Summary
U1 snRNP (U1), in addition to its splicing role, protects pre-mRNAs from drastic premature termination by cleavage and polyadenylation (PCPA) at cryptic polyadenylation signals (PASs) in introns. Here, a high throughput sequencing strategy of differentially expressed transcripts (HIDE-seq), mapped PCPA sites genome-wide in divergent organisms. Surprisingly, while U1 depletion terminated most nascent gene transcripts within ~1 kb, moderate functional U1 level decreases, insufficient to inhibit splicing, dose-dependently shifted PCPA downstream, eliciting mRNA 3′ UTR shortening and proximal 3′ exon switching characteristic of activated immune and neuronal cells, stem cells and cancer. Activated neurons’ signature mRNA shortening could be recapitulated by U1 decrease and antagonized by U1 over-expression. Importantly, we show that rapid and transient transcriptional up-regulation inherent to neuronal activation physiology creates U1 shortage relative to pre-mRNAs. Additional experiments suggest co-transcriptional PCPA counteracted by U1 association with nascent-transcripts, a process we term telescripting, ensuring transcriptome integrity and regulating mRNA length.
Introduction
Messenger RNAs in eukaryotic cells are produced from precursor transcripts (pre-mRNAs) by post-transcriptional processing. In metazoans, two processing reactions are particularly extensive and contribute most significantly to mRNA transcriptome diversity - splicing of introns and alternate cleavage and polyadenylation (Di Giammartino et al., 2011; Hartmann and Valcarcel, 2009; Wang et al., 2008). Splicing is performed by a spliceosome that assembles on each intron and is comprised predominantly of small nuclear RNPs (snRNPs), U1, U2, U4, U5 and U6 snRNPs, in equal stoichiometry (Nilsen, 2003; Wahl et al., 2009). U1 snRNP (U1) plays an essential role in defining the 5′ splice site (5’ss) by RNA:RNA base pairing via U1 snRNA’s 5′ nine nucleotide (nt) sequence. Using antisense morpholino oligonucleotide complementary to U1 snRNA’s 5′ end (U1 AMO) that interferes with U1 snRNP’s function in human cells, we observed accumulation of introns in many transcripts, as expected for splicing inhibition (Kaida et al., 2010). However, in addition, the majority of pre-mRNAs terminated prematurely from cryptic PASs in introns, typically within a short distance from the transcription start site (TSS). These findings indicated that nascent transcripts are vulnerable to premature cleavage and polyadenylation (PCPA) and that U1 has a critical function in protecting pre-mRNA from this potentially destructive process. We further showed that PCPA suppression is a separate, splicing-independent and U1-specific function, as it did not occur when splicing was inhibited with U2 snRNA AMO or the splicing inhibitor, spliceostatin A (SSA) (Kaida et al., 2007).
These observations were made by transcriptome profiling using partial genome tiling arrays, which provided limited information. Here, to define the parameters involved in PCPA and its suppression, we devised a strategy (HIDE-seq) to select and sequence only differentially expressed transcripts, identifying changes that occur upon U1 decrease to various levels and in different organisms. The sequence information obtained from HIDE-seq provided genome-wide PCPA maps and these, together with direct experiments, revealed that U1’s PCPA suppression is not only essential for protecting nascent transcripts, but is also a global gene expression regulation mechanism. Unexpectedly, PCPA position varied widely with the degree of U1 decrease, trending to usage of more proximal PASs with greater reduction. This yielded mRNAs with shorter 3′ untranslated regions (3′ UTRs) and alternatively spliced isoforms resulting from usage of more proximal alternative polyadenylation (APA) sites, characteristic of activated immune, neuronal, and cancer cells (Flavell et al., 2008; Mayr and Bartel, 2009; Niibori et al., 2007; Sandberg et al., 2008). We demonstrate that U1 decrease can recapitulate such specific mRNA changes that occur during neuronal activation. Indeed, we show that the rapid transcriptional up-regulation during neuronal activation is a physiological condition that creates U1 shortage relative to nascent transcripts. Furthermore, U1 over-expression inhibits activated neurons’ mRNA signature shortenings. We suggest that by determining the degree of PCPA suppression, U1 levels play a key role in PAS usage and hence mRNA length. We propose a model whereby U1 binds to nascent pre-mRNAs co-transcriptionally to explain how U1 shortage results in a corresponding loss of distal PASs suppression from the cleavage and polyadenylation machinery that is associated with the RNA polymerase II (polII) transcription elongation complex (TEC) (Das et al., 2007; Hirose and Manley, 1998; McCracken et al., 1997).
Results
HIDE-seq: A strategy for high throughput sequencing of only transcriptome differences
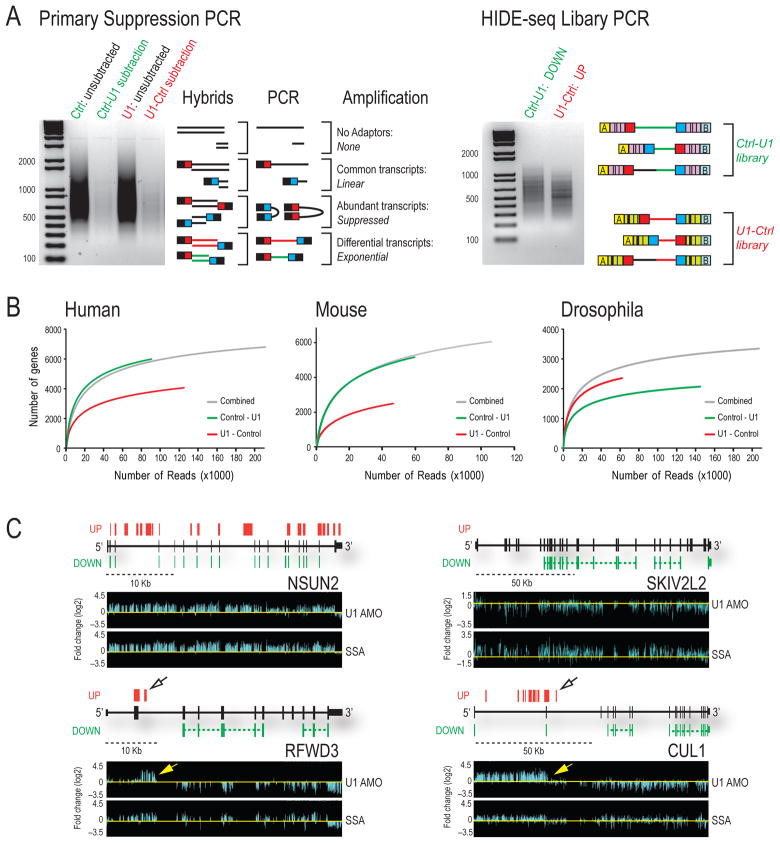
To identify transcriptome changes after U1 snRNP functional depletion, cDNA libraries were prepared from poly(A) RNA of human cells (HeLa) 8 hr after transfection with 15 nmole of U1 AMO (U1 depleted) or control AMO (Kaida et al., 2010). Each cDNA library was digested separately with three 4 bp restriction endonucleases to produce fragments of uniform length (~250 bp), and different adaptor oligonucleotides were ligated separately to the 5′ ends of the experimental cDNA for subsequent amplification. Subtractive hybridization and suppression PCR selectively amplified only the differentially expressed transcripts (Diatchenko et al., 1996; Gurskaya et al., 1996). Nested primers fused to 454 sequencer linkers and sample-specific barcodes were used to generate amplicons of subtracted libraries prepared in both U1-Control and Control-U1 directions for massive parallel sequencing in the same well, controlling for sample-to-sample variation (Figure 1A). The reciprocal sequence reads from bidirectional subtraction further enhanced the definition of transcriptome changes.
Figure 1. HIDE-seq identifies genome-wide transcriptome changes.
(A) Unsubtracted and subtracted libraries prepared in both directions (Control-U1, green; U1-Control, red) are shown (left gel) along with possible products of suppression PCR. Differentially expressed sequences are in red or green and common sequences in black. Nested PCR (right gel) was performed with primers containing barcodes (hashed lines) and 454 adaptors A and B (yellow and light blue). (B) Randomly sampled reads with greater than 90% coverage and 90% identity were plotted against affected genes for HeLa, NIH/3T3 and S2 from individual or combined (grey) subtractions. (C) HIDE-seq reads are above [U1-Control (UP)] and below [Control-U1 (DOWN)] gene structures in black (block = exon, thin block = UTR, horizontal line = intron). Spliced reads are connected by colored dashed lines whereas immediately adjacent reads separated by RsaI, HaeIII, and AluI sites are not. Log2 fold changes from GTA of U1 AMO or SSA-treated HeLa cells normalized to controls (bottom panels). Yellow and open arrows point to inferred PCPA sites. See also Table S1 and Supplemental Figure S1.
High throughput amplicon sequencing was performed using only 1/4–1/2 of a 454 sequencing plate (~300,000 reads) for extensive profiling (Table S1). Nearly 70% of HIDE-seq reads, averaging 150–400 nt, were unambiguously mapped (90% identity/90% coverage) to the genome (Table S1). Reads located in intergenic regions and sequences not unique to either of the subtraction directions were excluded from further analysis. Only a minor fraction (0.03%) was from ribosomal RNA as compared to >70% without subtraction, confirming the efficiency of the method and indicating the majority of reads are informative (Table S1). Importantly, as more reads were obtained the number of new affected genes discovered plateaued (Figure 1B), as did coverage within a gene (data not shown), suggesting that extensive coverage was achieved and allowing a lack of reads to indicate little or no sequence change at a given location.
Data from high density genomic tiling arrays (GTA) of human chromosomes 5, 7, and 16 for HeLa cells treated with U1 AMO under the same conditions used here (15 nmole) provided a comprehensive dataset with which to compare the HIDE-seq methodology (Kaida et al., 2010). HIDE-seq reads present at a given genomic locus were interpreted as being up or down in U1 AMO-treated cells relative to control, according to whether they came from the U1-Control (red) or the control-U1 (green) subtraction, respectively (Figure 1C). For GTA we used SSA as a reference for intron accumulation (e.g. NSUN2 and SKIV2L2) and to facilitate discovery of PCPA, represented by 5′ intron accumulation that terminates abruptly followed by a decrease in downstream signals (e.g. RFWD3 and CUL1). HIDE-seq readily identified the same transcriptome changes as GTA (Figure 1C). Sorted for chromosomes 5, 7 and 16, it captured the highly significant (≥2 fold change, ≥100 nt, p-value <0.01) intron accumulations detected by GTA in 85% (189/223) of the genes (Supplemental Figure S1a), a very high correspondence considering that RNAs came from separate biological experiments. HIDE-seq discovered 198 additional genes with intron accumulation on these same three chromosomes, 82% of which were confirmed by lower stringency GTA analysis, indicating HIDE-seq is highly reliable and sensitive. Moreover, HIDE-seq identified these and other differences genome wide, capturing subtle sequence changes such as exon skipping (Supplemental Figure S1b-c). Although HIDE-seq is not quantitative or necessarily complete, it represents an alternative and complementary approach to full transcriptome sequencing, providing a detailed snapshot of transcriptome differences at a fraction of the cost. Combined with a streamlined informatics pipeline we developed (http://www.upenn.edu/dreyfusslab), HIDE-seq is a simple and powerful strategy that is widely applicable with different sequencing platforms (e.g. Illumina; data not shown).
PCPA and its suppression by U1 are evolutionarily conserved
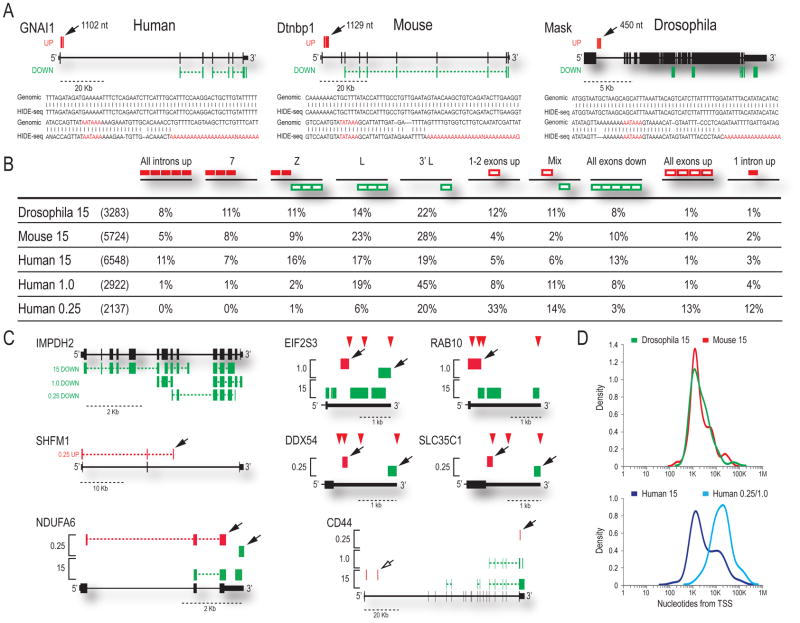
We used HIDE-seq to determine if PCPA occurs in divergent organisms following U1 depletion in mouse (3T3) and Drosophila (S2) cells (Figure 1B) with AMOs specific to each organism’s U1 5′-end sequence. As in humans, premature termination in introns was U1 snRNP-specific and was not a consequence of splicing inhibition (Supplemental Figures S1e-f). HIDE-seq in HeLa, 3T3, and S2 detected sequence differences in 6548, 5724, and 3283 genes, respectively, which could be classified into several patterns (Figure 2B). Accumulation of polyadenylated intron reads, followed by a decrease in downstream exon signals, provided the most direct evidence for PCPA (Figure 2A, Supplemental Figures S2–S4). This pattern, designated as Z (Figure 2B), demonstrated that PCPA typically occurred in intron 1, with little or no transcription beyond that point. We developed an algorithm to detect Z and related patterns, designated as L and 7, which represent PCPA events but either the upstream accumulation or the downstream decrease was not detected, respectively (Figure 2B). It is likely that differential stability of the various transcripts produced determines whether a Z, 7 or L pattern is observed. Collectively, these accounted for ~40% of the changes. In addition, other patterns consistent with PCPA were detected, such as all exons down (e.g. Figure 1C; SKIV2L2), likely resulting from very early PCPA, although the possibility of transcriptional down-regulation cannot be excluded. Several genes with all introns up (e.g. Figure 1C; NSUN2) also had polyadenylated intronic reads, indicating that these, too, do not entirely escape PCPA. Another pattern, 3′L, representing a major class at moderate U1 decrease as discussed later, is similar to L but the down reads (i.e., Ctrl-U1 direction) are near the 3′ end of the gene, indicative of transcript shortening. Together, these patterns account for 75–90% of the transcriptome changes in U1 depleted cells, and their remarkable similarity in the three organisms indicates that PCPA and U1’s function in its suppression are essential for formation of full-length transcripts for the majority of metazoan genes.
Figure 2. Transcriptome profiles show a conserved function of U1 snRNP in PCPA suppression.
(A) HIDE-seq profiles are shown along with sequences of poly(A) reads (location indicated by solid arrows) aligned to the genome for S2, NIH/3T3, and HeLa. Distance (nt) is relative to the upstream 5’ss with the PAS hexamer likely used and stretches of A’s added to HIDE-seq reads in red. See also Supplemental Figures S2–S4. (B) Illustrations of patterns observed (depicted above each column; e.g. Z: red up then green down) after U1 AMO with the percentage of each category reported for Drosophila (15), mouse (15) and human at three doses (15, 1.0, 0.25) compared to the total number of affected genes in parenthesis. Introns = solid red boxes; exons = empty boxes. (C) Individual examples of patterns described in Figure 2B are shown with brackets indicating results from more than one concentration. Solid arrows indicate reads ending in poly(A) tails; open arrows are inferred PCPA; red triangles are documented PASs. (D) Using reads with poly(A) tails terminating in introns (Figures S2–S5), the distribution of distances of PCPA relative to the TSS are plotted for Drosophila and mouse (top), and human at high and low U1 AMO (bottom).
Moderate U1 decrease elicits 3′UTR shortening and a switch to shorter mRNA isoforms
The sequencing data from complete U1 depletion indicated that PCPA occurred ~10–30 nt downstream of cryptic PASs that are similar to PASs found at the 3′ end of transcripts (Hu et al., 2005) (Figure 2A; Supplemental Figures S2–S4). These included both canonical (AAUAAA or AUUAAA) and rare or not previously described hexamers (Beaudoing et al., 2000; Tian et al., 2005), as well as U-rich and GU-rich elements that typically surround PASs. To identify PASs that may be particularly vulnerable to PCPA and assess the relevance of U1’s suppression under less drastic conditions, we decreased U1 levels by ~25% and 50%, with 0.25 and 1.0 nmole U1 AMO, respectively (Kaida et al., 2010). Moderate U1 decreases differed from those observed in U1 depletion in three major ways (Figure 2B). First, significantly fewer genes were affected (~35–50% compared to 15 nmole; Figure 2B). Second, there was no general intron accumulation, indicating that splicing was not inhibited (Figure 2B; e.g. All introns up, Z, or 7). Third, and most surprisingly, moderate U1 decrease shifted the PCPA positions towards much greater distances from the TSS. Representative examples of the major patterns from various U1 level decreases are shown in Figure 2C. The majority of changes at the moderate U1 decrease showed a 3′L pattern, reflecting decreases near the ends of genes (e.g. IMPDH2) and consisted mostly of decreases in distal 3′UTR reads (e.g. EIF2S3 and Supplemental Figures S5 and S6), suggesting 3′ UTR shortening in genes of a wide range of sizes. A widespread shift to usage of more proximal PASs in introns in the 3′ half of genes (e.g. SHFM1) was also observed. Many of the genes at 0.25 nmole U1 AMO that had shorter 3′UTR (e.g. NDUFA6), also had several exon signals up-regulated (Figure 2B), possibly due to elimination of miRNA targets in the 3′ UTR (Bartel, 2009). The CD44 gene (Figure 2C) illustrates the PCPA continuum and its U1 dose-dependence, having a shorter 3′UTR at 0.25, an L pattern at 1.0, and a Z pattern from PCPA in the first intron at 15, suggesting usage of more proximal PASs in the same gene with further decrease of U1.
The overall change in PCPA position with the degree of U1 level decrease is illustrated in Figure 2D. Upon U1 depletion in three organisms, most PCPA occurred ~1 kb from the TSS, typically in the first (48%) or one of the first introns (intron 2: 26%; intron 3: 11%). However, at lower U1 decreases, PCPA occurred at much greater distances from the TSS (~20 kb) (Figure 2D-lower panel, Figure S5, Table S2). In most, if not all cases, including transcripts that terminated within ~1 kb from the TSS, several strong predicted PASs, and many more non-canonical PASs were bypassed before the site of PCPA. Importantly, many of the PCPA sites coincided precisely with previously reported polyadenylated ESTs from a wide range of cell types and tissues (Supplemental Figures S2 and S5), suggesting that PCPA is a natural phenomenon that occurs under normal physiological conditions.
Loss of U1 PAS suppression promotes alternative splicing of proximal 3′ terminal exons
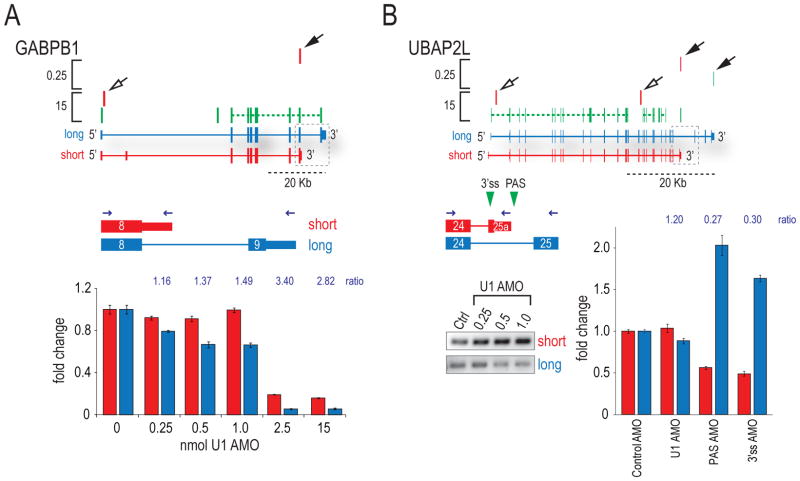
Two types of mRNA changes resulting from PCPA at low U1 AMO (Human 1.0 and 0.25) were detected, in addition to 3′UTR shortening (Supplemental Figure S6). First, PCPA in introns produced shorter mRNAs that lack the downstream exons and 3′UTR of the full-length transcript. In some of these the open reading frame (ORF) of the alternative terminal exon could potentially extend into the intron (Figure 3A), a scenario referred to as a “composite” or “bleeding” exon (Tian et al., 2007). Quantitative RT-PCR on RNA of cells transfected with a range of U1 AMO doses confirmed the shift to proximal polyadenylation predicted by HIDE-seq (Figure 3A). As expected, upon U1 depletion both short and long isoforms decreased, due to early PCPA or splicing inhibition, indicated by the Z patterns (e.g. GABPB1 2.5 and 15 nmole). At low U1 AMO, however, rather than an overall transcript level decrease, the amounts of the short forms remained relatively stable while the long forms decreased, causing the ratio of the short to the long isoform to increase (Figure 3A).
Figure 3. Moderate U1 reduction-induced PCPA regulates expression of short isoforms.
(A) HIDE-seq profile for GABPB1 at high and low U1 AMO are above diagrams of long (blue) and short (red) isoforms. RT-qPCR of short and long isoforms was performed on cDNA from cells transfected with a range of U1 AMO concentrations using a common forward and different reverse primers as indicated (blue arrows). Grey dotted boxes indicate region of transcript probed. Ratios of short to long are reported above histograms. (B) UBAP2L was probed by RT-PCR for short (due to 3′ exon switching) and long isoforms (gels) and by RT-qPCR on cDNA from cells treated with various AMOs. Green triangle = 3’ss and PAS AMOs, black arrows = poly(A) reads; open arrows = inferred PCPA.
A second type of polyadenylated read found in introns of the canonical transcript at low U1 AMO (solid arrows) mapped to the ends of exons that can be alternatively spliced mutually exclusively with the full-length gene’s terminal exon (“short” in Figure 3), an example of splicing-dependent APA (Edwalds-Gilbert et al., 1997; Zhang et al., 2005). The UBAP2L gene (Figure 3B) displayed a dose-dependent increase in the relative amount of the shorter isoform at low U1 AMO (RT-PCR gel inset). Interestingly, many 3′ exon switching cases, including UBAP2L, have been reported following immune cell activation and suggested to result from alternative splicing (Sandberg et al., 2008). However, we considered an alternative mechanism, in which PCPA occurs first and that it is this event that causes the upstream 5’ss to splice to an alternative 3’ss not utilized in the full-length transcript. To address this, we used AMOs to block either the 3’ss or the PAS of the short isoform and measured the levels of both isoforms by RT-qPCR (Figure 3B). As expected, the 3’ss AMO caused a switch to the long isoform by blocking alternative splicing. Interestingly, the PAS AMO also caused the level of the short isoform to drop drastically, while the long isoform doubled (Figure 3B; PAS AMO). Taken together, these data suggest that loss of U1 suppression of a PAS determines alternative splicing to a mutually exclusive terminal exon.
PCPA and its suppression is a 5′-3′ directional process
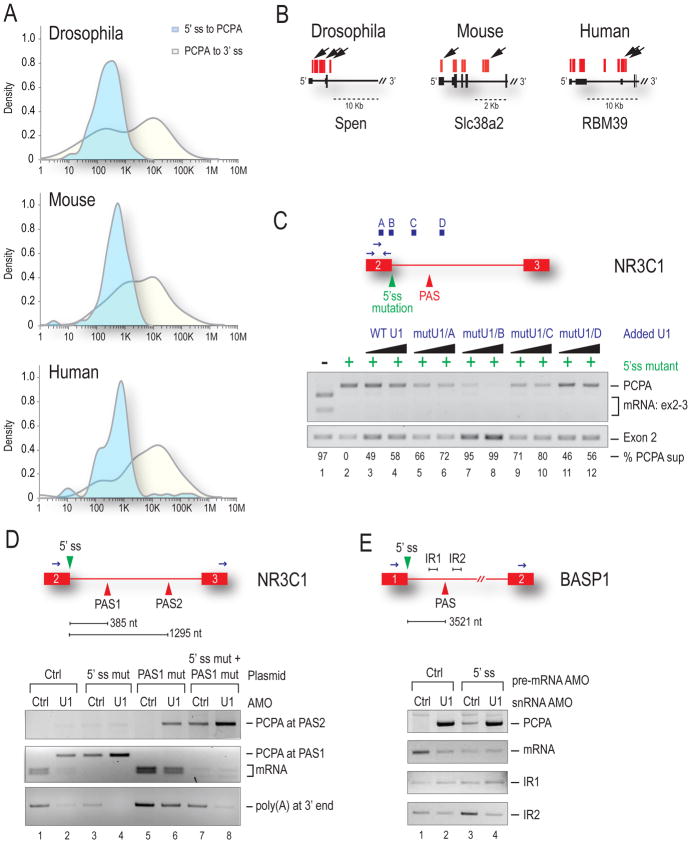
HIDE-seq provided information on PCPA position in introns, revealing that it occurred in all three organisms at a median distance of 500–1000 nt from the nearest 5’ss (Figure 4A, blue plot), but at variable and usually much greater distances from the nearest 3’ss (Figure 4A, yellow plot) (Supplemental Figures S2–S4, Table S2). In each organism we found cases of multiple PCPA sites within the same intron (e.g., Spen, RBM39) or same transcript (e.g., Spen, Slc38a2), demonstrating that there are several actionable PASs along a transcript (Figure 4B) and it is likely that U1 bound at the 5’ss and elsewhere in introns is required to suppress them.
Figure 4. U1 bound outside the 5’ss is required to suppress PASs >1kb away.
(A) Distribution of the log10 distance at which PCPA occurred was measured in each organism from the start of the poly(A) tail back to the upstream 5’ss (light blue) or downstream to the 3’ss (yellow). See also Supplemental Figures S2–S4 and Table S2. (B) Arrows indicate multiple PCPA sites per gene. (C) HeLa cells were transfected with WT NR3C1 mini-gene (lane 1) or one containing a 5’ss mutation (lanes 2–12) and increasing amounts of mutant U1 that base-pairs to one of four locations along the pre-mRNA (above in blue). PCPA in the intron was detected by 3′RACE with RT-PCR of exon 2 serving as the loading control. Percent suppression (PCPA to exon 2 ratio) was normalized to lane 2. (D) Control or U1 AMO were transfected along with WT NR3C1 mini-gene (lanes 1 & 2) or NR3C1 in which the PAS1 [385 nt] sequence in the intron was duplicated (PAS2) and placed 1295 nt downstream of the 5’ss. Mini-genes in which the 5’ss (lanes 3 & 4), PAS1 (lanes 5 & 6) or both (lanes 7 & 8) were mutated are indicated. 3′RACE was performed as in (C). (E) Control, U1 or an AMO to the 5’ss of the endogenous BASP1 gene were transfected as indicated. PCPA at the PAS 3.5 kb downstream of exon 1 was measured by 3′ RACE. RT-PCR on intronic sequences upstream (IR1) and downstream (IR2) of the PAS is shown.
To probe PCPA mechanism, we investigated if a splicing-defective U1 can suppress PCPA, using NR3C1 mini-gene constructs (Figure 4C) (Kaida et al., 2010). The WT mini-gene (lane 1) splices properly whereas a 5’ss mutation causes PCPA 385 nt into intron 2 (lane 2). PCPA was completely suppressed by a 5′ end mutated U1 complementary to the 5’ss (mutU1/B). Varying degrees of suppression were observed also for U1s tethered elsewhere, to the intron and upstream exon both in the vicinity of the cryptic PAS (~45–80%), and by increasing WT U1. This suggests that U1 can suppress PCPA even without being able to function in splicing and that U1 bound upstream does so more efficiently (e.g. mutU1/A and mutU1/C). We next constructed a NR3C1 mini-gene in which the actionable PAS in intron 2 was duplicated (Figure 4D). Transfection of this construct with U1 AMO (lane 2) caused PCPA at the first PAS (PAS1; 385 nt) and as previously reported, a 5’ss mutation caused PCPA (lane 3), but more occurred from this PAS in the presence of U1 AMO (lane 4) (Kaida et al., 2010). When PAS1 was mutated, PCPA now occurred from the second PAS (PAS2) located 1295 nt from the 5’ss, indicating that a downstream PAS(s) is also vulnerable and that PCPA occurs with 5′ to 3′ directionality. When both the 5’ss and PAS1 were mutated, some PCPA occurred from PAS2 (lane 7), indicating that protection from the 5’ss extends out to PAS2, but again U1 AMO elicited more PCPA (lane 8) suggesting that additional PCPA suppression is provided by U1 bound to sequences other than the 5’ss.
To address how far beyond a 5’ss U1 snRNP’s PCPA suppression extends, we studied the endogenous BASP1 gene which is PCPAed relatively far from the 5’ss, ~3.5 kb into the first intron (Figure 4E). Interestingly, little or no PCPA occurred with a 5’ss-blocking AMO (lane 3), while co-transfection with the 5’ss AMO and U1 AMO (lane 4) resulted in a substantial amount, suggesting that suppression by U1 from the 5’ss alone is insufficient. Additional controls confirmed that the 5’ss AMO was effective in inhibiting splicing (lanes 2–4: mRNA decreases and IR1 increases) and a cryptic 5’ss was not activated (Figure 4E). We conclude that at a distance of 3.5 kb, a PAS is outside the protective range of 5’ss-bound U1. Taken together, the HIDE-seq data and direct probing demonstrate the 5′ to 3′ directionality of the PCPA process and strongly suggest a need for U1 in excess of what is required for splicing.
U1 decrease recapitulates isoform shortening in activated neurons
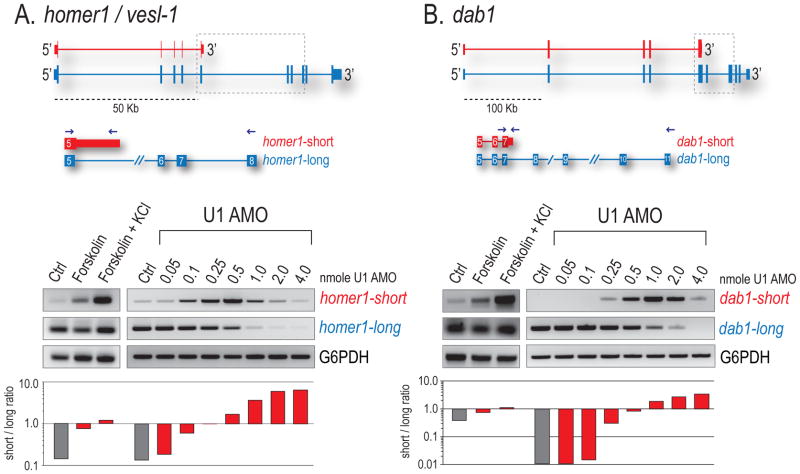
Examples consistent with the transcript shortening we describe have been characterized in activated neurons, particularly the homer-1/vesl-1 gene, which plays a critical role in synaptogenesis. Neuronal activation results in a rapid shift (< 6 hr) in the processing of the pre-mRNA encoding homer-1, from a full-length (L) to a shorter mRNA (S). Homer-S is produced by an extension of exon 5 and APA in the downstream intron to delete the C-terminal encoding exons (Niibori et al., 2007). Using rat PC12 cells, we show by RT-PCR that a range of low U1 AMO (0.25–0.5 nmole) dramatically increased the S form and caused a reciprocal dose-dependent decrease in homer-L, mirroring the switch seen upon neuronal activation with forskolin and KCl (Figure 5A). This shift in ratio of S/L (histogram) was evident even at 0.1 nmol, corresponding to an estimated 10% decrease in U1. Further U1 decrease (1.0–4.0 nmol) caused both forms to disappear, likely as a result of PCPA shifting closer to the TSS. In addition to homer-1, activity-dependent shortening of many other proteins critical for synaptogenesis has been described, including Dab1 (Flavell et al., 2008). Indeed, in mouse MN-1 cells, Dab1 also displayed a similar isoform shift with U1 decrease (Figure 5B). Our results suggest that U1 levels can regulate the size of mRNA isoforms produced from many genes.
Figure 5. Moderate U1 reduction recapitulates isoform switching upon neuronal activation.
Rat PC12 (A) and mouse MN-1 (B) neurons were stimulated with forskolin or forskolin/KCl for 3 hrs or separately transfected with control AMO or increasing amounts of U1 AMO. RT-PCR detecting short and long forms of Homer-1/Vesl-1 and Dab-1 are shown with histograms depicting the ratio of S/L. Grey dotted boxes indicate the region of transcript probed, blue arrows show primer locations, and G6PDH is a loading control.
Transcriptional up-regulation in activated neurons creates U1 shortage
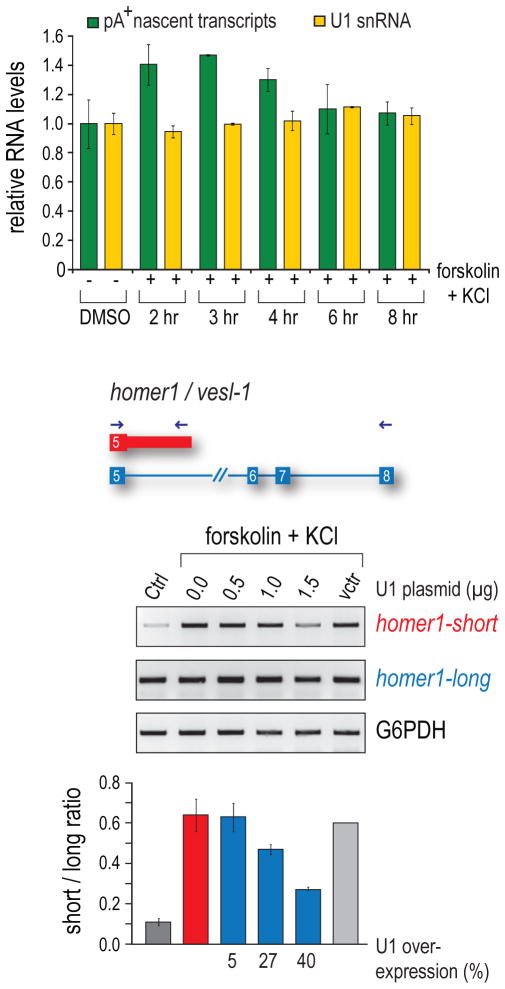
Two possible scenarios could potentially explain the shift to usage of more proximal PASs in activated cells if this was indeed due to loss of U1 PCPA suppression. Either U1 levels decreased or the amount of nascent transcripts that it needs to protect increased. To explore these possibilities under physiological conditions, we determined the amount of U1 and pre-mRNAs during neuronal activation (Figure 6A). Comparing control to activated PC12 cells, U1 (yellow) was quantified by RT-qPCR, and nascent transcripts (green) were pulse labeled with 5′6′-3H uridine for 30 min before RNA isolation and poly(A) RNA selection, followed by scintillation counting to determine the ratio of nascent mRNA to total RNA. This revealed a robust increase in transcriptional output of about 40–50% at 2–4 hrs post activation, whereas U1 levels did not change, creating a significant U1 shortage relative to its targets. This gap returned to baseline at 8 hrs, providing a built-in window of opportunity for rapid global gene expression regulation in response to external stimuli. Notably, Homer-S amounts coincided with the U1 shortage window, rising sharply after activation, peaking at 2–4 hours (~10 fold increase) and returning to near baseline levels at 6–8 hours. In contrast, the predominant form, Homer–L, remains mostly unchanged upon activation (data not shown).
Figure 6. U1 ratio to pre-mRNA determines immediate-early polyadenylation switch.
(A) PC12 were activated with forskolin/KCl or mock treated (DMSO) for the indicated times and labeled with 3H-uridine for 30 min prior to collection. Ratios of nascent transcripts (green) to total RNA were determined by scintillation and normalized to amounts of unlabeled total RNA. DMSO controls represent the average of 6 time points per experiment and activated values represent the average of two independent experiments (3 hr time point measured once). U1 snRNA (yellow) was measured by RT-qPCR at the same time points. (B) PC12 cells were transfected overnight (24 hrs) to over-express U1 (or empty vector) and activated with forskolin/KCl. Homer-1 isoforms were probed by RT-PCR 3 hrs after activation, as in Figure 5A. Histograms depict the ratio of S/L forms from biological repeat experiments with percent U1 over-expression reported below.
U1 over-expression inhibits mRNA shortening in activated neurons
While the U1 shortage creates an opportunity for proximal PASs to be used, the switch to shorter isoforms may not necessarily result specifically from loss of U1’s function. To address this, we asked if U1 over-expression could counteract the switch from Homer-L to Homer-S in activated neurons. PC12 cells were transfected with increasing amounts of a U1 snRNA expression vector or an empty vector 24 hr prior to activation. Exogenously expressed U1 quantified by RT-qPCR showed that over-expression (~40% of endogenous U1) prevented the homer-1 switch to the Homer-S isoform in a dose-dependent manner (Figure 6B). This suggests a role for U1 PCPA suppression in the neuronal activation pathway and demonstrates that altering U1 levels regulates gene expression.
Discussion
Partial genome tiling arrays previously showed that U1 protection is necessary to prevent drastic premature termination of the majority of nascent pol II transcripts by PCPA from cryptic PASs scattered throughout introns (Kaida et al., 2010). We refer to this activity as telescripting, as it is necessary for nascent transcripts to extend over large distances. Here, a rapid and versatile high throughput strategy for identifying transcriptome changes, HIDE-seq, and experiments based on the wealth of information it provided, yielded a much greater definition of U1 telescripting, revealing it is not only an evolutionarily conserved measure for ensuring transcriptome integrity, but also a robust mechanism for gene expression regulation. Surprisingly, PCPA position in a given gene varied depending on the amount of available U1. While U1 depletion inhibited splicing and caused PCPA, typically in the first intron, moderate U1 decreases (10–50%) did not inhibit splicing, but caused PCPA farther downstream, trending towards greater distances from the TSS with lesser U1 decrease. Significantly fewer, but still numerous genes were affected by moderate U1 decrease compared to U1 depletion and it was non-destructive, producing shorter mRNAs due to usage of alternative, more promoter proximal PASs rather than the normal PAS at the 3′ end of the full-length gene. Thus, telescripting can be modulated by decreasing available U1 over a large range without splicing being compromised, consistent with the idea that there is an excess of U1 over what is required for splicing. We suggest that telescripting provides a novel global gene expression regulation mechanism.
Our data demonstrate that the predominant transcriptome changes resulting from incomplete telescripting are various forms of mRNA shortening, including 3′UTR shortening and PCPA in introns. Thus, telescripting plays a major role in determining mRNA length and isoform expression. Indeed, several mRNAs of different lengths can be produced from the same gene by PCPA, corresponding to the degree of U1 decrease (e.g., Figure 3, GABPB1, UBAP2L). Importantly, PCPA can profoundly modulate protein expression levels and isoforms. While 3′ UTR shortening would not change the sequence of the encoded protein, it removes elements, such as microRNA- and hnRNP protein-binding sites, that could be critical for the mRNA’s regulation, including its stability, localization, and translational efficiency (Filipowicz et al., 2008; Huntzinger and Izaurralde, 2011). In contrast, PCPA in introns would produce an mRNA that encodes a protein lacking the C-terminus or containing a new C-terminal peptide, if the ORF extends into the terminal intron. An additional scenario associated with intronic PCPA is 3′ exon switching, also referred to as splicing-dependent APA, reflecting the general view that mechanistically, alternative splicing determines the PAS that is utilized. However, we showed that AMO masking of the alternative terminal exon’s PAS prevented its splicing, indicating that splicing into the alternative terminal exon depended on PCPA from this PAS (Figure 3B). Thus, PCPA can be the primary event in 3′ exon switching, revealing an unexpected splicing-independent role for U1 in alternative splicing regulation.
Several lines of evidence strongly suggest that U1 telescripting is a physiological phenomenon. First, over the entire range of U1 AMOs we used, numerous PCPA sites coincided precisely with previously detected polyadenylated transcripts, indicating that these cryptic sites are utilized naturally. These include ESTs representing short mRNA isoforms from a wide range of specimen (Supplemental Figures S2 and S5), of which some have been noted as APA from proximal PASs (Lou et al., 1996; Pan et al., 2006; Tian et al., 2007). Second, many of the mRNA shortening events resulting from loss of PCPA suppression are indistinguishable from the widespread mRNA shortening observed in activated T lymphocytes and neurons, proliferating cells and cancer cells (Flavell et al., 2008; Mayr and Bartel, 2009; Sandberg et al., 2008; Zhang et al., 2005). Notably, ~33% of the genes that undergo 3′ UTR shortening in activated T cells (Sandberg et al., 2008) are similarly affected by low U1 AMO in HeLa cells (data not shown), as are RAB10 and CCND1 (Figure 2C and Supplemental Figure S6), seen in cancer cells (Mayr and Bartel, 2009). Third, moderate U1 decrease recapitulated precisely hallmark switches to shorter isoforms in a neuronal activation model. Using native inducers (KCl and forskolin), we have shown that U1 decrease alone causes the same isoform switching in both homer-1 and Dab-1 in a dose-dependent manner. Representing the best-characterized example, the homer-1 gene switches to an isoform lacking the C-terminal domain-encoding exons, which antagonizes the full-length protein’s critical activity in synapse strengthening and long-term potentiation (Sala et al., 2003).
Having relied in our studies on deliberate U1 down-regulation, we considered whether there are physiological circumstances under which U1 levels could become deficient. Given U1’s abundance and very long half-life (Sauterer et al., 1988), it seemed unlikely that its levels would significantly decrease in absolute terms in the short timeframe in which mRNA shortening is observed. We considered an alternative scenario whereby U1 shortage relative to the targets it needs to protect in nascent transcripts could arise simply by an increase in transcriptional output of pre-mRNAs. Supporting this scenario, our measurements showed a rapid and transient increase in nascent transcripts of ~40–50% upon neuronal activation, while U1 levels showed little if any change (Figure 6A). This creates a significant U1 shortage relative to nascent pre-mRNAs, the magnitude of which is in the same range of the AMO experiments. This transcription-driven gap, detectable at 2–4 hr and returning to baseline at 6–8 hr after activation, creates a window of opportunity for APA to occur from proximal PASs due to the transient decrease in telescripting capacity. Importantly, the switch to shorter isoforms during neuronal activation could be antagonized by U1 over-expression in a dose-dependent manner. These data provide further evidence that U1 PCPA suppression is a built in PAS selection mechanism, and thus plays a major role in regulating gene expression during neuronal activation and potentially in response to other physiological stimuli.
The mechanism and factor(s) involved in the mRNA shortening in diverse activation conditions have not been identified (Flavell et al., 2008; Ji and Tian, 2009; Mayr and Bartel, 2009; Sandberg et al., 2008). However, the remarkable similarities they have with U1 shortage suggest that they also involve loss of U1 telescripting, at least during the initial (immediate/early) phase. Other factors have been described that could also cause a shift to proximal alternative PASs, particularly up-regulation of general polyadenylation and 3′ end processing factors, such as Cstf64 (Chuvpilo et al., 1999; Takagaki et al., 1996). However, this requires new protein synthesis and takes many (>18) hours (Shell et al., 2005), and therefore cannot explain the rapid switch to proximal PAS, in contrast to U1 shortage, which is immediate upon stimulation and occurs even in the presence of protein synthesis inhibition (data not shown) (Loebrich and Nedivi, 2009). The potential role of U1 and other factors at later times after cell activation and in other cells remains to be determined. Other means of creating U1 shortage, without transcription increase, can be envisioned, such as its sequestration in nuclear structures or by expression of other RNAs to which it could bind.
We propose a model for U1 telescripting that could explain its role in mRNA length regulation and isoform switching (Figure 7). We suggest that PCPA occurs co-transcriptionally by the same machinery that carries out normal 3′ end cleavage and polyadenylation in the terminal exon of the full-length gene, and is a byproduct of the coupling between transcription and this downstream process (Calvo and Manley, 2003; Dantonel et al., 1997; McCracken et al., 1997). CPA factors associate with the pol II TEC (Glover-Cutter et al., 2008; Hirose and Manley, 1998) close to the TSS and are therefore poised to process newly transcribed PASs with favorable sequence and structural features (actionable PASs) that it encounters throughout most of the length of the gene. This is normally prevented by U1 snRNP that binds to the nascent transcript. Previous studies have shown that U1 can inhibit polyadenylation of the normal PAS when tethered in its proximity in the terminal exon and in vitro (Ashe et al., 2000; Fortes et al., 2003; Gunderson et al., 1998; Vagner et al., 2000). U1 is recruited to nascent pol II transcripts, including intronless transcripts, by multiple interactions with the pre-mRNA and RNA processing factors as well as the transcriptional machinery (Brody et al., 2011; Das et al., 2007; Lewis et al., 1996; Lutz et al., 1996). In all three organisms we studied, PCPA was typically not detected in the first few hundred nucleotides, rising sharply thereafter and peaking ~1 kb from the TSS (Figure 2D). It is possible that actionable PASs upstream of this point are not utilized because CPA factors may have not yet associated with the TEC (Mayer et al., 2010; Mueller et al., 2004), which depends on pol II’s carboxyl terminal domain phosphorylation state (Buratowski, 2009). We suggest that this lag could serve (and may have evolved) to allow U1 binding before transcripts are exposed to CPA, to prevent their early destruction. Consistent with this distance from TSS consideration, PASs in the 5′UTR that are not PCPAed are functional when placed at the 3′ end of the gene (Guo et al., 2011).
Figure 7. A proposed model of co-transcriptional PCPA and its suppression by U1.
Cleavage and polyadenylation factors (CPSF) associate with pol II TEC and is thus poised for 3′-end cleavage and polyadenylation, but can also cause premature termination (PCPA) from cryptic PASs found throughout pre-mRNAs. U1 is recruited to nascent transcripts by multiple interactions, including base pairing of its 5′-end with cognate pre-mRNA sequences (5’ ss and other) and can suppress PCPA by inhibiting the pol II associated CPSF over < 1kb range. U1 shortage increases the probability of distal PASs remaining unprotected.
Our data also indicated that 5’ss bound U1 has a limited protective range of up to ~500–1000 nt and therefore would be insufficient to ensure telescripting through larger introns. Complete PCPA suppression could not be provided by U1 from the 5’ss alone even within this perimeter and plays almost no role in protecting more distal PASs, depending instead on additional U1 bound in introns (Figures 4D and 4E). Indeed, introns contain numerous U1 binding sites that do not function as 5’ss which we suggest serve to anchor U1 to protect introns from PCPA. Furthermore, we found that U1 with a mutated 5′ sequence that cannot function in splicing can still function in telescripting (Figure 4C), supporting the notion of two separate U1 roles. U1’s capacity to interact with nascent transcripts directly, even without base pairing (Patel et al., 2007; Spiluttini et al., 2010), enhances its association throughout the pre-mRNA, allowing it to scan the transcript and accelerate the rate with which it can find more stable base pairing sites, including the 5’ss. Importantly, PAS mutations cause PCPA from downstream intronic PASs (Figure 4D), suggesting a directional process, consistent with a co-transcriptional mechanism. We propose that U1 shortage causes transcript shortening because as co-transcriptional recruitment of U1 to nascent transcripts becomes limiting, it leaves distal PASs less protected, providing a built-in and U1 dose-dependent mechanism for mRNA length regulation and isoform switching.
Experimental Procedures
Cell treatments
HeLa, NIH/3T3, and S2 cells were transfected by electroporation using a Genepulser (Bio-Rad) or Nucleofector (Amaxa) with control or antisense morpholino (AMO) to U1 at 0.25, 1.0 and 15 nmole for 8 hrs as described (Kaida et al., 2010). HeLa transfection with gene-specific AMOs (15 nmole/7.5 μM) to PASs, 5′ and 3’ss was performed in the same manner. All AMO sequences synthesized by Gene Tools are listed in the Extended Experimental Procedures. SSA treatment was 100 ng/ml for 8 hrs. PC12 and MN-1 cells were stimulated with 20 μM forskolin and/or 50 mM KCl for 3 hrs (Impey et al., 1998) or transfected with U1 AMO for 8 hrs by Amaxa Nucleofector.
HIDE-seq library preparation
Subtracted cDNA libraries were prepared using Clontech’s PCR-Select cDNA Subtraction kit (Diatchenko et al., 1996; Gurskaya et al., 1996; Lukyanov et al., 1995) with several modifications listed below. Poly(A) RNA was reverse transcribed with random hexamers and custom oligo(dT) primers containing blunt end restriction sites to prepare individual cDNA pools. Following second strand synthesis, ds cDNAs were each digested separately with RsaI, HaeIII, or AluI, to produce independent libraries of the same samples. Each sample (control AMO or U1 AMO) served as the tester or the reference (i.e. forward and reverse subtractions). Ligation of adaptors, hybridization, and primary PCR (Figure 1A, left gel) steps were as described and detailed in the Extended Experimental Procedures. Nested PCR (Figure 1A, right gel) was performed on each library with primers containing 454 adaptors, barcodes, and linker molecules. Forward and reverse libraries were multiplexed, subjected to emulsion PCR, and bi-directionally sequenced using Titanium 454 chemistry.
Genomic tiling arrays, RT-PCR, Real time and 3’ RACE
Arrays were performed in triplicate for human chromosomes 5, 7 and 16 (Kaida et al., 2010). All primers, RNA preparation methods, conditions, and cell types are listed in the Extended Experimental Procedures. mRNA isoform and U1 snRNA levels determined by qPCR were normalized to G6PDH and 5s rRNA, respectively.
Mini-gene, mutant U1 transfections and U1 over-expression
The NR3C1 mini-gene and the 5’ss and PAS mutations contained therein have been described (Kaida, et al, 2010). To duplicate the PAS, the NR3C1 plasmid EcoRV-StuI fragment was reinserted into the original mini-gene at the EcoRV site. Modification of U1’s 5’ss binding domain and plasmid concentrations are listed in the Extended Experimental Procedures. 3′RACE products were digested with HindIII to distinguish PCPA and mRNA bands. PC12 were transfected with U1 snRNA in a pSiren-RetroQ expression vector (Clontech) driven by the native U1 promoter. RT-qPCR using probes specific for a 1 bp difference in transfected U1 was used to determine the extent of over-expression.
Tritiated uridine labeling of mRNA
PC12 cells were pulsed with 50 μCi of 5′6′-3H uridine in 500 μl media with 0.04 μg/ml actinomycin D for 30 min before collecting. Total RNA was isolated by Trizol and poly(A) RNA was selected on Oligotex beads (Qiagen). Radioactivity in each fraction was determined by scintillation counts and ratios of mRNA to total RNA were normalized to the concentration of total RNA determined on a Nanodrop spectrophotometer.
HIDE-seq Data analysis
A description of read alignments, the development of an algorithm to describe patterns found by HIDE-seq, the identification of poly(A) reads, and the calculation of density plots are in the Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory for helpful discussions, especially Drs. Pilong Li, Jeongsik Yong, Kazuhiro Fukumura, and Lauren Brady. We thank the University of Pennsylvania Genomics core facility for 454 sequencing and Dr. Bin Tian for help with PAS analysis. This work was supported by the Association Française Contre les Myopathies (AFM). GD is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashe MP, Furger A, Proudfoot NJ. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA. 2000;6:170–177. doi: 10.1017/s1355838200991957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10:261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci U S A. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- Guo J, Garrett M, Micklem G, Brogna S. Poly(A) signals located near the 5′ end of genes are silenced by a general mechanism that prevents premature 3′-end processing. Mol Cell Biol. 2011;31:639–651. doi: 10.1128/MCB.00919-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Diatchenko L, Chenchik A, Siebert PD, Khaspekov GL, Lukyanov KA, Vagner LL, Ermolaeva OD, Lukyanov SA, Sverdlov ED. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- Hartmann B, Valcarcel J. Decrypting the genome’s alternative messages. Curr Opin Cell Biol. 2009;21:377–386. doi: 10.1016/j.ceb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, Deloulme JC, Chan G, Storm DR. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- Loebrich S, Nedivi E. The function of activity-regulated genes in the nervous system. Physiol Rev. 2009;89:1079–1103. doi: 10.1152/physrev.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Gagel RF, Berget SM. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- Lukyanov KA, Launer GA, Tarabykin VS, Zaraisky AG, Lukyanov SA. Inverted terminal repeats permit the average length of amplified DNA fragments to be regulated during preparation of cDNA libraries by polymerase chain reaction. Anal Biochem. 1995;229:198–202. doi: 10.1006/abio.1995.1402. [DOI] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O’Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Niibori Y, Hayashi F, Hirai K, Matsui M, Inokuchi K. Alternative poly(A) site-selection regulates the production of alternatively spliced vesl-1/homer1 isoforms that encode postsynaptic scaffolding proteins. Neurosci Res. 2007;57:399–410. doi: 10.1016/j.neures.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Pan Z, Zhang H, Hague LK, Lee JY, Lutz CS, Tian B. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. Gene. 2006;366:325–334. doi: 10.1016/j.gene.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Patel SB, Novikova N, Bellini M. Splicing-independent recruitment of spliceosomal small nuclear RNPs to nascent RNA polymerase II transcripts. J Cell Biol. 2007;178:937–949. doi: 10.1083/jcb.200706134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci. 2003;23:6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SA, Hesse C, Morris SM, Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci. 2010;123:2085–2093. doi: 10.1242/jcs.061358. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Ruegsegger U, Gunderson SI, Keller W, Mattaj IW. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA. 2000;6:178–188. doi: 10.1017/s1355838200991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.