SUMMARY
RIG-I and MDA5 detect viral RNA in the cytoplasm and activate signaling cascades leading to the production of type-I interferons. RIG-I is activated through sequential binding of viral RNA and unanchored lysine-63 (K63) polyubiquitin chains, but how polyubiquitin activates RIG-I and whether MDA5 is activated through a similar mechanism remain unresolved. Here we showed that the CARD domains of MDA5 bound to K63 polyubiquitin and that this binding was essential for MDA5 to activate the transcription factor IRF3. Mutations of conserved residues in MDA5 and RIG-I that disrupt their ubiquitin binding also abrogated their ability to activate IRF3. Polyubiquitin binding induced the formation of a large complex consisting of four RIG-I and four ubiquitin chains. This hetero-tetrameric complex was highly potent in activating the antiviral signaling cascades. These results suggest a unified mechanism of RIG-I and MDA5 activation and reveal a unique mechanism by which ubiquitin regulates cell signaling and immune response.
INTRODUCTION
Innate immunity is an evolutionarily conserved form of host defense mechanism found in most multicellular organisms. The detection of pathogens by the innate immune system involves recognition of pathogen-associated molecular patterns (PAMPs) by a set of host pattern recognition receptors (PRRs) (Kawai and Akira, 2011). Following virus infection, viral RNA is recognized in the endosome in specialized cells by membrane-bound Toll-like receptors (TLRs), and in the cytosol of vertebrate cells by RIG-I like receptors (RLRs), which include RIG-I, MDA5 and LGP2 (Yoneyama et al., 2004). Upon viral recognition, RIG-I and MDA5 interact and activate the mitochondrial adaptor protein MAVS (also known as IPS-1, VISA or CARDIF), and initiate downstream signaling pathways to activate the transcription factors NF-κB and IRF3, which regulate the production of type-I interferons (e.g., IFNα and IFNβ) and proinflammatory cytokines (Rehwinkel and Reis e Sousa, 2010).
All three members of the RLRs share highly conserved domain structures, including a DExD/H box helicase domain and a C-terminal RNA binding domain (CTD). RIG-I and MDA5, but not LGP2, have at their N-termini two caspase activation and recruitment domains (CARDs) in tandem that mediate signaling to downstream adaptor proteins. RIG-I and MDA5 have been shown to play a key role in type-I IFN induction after RNA virus infection, while LGP2 is thought to play a regulatory role. Current models suggest that, in the absence of viral RNA, RIG-I adopts an auto-inhibited conformation. The binding of viral RNA, which contains 5’-triphosphate (5’-ppp) and certain duplex structures, to the CTD and helicase domains of RIG-I induces an ATP-dependent conformational change that exposes the CARDs (Kowalinski et al., 2011). RIG-I then translocates on the viral RNA, while the CARDs interact with the ubiquitin ligase TRIM25, which functions together with the ubiquitin E2 complex Ubc13-Uev1A to catalyze the synthesis of K63 polyubiquitin chains (Gack et al., 2007; Myong et al., 2009). We have recently found that free K63 polyubiquitin chains, which are not conjugated to any cellular protein, can bind to RIG-I CARD domains (Zeng et al., 2010), and that this binding is important for the activation of IRF3 and the induction of IFNβ. Thus, RIG-I activation requires sequential binding of viral RNA to the CTD followed by the binding of endogenous K63 polyubiquitin chains to the N-terminal CARDs. However, how polyubiquitin binding leads to RIG-I activation is an important question that remains unresolved.
While MDA5 is known to play crucial roles in immune responses against many RNA viruses, especially picornaviruses (Kato et al., 2006), little is known about how MDA5 is activated and how it activates the downstream pathways leading to IFN production. The C-terminus of MDA5 is distinct from the CTD of RIG-I and it does not bind 5’-pppRNA (Takahasi et al., 2009). Although long double-stranded RNA analogue poly[I:C] has been shown to induce IFNβ in a MDA5-dependent manner (Kato et al., 2008), the physiological ligand of MDA5 has not been defined. A previous study found that RIG-I, but not MDA5, is ubiquitinated by TRIM25, and that a putative ubiquitination site of RIG-I (K172) is not conserved in MDA5, raising the question of whether ubiquitination plays any role in MDA5 activation (Gack et al., 2007).
In this study, we showed that MDA5 can activate IRF3 in a cell-free system and that MDA5 CARDs bind to K63 polyubiquitin chains. Mutations in the conserved residues of MDA5 and RIG-I CARDs that disrupt their binding to ubiquitin chains also impaired their ability to activate IRF3 and induced IFNβ. Further, using RIG-I CARDs as a model, we demonstrate that K63 polyubiquitin binding induced oligomerization of RIG-I in vitro and in virus-infected cells. Irrespective of ubiquitin chain lengths, RIG-I forms a tetramer which is highly active in signaling to IRF3.
RESULTS
Ubiquitin-Dependent Activation of the MDA5 Signaling Cascade in a Cell-Free System
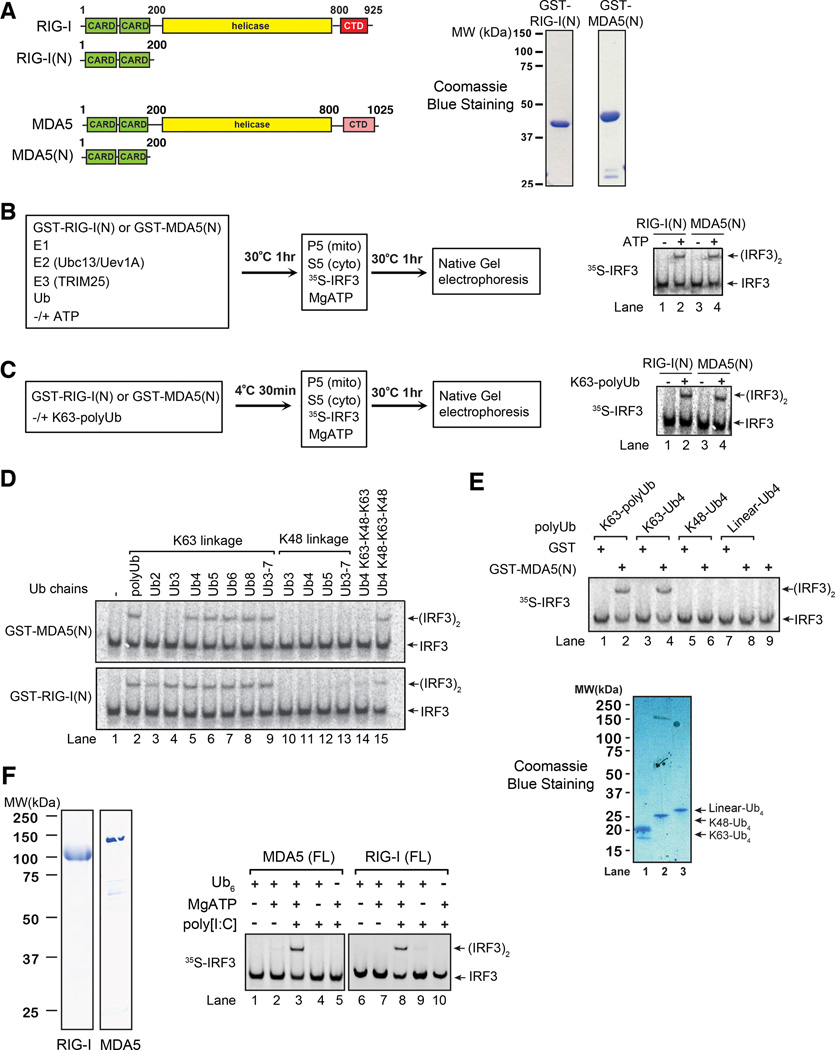
We have previously shown that RIG-I N-terminus containing the CARDs [RIG(N)] binds K63 polyubiquitin chains, and that this binding triggers a signaling cascade that leads to the activation of IRF3 and IKK in a cell-free system consisting of mitochondria and cytosolic extracts (Zeng et al., 2010). To investigate if MDA5 CARDs [MDA5(N)] could also activate IRF3 through a similar mechanism, we expressed GST-MDA5(N) in E. coli, purified it, and incubated it in a ubiquitination reaction containing E1, Ubc13-Uev1A (E2), TRIM25 (E3) and ubiquitin in the presence or absence of ATP (Figure 1A & 1B). Afterwards, the reaction mixtures were incubated with mitochondria (P5) and cytosolic extracts (S5) together with 35S-IRF3 and ATP, followed by native gel electrophoresis to detect IRF3 dimerization. Like RIG-I(N), MDA5(N) was capable of causing IRF3 dimerization following the ubiquitination reaction (Figure 1B). To determine if ubiquitination of RIG-I(N) or MDA5(N) was involved in its activation, we carried out the ubiquitination reaction as described in Figure 1B except that GST-RIG-I(N) or MDA5(N) was omitted from the reaction. The thio-modifying agent N-ethymaleimide (NEM) was added to the reaction mixture to inactivate E1 and E2, and then the reaction mixture, which contained mostly unanchored K63 polyubiquitin chains (Xia et al., 2009), was incubated with GST-RIG-I(N) or GST-MDA5(N). Both GST-RIG-I(N) and GST-MDA5(N) gained the ability to activate IRF3 after incubation with K63 polyubiquitin chains (Figure 1C). We have previously shown that unanchored K63 ubiquitin chains could directly activate RIG-I(N) (Zeng et al., 2010). Similarly, incubation of K63 chains containing four or more ubiquitin with GST-MDA5(N) led to IRF3 activation in the in vitro assay (Figure 1D). In contrast, K48 ubiquitin chains had no activity. Interestingly, Ub4 containing the K48-K63-K48 mixed linkage weakly stimulated GST-RIG-I(N) and GST-MDA5(N), whereas Ub4 containing the K63-K48-K63 linkage was inactive, hinting that the K63 linkage in the center of Ub4 may be important (Figure 1D, lanes 14–15). Linear Ub4, in which the N-terminal methionine of one ubiquitin is conjugated by the C-terminus of the preceding ubiquitin, did not activate MDA5(N) (Figure 1E).
Figure 1.
MDA5 activates IRF3 in a cell-free system through a ubiquitin-dependent mechanism. A. Diagrams of RIG-I and MDA5 (left); Coomassie blue stained gels of their N-terminal CARD domains expressed in and purified from E. coli (right). B. GST-RIG-I(N) or GST-MDA5(N) was incubated with ubiquitination components as shown in the diagram. Aliquots of the reaction mixtures were further incubated with mitochondria (P5) and cytosolic extracts (S5) together with 35S-IRF3 and ATP. IRF3 dimerization was analyzed by native gel electrophoresis. C. Similar to B., except that RIG-I(N) or MDA5(N) was incubated with polyubiquitination mixtures in which E1 and E2 had been inactivated by NEM. D & E. Similar to C., except that RIG-I(N) or MDA5(N) was incubated with unanchored ubiquitin chains of different lengths and linkages as indicated. F. Similar to C., except that full-length (FL) RIG-I or MDA5 was incubated with K63-Ub6, MgATP, and poly[I:C] as indicated. Shown on the left are Coomassie blue stained gels of RIG-I and MDA5 proteins. Results shown are representatives of three experiments.
Full-length MDA5 is activated by long poly[I:C] as well as by undefined ligands associated with certain RNA viruses such as those of picornaviridae (Kato et al., 2006). To determine if full-length MDA5 could lead to IRF3 activation in the in vitro reconstitution system, MDA5 expressed and purified from E. coli was tested. Similar to RIG-I, incubation of MDA5 with poly[I:C], ATP and K63 polyubiquitin chains led to strong activation of IRF3 in the cell-free system (Figure 1F).
RIG-I and MDA5 CARD Domains Bind K63 Polyubiquitin Chains
CARD domains belong to the death domain (DD) superfamily, which also includes the death domain (DD), death effector domain (DED), and pyrin domain (PYD) subfamilies (Park et al., 2007). Many proteins of the DD superfamily are involved in immunity, inflammation and cell death. Our finding that RIG-I CARDs bind to K63 polyubiquitin chains (Zeng et al., 2010) raises the question of whether the CARD or DD domains of some other proteins are also ubiquitin-binding domains (UBD). To begin to address this question, we carried out bioinformatics analysis of the CARD domain subfamily (Supplementary Figure S1A). CARD domain subfamily sequences were collected by PSI-BLAST. Clustering analysis based on sequence homology shows that the CARD domains of RIG-I (two CARD domains), MDA5 (two CARD domains) and MAVS form a distinct cluster that is distantly related to other CARD domains, such as those present in NOD1, NOD2 and RIP2, which are important in innate immunity. Although all DD superfamily domains share a conserved six-helical bundle fold, the CARD domain subfamily is also distinct from DD and DED subfamilies based on structural-based homology analysis (Supplementary Figure S1B).
Our previous study showed that both CARD domains of RIG-I are required for binding to K63 polyubiquitin chains and the single CARD of MAVS does not bind to these chains (Zeng et al., 2010). Similar to GST-RIG-I(N), GST-MDA5(N) bound to K63 polyubiquitin chains, but not linear or K48 polyubiquitin chains (Figure 2A and Supplementary Figure S1C). In contrast, the CARD of NOD1, tandem CARDs of NOD2, and tandem DEDs of caspase 8 did not exhibit detectable ubiquitin chain binding activity (Supplementary Figure S1D & S1E). Although these results do not exclude the possibility that some other proteins in the DD superfamily can bind ubiquitin chains, they suggest that the CARD domains of RIG-I and MDA5 are unique in their ability to bind specifically to K63 polyubiquitin chains.
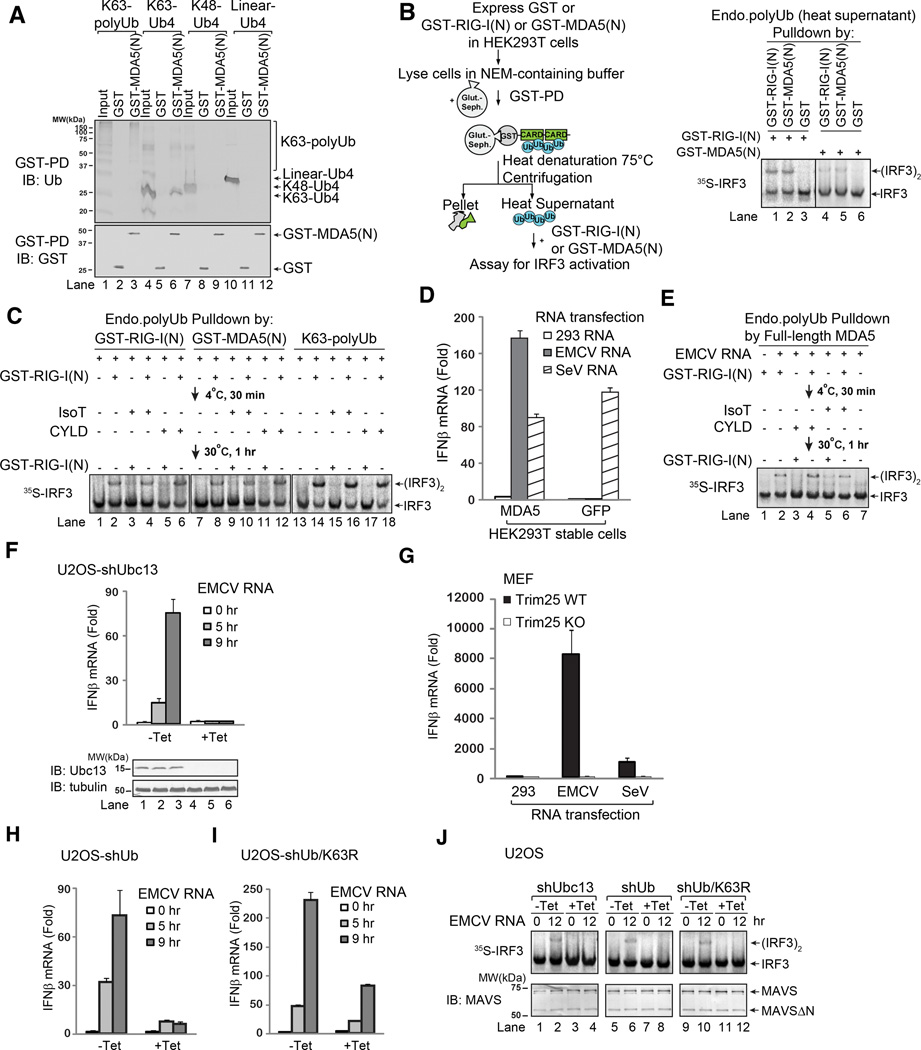
Figure 2.
K63 polyubiquitin chains bind to and activate MDA5. A. GST or GST-MDA5(N) was incubated with polyUb chains containing K63, K48, or linear linkage, and then pulled down with glutathione beads followed by immunoblotting with indicated antibodies. Input represents 10% of polyubiquitin used for GST pulldown (GST-PD) experiments. B. Endogenous polyUb chains in HEK293T cells were isolated using the procedure shown in the diagram (left). After centrifugation, the supernatant containing the ubiquitin chains was incubated with GST-RIG-I(N) or GST-MDA5(N), which were then incubated with mitochondria (P5) and cytosol (S5) to measure IRF3 activation. C. Endogenous polyUb chains associated with RIG-I(N) or MDA5(N) were isolated as in B., and then incubated with GST-RIG-I(N) followed by IsoT or CYLD treatment, or in reverse order, before IRF3 dimerization assay. Parallel experiments were carried out using free K63 polyUb chains as controls (lanes 13–18). D. HEK293T cells stably expressing GFP (control) or MDA5 were transfected with indicated RNA. IFNβ induction was measured by quantitative PCR (qPCR). E. Similar to B and C, except that endogenous polyUb chains associated with MDA5 were isolated from EMCV RNA-transfected HEK293T cells stably expressing MDA5-Flag. The polyUb chains were incubated with GST-RIG-I(N) and IsoT or CYLD in the indicated order, and then with mitochondria (P5) and cytosol (S5) to measure IRF3 dimerization. F. U2OS integrated with tetracycline-inducible shRNA against Ubc13 (shUbc13) were treated with or without tetracycline (Tet). IFNβ induction by EMCV RNA was measured by qPCR. G. WT or Trim25-deficient (KO) MEF cells were transfected with indicated RNA. IFNβ was measured by qPCR. H & I. Similar to F., except that U2OS cells stably expressing tetracycline-inducible shRNA against ubiquitin (shUb, left), and those in which endogenous ubiquitin was replaced with K63R ubiquitin (shUb/K63R, right), were transfected with EMCV RNA, followed by measurement of IFNβ by qPCR. J. RNA transfection of U2OS cells was carried out as in H and I, and then mitochondria (P5) were isolated and incubated with cytosolic extracts (S5) to measure IRF3 dimerization. Aliquots of the mitochondrial extracts were immunoblotted with a MAVS antibody. Results shown are representatives of two experiments.
To determine if MDA5 binds to endogenous, unanchored, K63-linked, polyubiquitin chains in cells, we used a previously developed method to isolate these polyubiquitin chains (Figure 2B, left) (Zeng et al., 2010). In this method, we overexpressed GST-MDA5(N) in HEK293T cells, prepared cell lysates in which most of the cellular deubiquitinating enzymes (DUBs) were inactivated by NEM, and then used glutathione-Sepharose to pull down GST-MDA5(N) as well as the associated endogenous ubiquitin chains. The isolated complexes were heated at 75°C to denature most of the proteins while sparing free ubiquitin chains which are relatively heat resistant. After centrifugation to remove the denatured proteins, the supernatant containing the ubiquitin chains was incubated with recombinant GST-RIG-I(N) or GST-MDA5(N) protein to measure IRF3 activation. As shown in Figure 2B, the heat-resistant supernatant from the cells expressing GST-RIG-I(N) or GST-MDA5(N), but not GST, contained the activity that, when incubated with fresh GST-RIG-I(N) or GST-MDA5(N) protein again, stimulated IRF3 dimerization in the in vitro assay. This activity was destroyed by the DUBs isopeptidase T (IsoT) and CYLD (Figure 2C), which specifically cleave unanchored and K63 linked polyubiquitin chains, respectively (Reyes-Turcu et al., 2006; Xia et al., 2009). Preincubation of the heat-resistant supernatant with GST-RIG-I(N) rendered the activity resistant to IsoT and CYLD, indicating that GST-RIG-I(N) protected unanchored K63 polyubiquitin chains from degradation by the DUBs. Taken together, these results show that MDA5 CARDs bind endogenous unanchored K63 polyubiquitin chains in cells.
EMCV RNA Induces the Association of MDA5 with Endogenous Polyubiquitin Chains
Encephalomyocarditis virus (EMCV) is a picornavirus known to induce type-I interferons through MDA5 (Kato et al., 2006). Infection of murine embryonic fibroblasts (MEF) with EMCV induced IFNβ and this induction was abolished by the knockdown of MDA5 with a lentiviral shRNA vector (Supplementary Figure S1F). We found that total RNA from EMCV-infected HEK293T cells induced IFNβ much more strongly (~300 fold higher) than EMCV infection (Supplementary Figure S1G), suggesting that some viral proteins might inhibit IFNβ induction by the viral RNA. shRNA against MDA5 blocked IFNβ induction by EMCV RNA, but not Sendai virus (SeV) RNA, which is known to activate RIG-I. HEK293T cells stably expressing MDA5, but not GFP control, also produced IFNβ in response to transfection of EMCV RNA (Figure 2D). To determine if EMCV RNA stimulates the association of unanchored K63 polyubiquitin chains with full-length MDA5 in cells, we immunoprecipitated MDA5 from HEK293T cells transfected with EMCV RNA, prepared the heat-resistant supernatant as described above, and then measured its activity in the presence or absence of GST-RIG-I(N) (Figure 2E). The supernatant prepared from EMCV-transfected cells contained IRF3-stimulatory activity that is sensitive to IsoT or CYLD treatment but protected from these DUBs by preincubation with GST-RIG-I(N). These results suggest that MDA5 associates with endogenous unanchored K63 polyubiquitin chains in response to stimulation with EMCV RNA.
K63 Polyubiquitination is Required to Activate the MDA5 Pathway
To further investigate the role of K63 polyubiquitination in MDA5 activation by EMCV RNA in cells, we used a previously established human U2OS cell line in which the K63-specific ubiquitin E2 Ubc13 could be efficiently knocked down using tetracycline-inducible shRNA (Xu et al., 2009). Ubc13 depletion blocked IFNβ induction by EMCV RNA (Figure 2F). Similarly, MEF cells from Trim25 knockout mice failed to induce IFNβ in response to EMCV RNA transfection (Figure 2G). Previously we also established U2OS cell lines in which endogenous ubiquitin could be knocked down by shRNA and replaced with wild type or K63R mutant ubiquitin in a tetracycline-inducible manner (Xu et al., 2009). IFNβ induction by EMCV RNA was largely blocked in cells expressing ubiquitin shRNA and significantly impaired in cells expressing K63R ubiquitin (Figure 2H & 2I). To test if MAVS activation is impaired by a defect in K63 polyubiquitination, we isolated the mitochondria from EMCV RNA transfected cells and tested their ability to stimulate IRF3 dimerization in the in vitro assay. Figure 2J shows that mitochondria from cells expressing shRNA against Ubc13 or Ub and those in which endogenous ubiquitin was replaced with K63R ubiquitin failed to activate IRF3. Taken together, these results suggest that MAVS activation by MDA5 in response to EMCV RNA is dependent on K63 polyubiquitination.
Both CARD Domains of RIG-I and MDA5 are Important for Polyubiquitin Binding
To delineate the regions in RIG-I and MDA5 that are important for ubiquitin binding and IRF3 activation, we generated a series of mutants with progressive deletion of amino acids from the N-terminus of the first CARD or the C-terminus of the second CARD (Supplementary Figure S2A & S2C). These mutants were expressed in E. coli as GST fusion proteins and affinity purified. The purified proteins were tested for their binding to K63 polyubiquitin chains and their ability to activate IRF3 in the cell-free system (Figures S2B & S2CD). Overall, with both RIG-I(N) and MDA5(N), there was a good correlation between their ability to bind K63 ubiquitin chains and their ability to activate IRF3. In particular, a RIG-I fragment containing residues 6–188 was capable of binding to K63 polyubiquitin chains and activating IRF3, and further removal of 2 amino acids from the C-terminus of the second CARD (6–186) abolished both activities (compare lanes 10 & 11; Figure S2B). The truncation of the first 7 amino acids (8–190) also significantly impaired RIG-I’s activities (compare lanes 7 & 8 in Figure S2B). Similarly, the deletion of just 5 amino acids from the second CARD of MDA5 destroyed its ubiquitin binding as well as its ability to activate IRF3 (compare lanes 2 & 3 in Figure S2D).
Polyubiquitin Binding is Essential for RIG-I and MDA5 to Activate IRF3 and Induce IFNβ
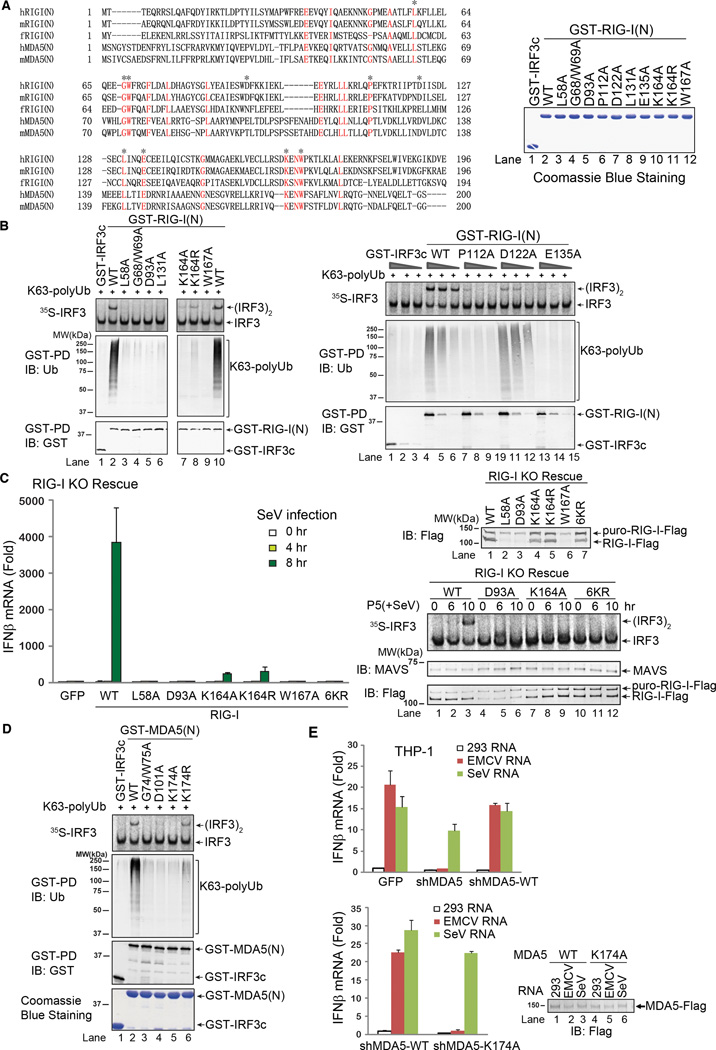
To further define the relationship between ubiquitin binding and IRF3 activation by RIG-I and MDA5, we sought to identify key residues in the CARD domains that are important for their functions. Sequence alignment of the CARD domains of RIG-I from human, mouse and zebrafish, as well as those of human and mouse MDA5, revealed several highly conserved residues (Figure 3A). These residues were mutated (highlighted by asterisk) and GST fusion proteins containing these mutations were expressed in E. coli and affinity purified. Strikingly, among 10 point mutants of RIG-I(N) tested, 8 mutations (L58A, G68/W69A, D93A, L131A, K164A, W167A, P112A and E135A) abolished RIG-I(N)’s binding to K63 polyubiquitin chains as well as its ability to activate IRF3 (Figure 3B). One mutation, K164R, led to reduced ubiquitin chain binding as well as weakened IRF3 activation (Fig. 3B, lane 8 on the left panel). Another mutant, D122A, bound normally to ubiquitin chains but had reduced ability to activate IRF3 (Fig. 3B, lanes 10–12 on the right panel). This result suggests that after RIG-I binds to K63 polyubiquitin chains, the D122A mutation impairs its ability to activate the downstream signaling cascade, possibly by affecting the interaction between RIG-I and MAVS.
Figure 3.
Ubiquitin binding is essential for RIG-I and MDA5 activation. A. Sequence alignment of the N-termini of RIG-I and MDA5 from human (h) and mouse (m) as well as zebrafish (f) RIG-I N-terminus. Asterisks indicate the residues to be mutated in this study. Shown on the right are the GST-RIG-I(N) mutant proteins expressed in and purified from E. coli. B. Point mutants of RIG-I(N) were incubated with K63 polyUb and then analyzed by GST pull-down and IRF3 dimerization assays. C. Full-length RIG-I WT and mutants were stably expressed in RIG-I knockout (KO) MEF cells. The reconstituted cells were infected with SeV for the indicated time, and then IFNβ was measured by qPCR (left). Mitochondrial fractions (P5) were prepared from the virus-infected cells, and MAVS activity was measured in IRF3 dimerization assay (bottom right). The RIG-I proteins in the reconstituted cells were immunoblotted with a RIG-I antibody (upper right). The upper band denotes a fusion protein in which RIG-I was not cleaved from the puromycin resistance gene product due to incomplete self-cleavage of the 2A peptide in the lentiviral vector. D. Point mutants of MDA5(N) were tested for K63 polyUb binding and IRF3 activation as in B. E. THP-1 cells stably expressing a lentiviral shRNA vector targeting MDA5 or those in which endogenous MDA5 was replaced with WT or K174A MDA5 were transfected with indicated RNA, and then IFNβ induction was measured by qPCR. Results shown are representatives of two experiments.
The crystal structures of RIG-I and its double-stranded RNA bound forms have been solved (reviewed by O'Neill and Bowie, 2011). These structures show that the tandem CARDs of RIG-I interact with each other (Kowalinski et al., 2011). Furthermore, the second CARD interacts with a unique insertion domain (HEL2i) between helicase domains 1 and 2; this interaction may prevent the binding of CARDs to polyubiquitin chains in the absence of viral infection. Interestingly, modeling of human RIG-I CARDs based on the structure of the duck RIG-I CARDs shows that several residues whose substitutions impair ubiquitin binding are mapped to the surface of CARD2 or at the junction between CARD1 and CARD2 (Supplementary Figure S2E & S2F). These residues include D93, P112, D122, E135, K164 and K172.
We have recently shown that MAVS undergoes a massive prion-like aggregation on the mitochondrial membrane in response to viral infection of cells or upon the contact of mitochondria with the RIG-I complex containing K63 polyubiquitin chains in vitro (Hou et al., 2011). In the presence of K63 polyubiquitin chains, the aggregation of MAVS induced by WT GST-RIG-I(N) was detected by semi-denaturing detergent agarose gel electrophoresis (SDD-AGE), which is commonly used for detecting prion-like aggregates (Supplementary Figure S2G) (Alberti et al., 2009). In contrast, RIG-I mutants containing D93A or six-lysine mutations (K99, 169, 172, 181, 190 & 193; termed 6KR), both of which are defective in ubiquitin binding, failed to induce MAVS aggregation. The D122A mutant also had a greatly reduced ability to cause MAVS aggregation, which likely explains the reduced ability of this mutant to activate IRF3 despite its normal binding to K63 polyubiquitin (Figure S2G, lanes 11–14).
To determine if mutations that abrogate the ubiquitin binding of RIG-I also affect its ability to induce IFNβ in cells, we used a lentiviral vector to express full-length RIG-I or its mutated forms in RIG-I-deficient MEFs (Figure 3C). The lentiviral vector contains RIG-I fused to the puromycin-resistant gene through a picornavirus self-cleaving 2A peptide. The majority of RIG-I was cleaved from the puromycin resistant gene product, although some fusion protein remained (Figure 3C, upper right). Nevertheless, the expression of WT RIG-I rescued IFNβ induction in response to SeV infection. In contrast, L58A, D93A W167A and 6KR mutants failed to induce IFNβ. K164A and K164R mutants were severely defective in inducing IFNβ, consistent with the reduced ability of these mutants in activating IRF3 (Figure 3B & 3C). Crude mitochondria isolated from cells reconstituted with WT RIG-I were able to activate IRF3 in the in vitro assay if the cells were infected with SeV for 10 hours (Figure 3C, lower right). In contrast, mitochondria from cells reconstituted with D93A, K164A and 6KR mutants of RIG-I had no activity. Control experiments showed that K164A and 6KR RIG-I mutants had ATPase activity similar to that of WT RIG-I (data not shown), suggesting that these mutations do not cause global misfolding or inactivation of RIG-I. Collectively, these results indicate that ubiquitin binding of RIG-I is essential for IFNβ induction by SeV.
Overexpression of GST-RIG-I(N) in HEK293T cells led to a strong induction of IFNβ-luciferase reporter, providing a simple assay to rapidly screen the IFNβ-inducing activity of a large panel of RIG-I CARD mutants (Supplementary Figure S2H). Importantly, all of the ubiquitin-binding defective mutants of RIG-I failed to induce IFNβ in this assay. These mutants also failed to pull down endogenous polyubiquitin chains from transfected cells (Figure S2I). Similar to the in vitro experiments, RIG-I(N)-D122A bound to endogenous ubiquitin chains, but its ability to induce IFNβ was reduced, suggesting a defect at a step downstream of ubiquitin binding (Figures S2H and S2I, left). Three mutants, K146R, E165A and N166A, which bound to polyubiquitin chains normally, were also capable of inducing IFNβ (Figure S2H and S2I, right). Apparently, these mutants were also covalently modified by ubiquitin chains. In general, there was a good correlation between RIG-I ubiquitination and IFNβ induction. However, we have previously shown that the removal of ubiquitin chains from RIG-I by a deubiquitination enzyme (viral OTU) does not impair the ability of RIG-I to activate IRF3 (Zeng et al., 2010). Furthermore, MDA5 does not contain a conserved lysine known to be ubiquitinated in RIG-I and there is no evidence that MDA5 is ubiquitinated (Gack et al., 2007). We found mutations that abrogated the ability of MDA5(N) to bind K63 polyubiquitin chains also impaired its ability to activate IRF3 in vitro and induce IFNβ in vivo (Figure 3D and Supplementary Figure S2J and S2K). Moreover, WT MDA5, but not the ubiquitin-binding defective mutant K174A, was able to rescue IFNβ induction by EMCV RNA in the human monocytes THP-1, which had been depleted of endogenous MDA5 by shRNA (Figure 3E). Taken together, these results demonstrate that the binding to K63 polyubiquitin chains is essential for RIG-I and MDA5 to activate IRF3 and induce IFNβ.
Ubiquitin Binding Rescues the Activity of RIG-I CARD Domain Mutants
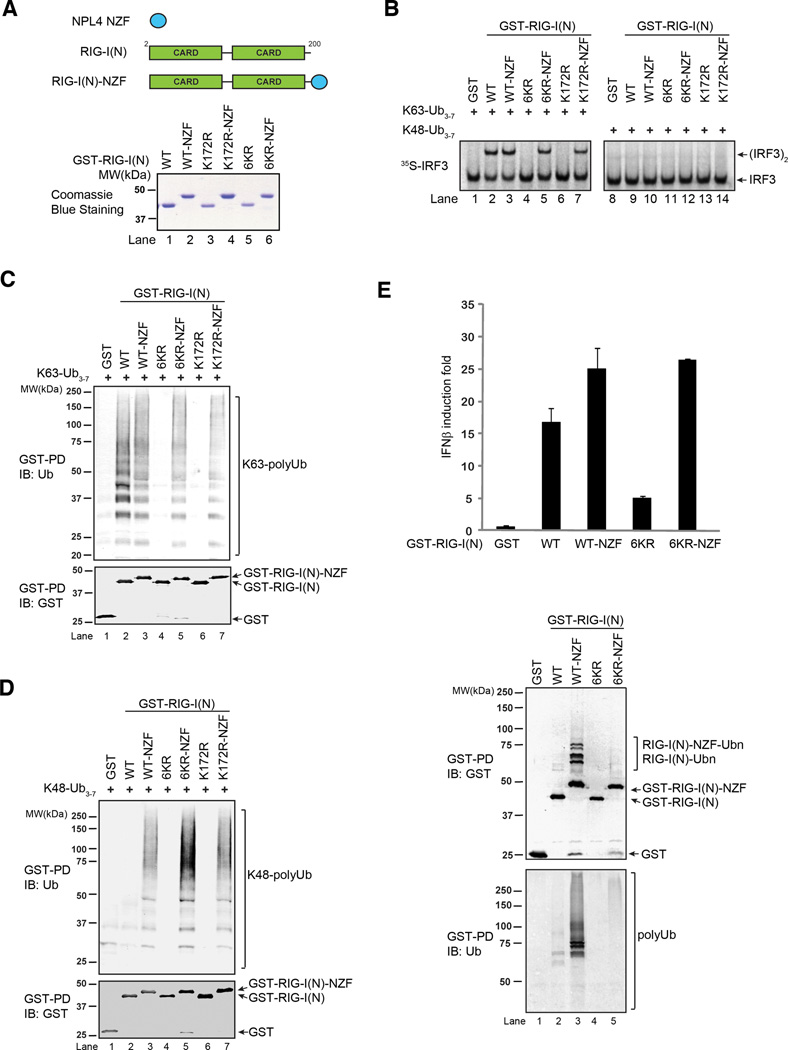
Previous studies have shown that K172R mutation of RIG-I impairs its ubiquitination and IFNβ induction (Gack et al., 2007). However, subsequent studies have found that this mutation also abrogates the binding of RIG-I to polyubiquitin chains (Zeng et al., 2010). To determine whether ubiquitin binding or ubiquitination of RIG-I is important for its signaling functions, we fused RIG-I CARD mutants to the novel zinc finger (NZF) domain of NPL4 (Kanayama et al., 2004; Meyer et al., 2002), which is known to bind polyubiquitin chains (Figure 4A). Remarkably, the NZF domain rescues the ability of RIG-I(N)-K172R to activate IRF3 in the presence of K63, but not K48, polyubiquitin chains (Figure 4B, compare lanes 6 and 7). The activity of RIG-I(N) 6KR mutant was also restored by NZF fusion (Figure 4B, compare lanes 4 and 5). In contrast, a RIG-I( N) mutant containing 14-lysine mutations (14KR) was inactive even after NZF fusion, indicating that mutations of some of the lysines affect RIG-I structure or functions (Supplementary Figure S3A & S3B). This experiment also rules out the remote possibility that the NZF domain alone could activate the RIG-I pathway. Interestingly, the NZF domain restores the binding of K172R and 6KR to both K63 and K48 polyubiquitin chains (Figure 4C & 4D), suggesting that even when K48 ubiquitin chains are forced to bind to RIG-I, they cannot activate the pathway. The NZF domain also restores the ability of RIG-I(N)-6KR and K172R to induce IFNβ-luciferase in HEK293T cells (Figure 4E, top; Supplementary Figure S3C). GST pull down from these cells revealed that RIG-I(N)-6KR-NZF and RIG-I(N)-K172R-NZF were able to pull down endogenous polyubiquitin chains, whereas their ubiquitination was much weaker than that observed for RIG-I(N)-NZF (Figure 4E, bottom; Supplementary Figure S3D). These results provide the direct evidence that polyubiquitin binding by RIG-I is critical for its signaling functions.
Figure 4.
Rescue of ubiquitination-defective mutants of RIG-I with a heterologous ubiquitin-binding domain. A. Diagram depicting the NPL4 novel zinc finger (NZF) ubiquitin binding domain and its fusion to RIG-I(N). The bottom panel shows GST-RIG-I(N) mutants and the NZF fusion proteins. B. Indicated proteins were incubated with K63- or K48-ubiquitin chains, and then analyzed for their ability to activate IRF3 dimerization in vitro. C & D. GST-RIG-I(N) mutant proteins were incubated with K63- (C) or K48- (D) linked ubiquitin chains followed by GST pull-down assays. E. RIG-I(N)-NZF fusion rescues the ability of 6KR to activate IFNβ reporter in vivo. Expression vectors for indicated proteins were transfected into HEK293T cells with IFNβ-luciferase reporter. Cell lysates were assayed for luciferase activity (top) or pulled down with glutathione Sepharose followed by immunoblotting (bottom). Results shown are representatives of two experiments.
Ubiquitin-Induced Oligomerization Activates RIG-I and MDA5
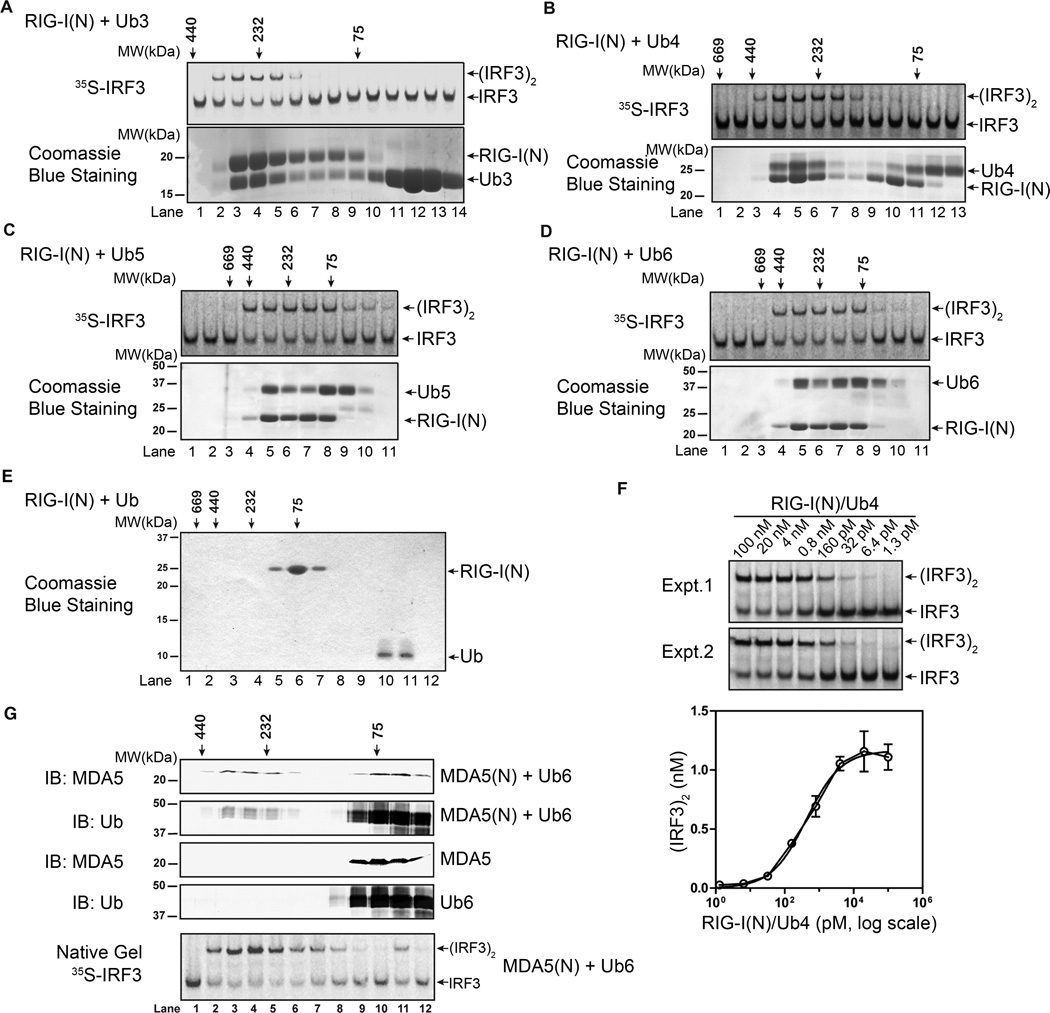
To understand how polyubiquitin binding activates RIG-I, we performed gel filtration analysis of RIG-I(N) following its incubation with K63 ubiquitin chains of different lengths. In these experiments, RIG-I(N) did not contain the GST tag, which could have confounded the results due to GST-mediated dimerization. K63-Ub3, -Ub4, -Ub5 and -Ub6, but not Ub, caused a shift of RIG-I(N) into high molecular weight (HMW) fractions (Figure 5A–E). Furthermore, the HMW complexes containing RIG-I(N) and ubiquitin chains are highly potent in activating IRF3 in the cell-free system. Titration experiments using the RIG-I(N)/Ub4 complex (lane 5 in Figure 5B) showed that the half maximal effective concentration (EC50) of this complex was ~404 pM (Figure 5F; this is likely an underestimate as it does not take into account of the dissociation of the complex during the reaction). Incubation of MDA5(N) with K63-Ub6 also caused the formation of a larger complex composed of both MDA5(N) and K63-Ub6, which strongly activated IRF3 in the in vitro assay (Figure 5G).
Figure 5.
Polyubiquitin binding induces oligomerization of RIG-I and MDA5 CARD domains. A–D. RIG-I(N) (no GST tag) was incubated with K63-linked ubiquitin chains of indicated lengths, and then fractionated by Superdex-200 gel filtration column. Aliquots of the fractions were analyzed in IRF3 activation assays (top), and SDS-PAGE followed by Coomassie Blue staining (bottom). E. RIG-I(N) was incubated with ubiquitin, and then analyzed by Superdex-200 gel filtration column followed by SDS-PAGE and Coomassie blue staining. F. Varying concentrations of RIG-I(N)/Ub4 complex (lane 5 in Figure 5B) were tested in IRF3 dimerization assays. Signal intensity on native gel was quantified by ImageQuant. Results from duplicated experiments are shown. G. MDA5(N) and K63-Ub6, separate or mixed, were analyzed by Superdex-200 gel filtration column. Aliquots of the fractions from indicated samples were analyzed by immunoblotting. The fractions from the mixture of MDA5 and K63-Ub6 were also assayed for their ability to stimulate IRF3 dimerization (bottom). Results shown are representatives of two experiments.
We estimated the stoichiometry of RIG-I(N):Ub chain complex by comparing the intensity of Coomassie blue-stained bands in the complex eluted from the gel filtration column with that of RIG-I(N) and Ub chains mixed at 2:1, 1:1 and 1:2 ratios (Supplementary Figure S4). Such estimates, although imprecise, revealed that RIG-I(N) and Ub chains had a stoichiometry of approximately 1:1 irrespective of the Ub chain lengths [ratio of RIG-I(N) to Ub3: ~0.8:0.7; to Ub4: 1.4:1.4; to Ub5: 1.2:1.1; to Ub6: 1.1: 0.9].
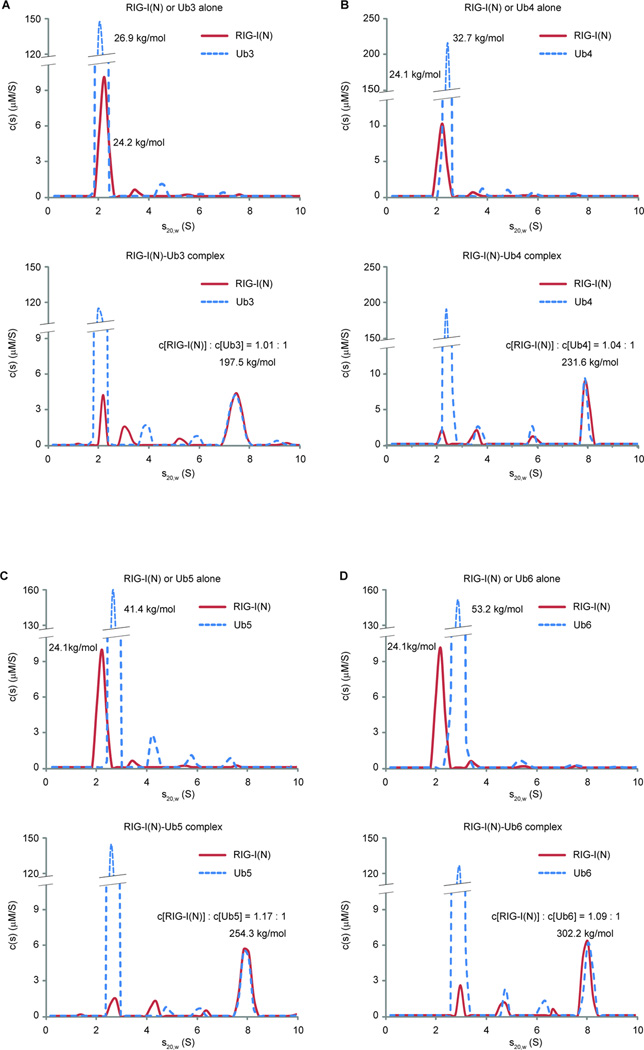
Polyubiquitin Binding Promotes the Formation of RIG-I Tetramer
To more precisely measure the stoichiometry and sizes of the RIG-I:Ub chain complexes, we performed sedimentation velocity analytical ultracentrifugation (AUC) (Figure 6). Calculation of sedimentation coefficients and frictional ratios of RIG-I(N) and K63-Ub3 alone revealed their molar masses of 24.2 kg/mol and 26.9 kg/mol, respectively, consistent with their theoretical values (Figure 6A, top; see also Experimental Procedures). In the sample where RIG-I(N) was incubated with a large molar excess of K63-Ub3, a faster-sedimenting population appeared with a molar mass of 197.5 kg/mol (Figure 6A, bottom). Multisignal sedimentation velocity analysis (Padrick et al., 2010) showed that RIG-I(N) and Ub3 in this complex had a stoichiometry of 1:1, consistent with gel filtration experiments (Supplementary Figure S4). Thus, the molar mass and stoichiometry measurements revealed that RIG-I(N) and Ub3 likely formed a complex containing 4 molecules of RIG-I(N) and 4 molecules of Ub3. Similar analyses using K63-Ub4, -Ub5 and -Ub6 suggest that RIG-I(N) forms complexes with these ubiquitin chains with a stoichiometry of approximately 4:4 irrespective of chain lengths (Figure 6B–D). The distribution of species during the sedimentation process showed few intermediate species, suggesting that RIG-I(N) and the Ub chains form hetero-tetrameric complexes in a highly cooperative manner.
Figure 6.
K63 polyubiquitin chains induce the formation of RIG-I tetramer. A. Analytical ultracentrifugation experiments of RIG-I(N) and K63-Ub3, alone (upper panel) or together (bottom panel), were performed. Absorbance at 280 nm and Rayleigh interferometry results were analyzed using SEDPHAT program. Sedimentation coefficient distribution of either RIG-I(N) or K63-Ub3 is shown on the same graph for comparison. B–D. Similar to A., RIG-I(N) and K63 ubiquitin chains of indicated lengths, alone or together, were analyzed by analytical ultracentrifugation. Calculated molar masses and indicated molar ratios are shown for indicated peaks. Results shown are representatives of three experiments.
RNA and Polyubiquitin Induce Oligomerization and Activation of Full-length RIG-I
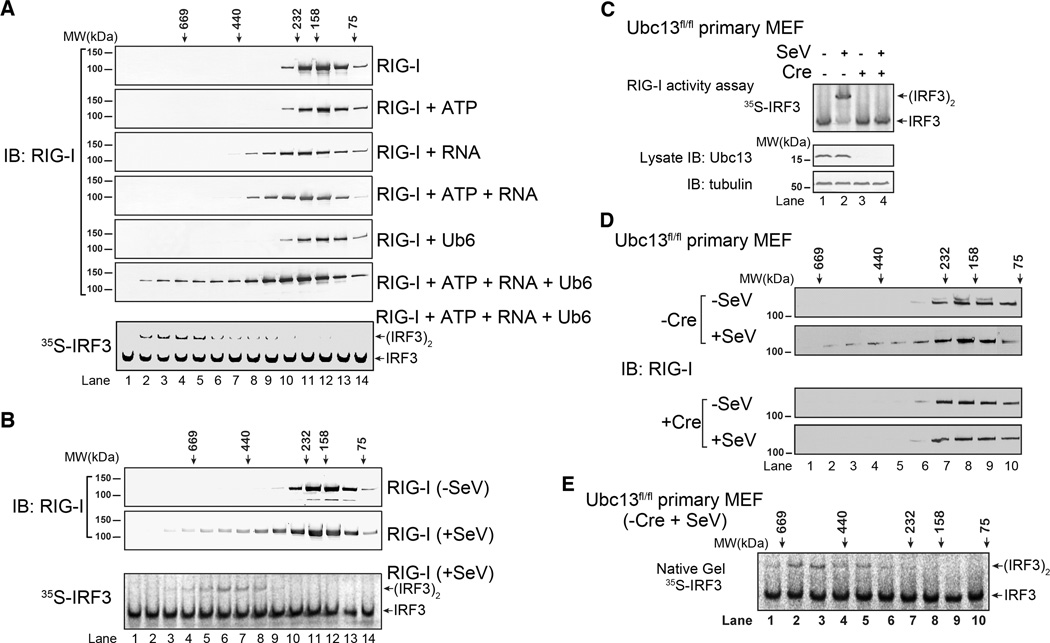
Full-length RIG-I can be activated in vitro when incubated with RNA ligands and K63 polyubiquitin chains in the presence of ATP (Zeng et al., 2010). To determine if full-length RIG-I forms oligomers after binding to K63 polyubiquitin chains, we carried out gel filtration analyses using purified RIG-I and K63-Ub6 (Figure 7A; Supplementary Figure S5A). RIG-I alone eluted from the column with the peak fraction corresponding to molecular weight between 75 kDa and 158 kDa (lane 12 in Figure 7A; Figure S5B), suggesting that it is a monomer. Incubation with 5’-pppRNA (containing 135 nucleotides) caused a shift in RIG-I elution peak to fractions corresponding to ~230 kDa, which may represent a RIG-I dimer bound to the RNA (lanes 10–11 in Figure 7A; Figure S5B). Incubation with K63-Ub6 alone did not change RIG-I distribution, because polyubiquitin binding to RIG-I requires both RNA and ATP (Zeng et al., 2010). Interestingly, when incubated with RNA, K63-Ub6 and ATP, some RIG-I shifted to HMW fractions that were larger than 400 kDa (Figure 7A, lanes 2–6). A small fraction of K63-Ub6 also co-migrated with RIG-I to the HMW fractions when the mixture contained RNA and ATP (Figure S5C, left). Although the signal detected with the ubiquitin antibody was weaker than that detected with the RIG-I antibody, semi-quantitative immunoblotting suggests an approximately equal molar ratio of RIG-I to K63-Ub6 in the HMW complexes (Figure S5C, right). The fractions with molecular weights larger than 600 kDa were most potent in activating IRF3 in the in vitro assay (Figure 7A and Figure S5C, bottom panels). Thus, the active HMW complexes could be composed of RIG-I tetramers with bound RNA and Ub6.
Figure 7.
RNA and polyubiquitin chains induce oligomerization of full-length RIG-I. A. RIG-I was incubated with ATP, RNA and/or K63-Ub6 as indicated. The mixtures were fractionated using Superdex-200 gel filtration column. Aliquots of the fractions were analyzed by immunoblotting and IRF3 dimerization assay as indicated. Only RIG-I incubated with ATP, RNA and K63-Ub6 had IRF3 stimulatory activity (bottom, and data not shown). B. HEK293T cells stably expressing RIG-I-Flag were infected with SeV or uninfected. RIG-I-Flag was affinity purified, and analyzed by gel filtration using Superdex-200 column. Aliquots of the fractions were analyzed by immunoblotting and in vitro IRF3 dimerization assay. C. Ubc13fl/fl primary MEF cells were infected with a lentiviral vector expression RIG-I-Flag, then the Ubc13 gene was deleted with Cre recombinase or left intact (+ or − Cre). The cells were infected with SeV, and then RIG-I was affinity purified and tested for its ability to activate IRF3 dimerization (top). The efficiency of Ubc13 depletion was verified by immunoblotting (bottom). D. WT and Ubc13-deleted MEF cells (−/+ Cre) were infected with SeV or not infected, and then RIG-I was affinity purified and fractionated on a Superdex-200 column followed by immunoblotting. E. The fractions from RIG-I isolated from the virus-infected WT MEFs as shown in D (−Cre, + SeV) were analyzed for their ability to stimulate IRF3 dimerization. Results shown are representatives of two experiments.
To determine if RIG-I forms oligomers in cells in response to viral infection, we established a HEK293T cell line stably expressing RIG-I-Flag, and purified RIG-I-Flag from these cells infected with SeV or uninfected. The RIG-I protein was fractionated by gel filtration chromatography (Figure 7B). RIG-I from uninfected cells eluted from the column with an elution profile suggestive of a monomer (lanes 11–12). While the majority of RIG-I from the virus-infected cells also eluted as a monomer that was incapable of activating IRF3, a small fraction eluted as HMW complexes that potently activated IRF3 (lanes 3–8). The heterogeneity of these active complexes may reflect RIG-I tetramers bound to heterogeneous viral RNA and ubiquitin chains in cells.
We performed two sets of experiments to investigate the role of ubiquitination in RIG-I oligomerization in cells in response to viral infection. First, we obtained primary MEFs from Ubc13 floxed/floxed mice and deleted the Ubc13 gene using a lentivirus expressing the Cre recombinase (Figure 7C–7E). To facilitate the isolation of the RIG-I complex, these cells were also engineered to stably express full-length RIG-I-Flag. The isolated RIG-I complex from SeV-infected cells strongly activated IRF3 in the in vitro reconstitution assay, but this activity was abolished in cells expressing Cre, which depleted Ubc13 (Figure 7C). Importantly, when analyzed by gel filtration, a fraction of RIG-I isolated from SeV-infected cells eluted from the column as HMW complexes (Figure 7D). Only these complexes, but not the lower molecular weight forms of RIG-I, were capable of activating IRF3 (Figure 7E). Depletion of Ubc13 prevented the formation of the HMW RIG-I complexes when cells were infected with SeV (Figure 7D, lower panel). These results indicate that Ubc13 is essential for RIG-I oligomerization in virus-infected cells, implying that K63 polyubiquitination is important in this process.
In the second set of experiments, we reconstituted RIG-I knockout MEFs with WT or ubiquitin-binding defective mutants of RIG-I (K164A and 6KR; Supplementary Figure S5D). SeV infection of cells reconstituted with WT RIG-I led to the formation of HMW RIG-I complexes capable of activating IRF3. In contrast, neither K164A nor 6KR RIG-I was able to form HMW complexes in virus-infected cells. Thus, the ability of RIG-I to bind K63 polyubiquitin chains is important for its oligomerization and activation in response to viral infection.
DISCUSSION
In this study, we established a cell-free system in which MDA5 led to the activation of IRF3 in a manner that depends on binding to poly[I:C] and K63 polyubiquitin chains. We showed that MDA5 CARD domains bind to K63 polyubiquitin chains and that this binding is important for IRF3 activation. We have identified several conserved residues within MDA5 and RIG-I CARD domains that are required for K63 polyubiquitin chain binding. Mutations that disrupt ubiquitin binding also impair the ability of RIG-I and MDA5 to activate IRF3 and induce IFNβ. Collectively, these results provide a unified mechanism of RIG-I and MDA5 signaling that involves the binding of K63 polyubiquitin chains through the tandem CARD domains.
We have carried out an extensive bioinformatics analysis of proteins harboring known and putative domains belonging to the death domain superfamily, including the CARD domains. We grouped these proteins based on their sequence homology and available structural information. We have tested several CARD domains for their ability to bind K63 polyubiquitin chains, but so far only closely related RIG-I and MDA5 tandem CARD domains have the ubiquitin binding activity. This limited analysis by no means rules out the possibility that some other domains of the DD superfamily could be functional ubiquitin binding domains. Nevertheless, it appears that the CARD domains of RIG-I and MDA5 have acquired the new ubiquitin-binding function and utilize the labile ubiquitin chains as an endogenous ligand to regulate their signaling functions.
Several lines of evidence strongly suggest that unanchored, rather than substrate-anchored, ubiquitin chains activate RIG-I and MDA5 in vitro and in cells: 1) unanchored K63 ubiquitin chains containing more than two ubiquitin directly bind and activate the CARD domains of RIG-I and MDA5 (here and Zeng et al, 2010); this binding in full-length RIG-I and MDA5 is regulated by RNA binding. In contrast, when ubiquitin chains are conjugated to protein targets such as TRIM25 or TRAF6, they fail to activate RIG-I or MDA5 (Zeng et al., 2010; Jiang X. and Chen, Z., data not shown); 2) a large collection of point mutations in the CARDs of RIG-I and MDA5 that disrupt their binding to K63 ubiquitin chains also impair their ability to activate IRF3 in vitro and to induce IFNβ in cells, indicating the importance of polyubiquitin binding by RIG-I and MDA5; 3) although a very small fraction of RIG-I is ubiquitinated in cells, removal of the ubiquitin chains using a viral deubiquitination enzyme does not impair the ability of RIG-I to activate IRF3 (Zeng et al., 2010); 4) a RIG-I mutant that can bind to ubiquitin but cannot be ubiquitinated was capable of inducing IFNβ in cells, suggesting that covalent ubiquitination of RIG-I is dispensable for its function (Zeng et al., 2010); 5) MDA5 does not contain a conserved lysine for ubiquitination and there is no evidence that MDA5 is ubiquitinated in cells; 6) fusion of the ubiquitination-defective mutant of RIG-I and MDA5 to a heterologous ubiquitin binding domain (NZF) restores IRF3 activation in vitro and IFNβ induction in cells; 7) both RIG-I and MDA5 bind to endogenous unanchored K63 polyubiquitin chains, which can be isolated from human cells and shown to potently activate the RIG-I pathway (here and in Zeng et al, 2010). Despite these evidence, we cannot rule out the possibility that an unknown ubiquitination target may activate RIG-I or MDA5 in certain signaling pathway.
Our previous study shows that unanchored K63 polyubiquitin chains generated by TRAF6 can activate TAK1 and IKK in the IL-1 pathway (Xia et al., 2009). This raises the question of how these ubiquitin chains achieve specificity in regulating distinct signaling pathways. A possible solution to this question lies in the fact that unanchored K63 ubiquitin chains are rapidly disassembled by abundant deubiquitination enzymes in cells. Thus, the ubiquitin chains may be delivered locally from the chain generator (E3s such as TRIM25 or TRAF6) to the receptor (e.g, RIG-I or TAB2) to initiate signaling; excess ubiquitin chains are degraded. As TRIM25 and TRAF6 are recruited to RIG-I and IL-1 receptor complex in response to virus infection and IL-1 stimulation, respectively, distinct signaling cascades are activated. Such temporal regulation and compartmentalization is a hallmark of other intracellular signaling molecules, such as cAMP and calcium, which are known to bind multiple cellular targets but regulate distinct signaling pathways in response to specific ligands.
Importantly, we demonstrated that the binding of K63 polyubiquitin chains leads to the formation of RIG-I tetramer, which is highly potent in activating MAVS and the downstream pathway. To our knowledge, this is the first example of ubiquitin binding leading to protein oligomerization. We also observed oligomerization of full-length RIG-I that is dependent on RNA, ATP and K63 polyubiquitin chains. Further, virus infection leads to the formation of a HMW RIG-I complex, and only this complex, but not the lower molecular weight forms of RIG-I, is capable of activating IRF3 in the cell-free system. Based on these results, we propose that sequential binding of RIG-I to viral RNA and endogenous K63 polyubiquitin chains leads to its oligomerization and subsequent activation. The oligomerization of RIG-I likely promotes the formation of CARD domain clusters, which presumably interact with the CARD domain of MAVS on the mitochondrial surface, leading to MAVS aggregation and activation.
Interestingly, we found that the stoichiometry of the RIG-I – ubiquitin chain complex is 4:4 irrespective of ubiquitin chain length, as long as the ubiquitin chains contain more than two ubiquitins linked through K63. It is tempting to speculate that individual ubiquitin in a chain bind to one or both of the tandem CARD domains, and the longer ubiquitin chains may actually ‘chain’ or ‘cross-link’ different RIG-I CARD domains together. Alternatively, the binding of K63 ubiquitin chains may induce a conformational change of the RIG-I CARD domains to promote their intermolecular interactions, resulting in their oligomerization. High-resolution structural studies are required to understand how ubiquitin chains bind to RIG-I CARD domains, how the oligomerization occurs, and how it terminates to produce RIG-I tetramers. The formation of HMW RIG-I complex is also detected in virus-infected cells and only these complexes are capable of activating IRF3. Importantly, deletion of Ubc13 and a mutation that disrupts ubiquitin binding of RIG-I abolished the formation of the HMW RIG-I complex, strongly suggesting that K63 polyubiquitin chains are important for the activation of RIG-I, probably by inducing its oligomerization. Further work is needed to determine the composition and stoichiometry of the active RIG-I complex isolated from virus-infected cells.
It remains to be determined how RIG-I oligomers become competent to activate MAVS. We have been unable to detect stable interaction between the CARD domains of RIG-I and that of MAVS in the presence or absence of K63 polyubiquitin chains. Interestingly, we recently found that incubation of sub-stoichiometric amounts of RIG-I – ubiquitin chain complex with mitochondria causes very rapid aggregation and activation of MAVS on the mitochondrial membrane through a prion-like mechanism (Hou et al., 2011). The aggregated forms of MAVS are highly potent in activating IKK and TBK1, resulting in robust production of type-I interferons. Virus infection causes a nearly complete conversion of full-length MAVS into the aggregate forms, whereas only a small fraction of RIG-I forms active HMW complexes after virus infection. Thus, MAVS CARD is prone to form prion-like aggregates whereas RIG-I CARDs tend to form a defined oligomer (tetramer in the presence of ubiquitin chains). We hypothesize that the RIG-I – ubiquitin chain complex acts like a catalyst in that it transiently contacts and induces the aggregation of the MAVS CARD, which in turn interacts with other MAVS to form functional aggregates. This mechanism allows for a rapid amplification of the signaling cascade upon detection of viral RNA by RIG-I and MDA5, leading to a robust antiviral immune response.
EXPERIMENTAL PROCEDURES
IRF3 Activation (Dimerization) Assay
Biochemical assays for IRF3 activation with cytosolic extract (S5) and crude mitochondria (P5) of cultured cells were described previously (Zeng et al., 2009). Briefly, hypotonic buffer [10 mM Tris-HCl (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, and protease inhibitor cocktail] was used to make S5, and isotonic buffer (hypotonic buffer plus 0.25 M D-Mannitol) was used to make P5. Cells were homogenized in appropriate buffers, and centrifuged at 1,000 g for 5 minutes to pellet nuclei. Post-nuclear supernatant was further centrifuged at 5,000 g for 10 minutes to separate P5 and S5. P5 was washed once with isotonic buffer. For IRF3 pathway activation by RIG-I or MDA5, proteins to be tested were added to reaction mixture containing, 0.5 mg/ml P5, 3 mg/ml S5, 1xMgATP buffer [20 mM Hepes-KOH (pH7.4), 2 mM ATP, 5 mM MgCl2], and 35S-IRF3. After incubation at 30°C for 1 hour, samples were centrifuged at 20,000 g for 5 minutes and supernatants were subjected to native PAGE to visualize IRF3 dimerization by autoradiography using PhosphorImager (GE Healthcare). ImageQuant was used to quantify signal intensity.
Biochemical Assay for RIG-I and MDA5 Activation in vitro
For RIG-I(N) or MDA5(N) activation by ubiquitination, RIG-I(N) or MDA5(N) was subjected to ubiquitination reaction, and then 1 µl of the reaction mixture was used in IRF3 activation assay described above. For RIG-I(N) or MDA5(N) activation by ubiquitin chains, 1 µl mixture containing 50 ng of RIG-I(N) or MDA5(N) and 50 ng of ubiquitin chain was incubated at 4°C for 30 minutes, and then tested in IRF3 activation assay. Five-fold serial dilution was used in titration experiments. For standard activation assays of full-length RIG-I or MDA5, 1 µl mixture containing 50 ng of RIG-I or MDA5, 50 ng poly[I:C] or 5’-pppRNA, 50 ng ubiquitin chains, in the presence of 1xMgATP Buffer, was incubated at 4°C for 30 minutes, and then used in IRF3 activation assay.
Analytical Ultracentrifugation
Multisignal Sedimentation Velocity (SV) experiments were carried out as described in (Padrick et al., 2010). Briefly, concentrated stocks of RIG-I(N), K63-Ub3, -Ub4, -Ub5, and -Ub6 were prepared in 20 mM Tris-HCl (pH7.5), 20 mM NaCl, 0.5 mM TCEP, and diluted in the same buffer to final concentrations of 4–8 µM for RIG-I(N) or 20–40 µM for K63-Ub chains, separate or in combination. Samples were allowed to equilibrate for 16 hours at 4°C, and loaded into centrifugation cells equipped with charcoal-filled Epon centerpieces and sapphire windows; the cells were allowed to equilibrate at 20°C for two hours before centrifugation. 390 µl of sample in the sample sector and the same volume of buffer without proteins in the reference sector were centrifuged with Beckman Optima XL-I analytical ultracentrifuge in an An50Ti rotor at 50,000 rpm or 35,000 rpm, at 20°C, until all components apparently sedimented to the bottom of the cell. UV absorbance at 280 nm (ABS) and Rayleigh interferometry (IF) data were collected simultaneously using the Beckman control software, and analyzed with SEDPHAT using multiwavelength model (Balbo et al., 2005). Molar signal increments (ε) of all components were calculated (εIF) or refined by SEDPHAT from experimental data (εABS), shown in Table 1. All values for buffer densities (ρ), buffer viscosities (η), and partial specific volumes (ν̄) of the proteins were estimated using SEDNTERP.
Table1.
Fixed IF signal increments and refined ABS signal increments used in this study.
| Protein | εIF (fringes/M·cm) | εABS (AU/M·cm) |
|---|---|---|
| RIG-I(N) | 64784.8 | 45217.9 |
| Ub3 | 70877.5 | 3841.26 |
| Ub4 | 94381.4 | 5132.05 |
| Ub5 | 117885.3 | 6220.40 |
| Ub6 | 141389.1 | 7329.04 |
εIF = 2.75Mc (Mc is the calculated molar mass)
RIG-I Oligomerization Analysis by Gel Filtration
For RIG-I in vivo oligomerization experiments, HEK293T cells stably expressing RIG-I-Flag (Chiu et al., 2009) were infected with SeV or mock treated for 16 hours. For MEF cells stably expressing RIG-I-Flag, cells were infected with SeV or mock treated for 10 hours. Cell lysates were prepared in Lysis Buffer [20 mM Tris-Cl (pH7.5), 150 mM NaCl, 0.5% NP40, 1 mM DTT, protease inhibitor cocktail]. RIG-I was immunoprecipitated from cell lysates with anti-Flag M2-agarose at 4°C for 2 hours. The agarose beads were washed three times with Lysis Buffer, and RIG-I was eluted with 0.2 mg/ml Flag peptide in Buffer A [20 mM Tris-Cl (pH7.5), 1 mM DTT]. For RIG-I in vitro oligomerization experiments, RIG-I (0.1 mg/ml) was incubated in various combinations with ATP (provided as 1xMgATP Buffer), 5'-pppRNA (0.1 mg/ml), and K63-Ub6 (1 mg/ml) in Buffer B [20 mM Tris-HCl (pH7.5), 100 mM NaCl, 0.5 mM TCEP] at 4°C for 30 minutes. 50 µl of each sample was subjected to Superdex-200 gel filtration analysis with Buffer B as the elution buffer.
For RIG-I(N) oligomerization experiments, RIG-I(N) (0.5 mg/ml) was incubated with K63-Ub3, -Ub4, -Ub5 or -Ub6 (0.5 mg/ml) at 4°C for 30 minutes. For MDA5(N) oligomerization experiments, MDA5(N) (0.05 mg/ml) was incubated with K63-Ub6 (2 mg/ml) at 4°C for 30 minutes. 50 µl of each mixture was loaded onto Superdex-200 gel filtration column with Buffer A. Gel filtration fractions were used for immunoblotting and IRF3 activation assay. For semi-quantification, fractions containing RIG-I(N):Ub chain complexes were subjected to SDS-PAGE alongside purified RIG-(N) and Ub chains mixed at defined ratio, followed by Coomassie Blue staining. Gel band intensities were quantified with ImageQuant.
Supplementary Material
HIGHLIGHTS.
Like RIG-I, MDA5 activates IRF3 in a cell-free system
Both RIG-I and MDA5 CARD domains bind K63 polyubiquitin chains and activate IRF3
Polyubiquitin binding is required for the activation of RIG-I and MDA5
Polyubiquitin binding induces the formation of a highly active RIG-I tetramer
ACKNOWLEDGMENTS
We thank Dr. Jae Jung (University of Southern California) for providing some of the RIG-I mutant plasmids and Trim 25-deficient MEFs, and Dr. Shizuo Akira (Osaka University) for the Ubc13fl/fl mice. This work was supported by grants from NIH (RO1-GM063692 and RO1-AI093967), Cancer Prevention and Research Institute of Texas (RP110430) and the Welch Foundation (I-1389).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures and seven figures.
REFERENCES
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc Natl Acad Sci U S A. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The Powerstroke and Camshaft of the RIG-I Antiviral RNA Detection Machine. Cell. 2011;147:259–261. doi: 10.1016/j.cell.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Deka RK, Chuang JL, Wynn RM, Chuang DT, Norgard MV, Rosen MK, Brautigam CA. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal Biochem. 2010;407:89–103. doi: 10.1016/j.ab.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Inagaki F. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.