalances in translation regulation or translation-independent functions of individual RPs22,23.
Previous studies have found that SIRT7 promotes ribosomal RNA transcription24, although this function appears to be specific to cell-type or experimental conditions (Supplementary Fig. 20). Whether such an activity functions in parallel to, or as a consequence of, SIRT7’s role in RP gene repression remains to be elucidated, but may suggest a broad role for SIRT7 in coordinating the cellular translation machinery. Interestingly, RP gene deletions and inhibition of translation have also been linked to lifespan extension in numerous model organisms, including mammals24,25, suggesting that gene repression by SIRT7 might also influence ageing-related cellular processes. Consistent with this hypothesis, one strain of Sirt7-deficient mice exhibits cardiac defects and shortened lifespan8, although this phenotype appears to depend on genetic background26. Future work should shed light on the potential role of SIRT7 in ageing-associated pathologies and lifespan determination.
METHODS SUMMARY
Histone deacetylation assays
In vitro histone deacetylation assays were performed as previously described28 using acetylated peptides or poly-nucleosomes purified from HeLa cells as substrate. Recombinant human SIRT7 protein was purified from baculovirus-infected insect cells as described8.
ChIP-qPCR and mRNA analysis
ChIP was performed as previously described29, except that the Qiagen PCR purification kit was used for DNA purification (Qiagen). Whole mRNA was purified from cells using the RNEasy Mini Kit (Qiagen). Quantitative RT-PCR was performed using the Universal ProbeLibrary System with a LightCycler 480 II (Roche), or using Taqman Gene Expression Assays on a 7300 Real Time PCR machine (Applied Biosystems). RNA from patient-matched tumour and unaffected control tissues was purchased from Ambion.
Tumour xenograft experiments
Equal numbers of U251 cells expressing luciferase and either control (pSR) or SIRT7 knockdown vectors (upper quadrants: 4 × 106 pSR or S7KD1 cells; lower quadrants: 8 × 106 pSR or S7KD2 cells) were implanted on the backs of RAG knockout mice. Tumour growth was monitored using calipers and visualized using a bioluminescence-based IVIS system (Caliper LifeSciences).
Full methods and associated references are available in the online version of this paper at www.nature.com/nature.
METHODS
Cell culture, RNAi and viral transduction
Human 293T, HT1080, U251 and U2OS cell lines were acquired from the ATCC, and DU145 cells were a gift from P. Khavari. These cells were cultured in Advanced DMEM (Invitrogen) supplemented with penicillin-streptomycin (Invitrogen), GlutaMAX-1 (Invitrogen), and 10% newborn calf serum. K562 cells were cultured in RPMI (Invitrogen) supplemented with penicillin-streptomycin (Invitrogen), GlutaMAX-1 (Invitrogen), and 10% newborn calf serum. IMR90 cells were cultured in DMEM/F12 (Invitrogen) containing penicillin-streptomycin, GlutaMAX-1, and 10% fetal bovine serum. Retroviral transduction was performed as previously described28. SIRT7 knockdown target sequences are as follows: S7KD1, 5'-CACCTTTCTGTGAGAACGGAA-3'; S7KD2, 5'-TAGCCATTTGTCCTTGAGGAA-3', S7KD3, 5’-GCCTGAAGGTTCTAAAGAA-3’, S7KD4, 5’-GAACGGAACTCGGGTTATT-3’. ELK4 knockdown target sequences are as follows: ELK4 KD1, 5’-CGACACAGACATTGATTCA-3’; ELK4 KD2, 5’-GAGAATGGAGGGAAAGATA-3’, as previously described20. ELK1 knockdown target sequence: GATGTGAGTAGAAGAGTTA. GABPα knockdown target sequence: TGAAGAAGCTCAAGTGATA. HDAC1 knockdown target sequences: HDAC1 KD1 5’- AGAAAGACCCAGAGGAGAA-3’, HDAC1 KD2 5’-GCAAGCAGATGCAGAGATT-3’. Double-stranded siRNAs were purchased from Thermo Scientific. For retroviral packaging, 293T cells were co-transfected with pVPack-VSV-G, pVPack-GP (Stratagene) and the SIRT7 knockdown or pSUPERretro control constructs, and viral supernatant was harvested after 48 hours. For transduction, cells were incubated with virus-containing supernatant in the presence of 8 µg/mL polybrene. After 48 hours, infected cells were selected for 72 hours with puromycin (2 µg/mL) or hygromycin (200 µg/mL). Antibodies and PCR primer details are provided in Supplementary Tables 7 and 8. Adenovirus expressing the small E1A gene alone was generated and used to infect IMR90 cells using the Virapower Adenovirus System (Invitrogen) per the manufacturer’s instructions. Anchorage-independent growth was measured as previously described30. Annexin V analysis was performed using the FITC Annexin Apoptosis Detection Kit (BD Pharmingen), on S7KD and control U2OS cells cultured in 1% serum. For cell cycle analysis, S7KD and control HT1080 cells were pulsed with 33µM BrdU, fixed in 75% ethanol in PBS, stained with FITC mouse anti-BrdU (BD Pharmingen) and propidium iodide, as previously described28. Flow cytometry data were acquired using a FACS LSRFortessa flow cytometer and FACS Diva software (BD Biosciences), and analyzed with CellQuest-Pro software (BD Biosciences). For analysis of H3K18Ac in E1A expressing cells, HT1080 cells were treated with control or SIRT7 siRNAs for 24 hours, then transfected with control (empty vector) or E1A-expressing vectors. Forty-eight hours after siRNA transfection, extracts were prepared and analyzed by western blot. Relative levels of H3K18Ac (rel. H3K18Ac) were determined by quantifying H3K18Ac western blot band intensities using ImageJ software, and normalizing to total H3 band intensities. Samples expressing E1A were set relative to their matched control.
Biochemical fractionation and co-immunoprecipitations
Samples enriched for cytoplasmic, nucleoplasmic, and chromatin fractions were prepared as previously described31. Co-immunoprecipitations were performed as previously described32, except that one 150 mm plate of cells was used per IP, Protein A/G beads (Sigma) were used instead of FLAG-resin, and elution was performed by boiling beads in Laemmli loading buffer.
Histone deacetylation assays
In vitro histone deacetylation assays were performed as previously described28. Purification of human SIRT7 protein from baculovirus-infected insect cells was described previously8. Calf thymus histones were obtained from Roche, and poly-nucleosomes were purified from HeLa cells as previously described33. Histone peptides were synthesized at the Yale W. M. Keck peptide synthesis facility, and liquid chromatography mass spectrometry was performed at the Stanford University Vincent Coates Foundation Mass Spectrometry Laboratory. To determine histone acetylation levels in cells, 293T cells were transiently transfected with pcDNA 3.1 vectors containing FLAG-tagged wild-type SIRT7, the SIRT7-HY catalytic mutant, or an empty vector. Whole-cell lysates were harvested after 48 hours. Western blot analysis of histone acetylation levels was performed with modification-specific antibodies.
Quantitative mass spectrometry
Acid-extracted total histones were subjected to chemical derivatization using D0-proionic anhydride and digestion with trypsin at a substrate to enzyme ratio of 10:1 for 6 hours at 37°C as previously described34. An additional round of propionylation was performed on the digested peptides, with one sample being derivatized with the same D0-propionic anhydride reagent, and the other being derivatized with D10-propionic anhydride for quantitative proteomics as previously described35. D10-propionic anhydride introduces a 5 Da shift by derivatization of the free N-termini of all peptides generated from the trypsin digest. Equal amounts of both samples as quantified earlier by a Bradford assay were mixed together, and digested peptides were desalted using homemade STAGE tips as reported earlier36. Desalted peptides were loaded onto fused silica microcapillary column (75 µm) packed with C18 resin constructed with an ESI tip through an Eksigent AS-2 autosampler (Eksigent Technologies Inc., Dublin, CA) at a rate of ~200 nL/minute. Peptides were eluted using a 5–35% solvent B gradient for 60 minutes (solvent A= 0.1 M acetic acid, solvent B = 70% MeCN in 0.1 M acetic acid). Nanoflow LC-MS/MS experiments were performed on an Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA) taking a full mass spectrum at 30,000 resolution in the Orbitrap and seven data-dependent MS/MS spectra in the ion trap. All MS and MS/MS spectra were manually verified and quantified.
ChIP-sequencing and computational analysis
ChIP for ChIP-sequencing analysis was performed as previously described37. Four ChIP samples were sequenced using Illumina Solexa Genome Analyzer II single end sequencing protocol, including two SIRT7 replicates and two input control replicates. Sequencing adapters and low-quality reads were removed, and the trimmed reads were aligned to human reference genome hg18 by GAII data processing pipeline, allowing up to 2 mismatches. The biological replicates of SIRT7 ChIP-sequencing were first analyzed individually to measure the reproducibility. The result indicated that the two biological replicates were very similar and met all the NIH ENCODE data quality guidelines for high reproducibility (Supplementary Table 4). The uniquely mapped reads from replicates of SIRT7 and input control samples were pooled respectively and processed by MACS (version 1.3.6)38 to generate the whole-genome ChIP-sequencing profiles, with the “--diag” option enabled for the sequencing depth saturation test. Clonal reads were automatically removed by MACS. In total 276 SIRT7 binding sites were identified with p-value cut off 1e-8. The wig files were normalized to 10,000,000 total tag number and converted into bigwig format for visualization. The SIRT7 target genes were identified by detecting the SIRT7 binding peaks within 3 kb upstream to 3 kb downstream of transcription start sites (TSS’s) of RefSeq genes using CEAS35. In total, 253 target genes were identified, including 241 protein-coding genes. The GO analysis was performed using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov) 39,40.
The cancer gene association study was performed using the Oncomine database (http://www.oncomine.org). For identification of ELK4 ChIP-sequencing target genes, the ChIP-sequencing reads from O'Geen et al. (GSE24685)16, were remapped to hg18, and peaks were called using MACS (Supplementary Table 9). The target genes were identified by searching for ELK4 peaks 3 kb up- and downstream of TSS’s.
De novo motifs with sizes from 6 to 15 nucleotides were searched within SIRT7 binding sites using MDModule41, with repetitive regions masked and running parameters “-s 100 -t 50”. The top 50 detected de novo motifs (top 5 of each motif size) were recorded and compared with JASPAR motif database using STAMP with default settings42. The position weight matrix of the ELK4 motif (Supplementary Table 6) was remapped to the identified SIRT7 peaks using cisgenome34, with parameter “–r 30”.
ChIP and mRNA analysis
Cells were prepared for ChIP as previously described29, with the exception that DNA was washed and eluted using a PCR purification kit (Qiagen) rather than by phenol-chloroform extraction. Whole mRNA was purified from cells using the RNEasy Mini Kit (Qiagen). Quantitative RT-PCR was performed using the Roche Universal ProbeLibrary System with a LightCycler 480 II (Roche), or using Taqman Gene Expression Assays (Applied Biosystems) on a 7300 Real Time PCR machine (Applied Biosystems). Pre-rRNA custom primer-probe mix was generated by Applied Biosystems using human pre-rRNA DNA sequence. RNA from patient-matched tumour and unaffected control tissues was purchased from Ambion.
Tumour xenograft experiments
Equal numbers of U251 cells expressing luciferase and either control (pSR) or SIRT7 knockdown (S7KD) vectors (upper quadrants: 4 × 106 pSR or S7KD1 cells; lower quadrants: 8 × 106 pSR or S7KD2 cells) were implanted on the backs of RAG knockout mice. Tumour growth was monitored using calipers and visualized using a bioluminescence-based IVIS system (Caliper LifeSciences).
Supplementary Material
Acknowledgments
We thank M. Snyder and colleagues for high-throughput sequencing (conducted as part of the ENCODE consortium), and members of the Chua and Gozani labs for helpful discussions and comments on the manuscript. This work was supported by grants from the NIH to K.F.C. (K08 AG028961, R01 AG028867), W.L. (U01DA025956), O.G. (R01 GM079641), K.S. (GM 30186, HG 4558), and B.A.G. (DP2OD007447); from the NSF to B.A.G. (CAREER Award, CBET-0941143); from the Department of Defense to W.L. (PC094421); from the Cancer Prevention and Research Institute Of Texas (CPRIT) to W.L. (RP110471-C3); from the Department of Veterans Affairs to K.F.C. (Merit Award); and by fellowship awards to M.F.B. (ARCS Scholarship and Mason Case Graduate Fellowship), R.I.T. (NIH training grant 1018438-142 PABCA), L.T. (American Italian Cancer Foundation Post-doctoral Research Fellowship), S.P. (NIH training grant 3T32DK007217-36S1) and N.L.Y. (NIH F32 NRSA). W.L. is a recipient of a Duncan Scholar Award. K.F.C. is a Paul Beeson Scholar and an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Supplementary Information is available with the online version of this paper at www.nature.com/nature.
Author Contributions
M.F.B. and K.F.C. conceived the project and, together with E.M. and O.G., designed the experiments. M.F.B. and E.M. performed and interpreted molecular and cell biology experiments, and M.F.B. and K.F.C. wrote the manuscript with input from co-authors. Y.X., K.C. and W.L. performed the bioinformatic analyses and wrote the corresponding manuscript sections. Z.M. and K.S. designed and performed the SIRT7 ChIP-sequencing experiments. L.T. performed and interpreted the experiments in Supplementary Fig. 17; B.G. and N.L.Y. performed the quantitative mass spectrometry in Fig. 1g; M.K. performed the mouse xenograft experiments in Fig. 4; R.T and S.P. generated constructs for various experiments and provided technical assistance. M.F.B., E.M., and Y.X. made independent contributions to the work.
Author Information
Reprints and permissions information can be obtained at www.nature.com/reprints. SIRT7 ChIP-sequencing data have been deposited in the NIH Gene Expression Omnibus (GEO) under accession number GSE28149. In addition, raw and processed data are also available on our project website http://dldcc-web.brc.bcm.edu/lilab/SIRT7/.
REFERENCES
- 1.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Ferrari R, et al. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwitz GA, et al. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manuyakorn A, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seligson DB, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vakhrusheva O, et al. Sirt7 Increases Stress Resistance of Cardiomyocytes and Prevents Apoptosis and Inflammatory Cardiomyopathy in Mice. Circ Res. 2008 doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 8.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassig CA, et al. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci U S A. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steeg PS, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 12.Leal JF, et al. Cellular senescence bypass screen identifies new putative tumor suppressor genes. Oncogene. 2008;27:1961–1970. doi: 10.1038/sj.onc.1210846. [DOI] [PubMed] [Google Scholar]
- 13.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- 15.Kaikkonen S, Makkonen H, Rytinki M, Palvimo JJ. SUMOylation can regulate the activity of ETS-like transcription factor 4. Biochim Biophys Acta. 2010;1799:555–560. doi: 10.1016/j.bbagrm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 16.O'Geen H, et al. Genome-wide binding of the orphan nuclear receptor TR4 suggests its general role in fundamental biological processes. BMC Genomics. 2010;11:689. doi: 10.1186/1471-2164-11-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashraf N, et al. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95:1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Nigris F, et al. Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. Br J Cancer. 2002;86:917–923. doi: 10.1038/sj.bjc.6600156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frye R. "SIRT8" expressed in thyroid cancer is actually SIRT7. Br J Cancer. 2002;87:1479. doi: 10.1038/sj.bjc.6600635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makkonen H, et al. Identification of ETS-like transcription factor 4 as a novel androgen receptor target in prostate cancer cells. Oncogene. 2008;27:4865–4876. doi: 10.1038/onc.2008.125. [DOI] [PubMed] [Google Scholar]
- 21.Braithwaite AW, et al. Adenovirus-induced alterations of the cell growth cycle: a requirement for expression of E1A but not of E1B. J Virol. 1983;45:192–199. doi: 10.1128/jvi.45.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amsterdam A, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebert BL, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford E, et al. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 26.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008 doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- 30.Chua KF, et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung T, et al. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia BA, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plazas-Mayorca MD, et al. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–5374. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 37.Moqtaderi Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 40.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 42.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.