Abstract
Background
Leprosy was the first disease classified according to the thymus derived T-cell in the 1960s and the first disease classified by the cytokine profile as intact interferon-γ (IFNγ) and interleukin-2 (IL2) or TH1 (tuberculoid) and deficient IFNγ and IL2 or TH2 (lepromatous), in the 1980s.
Objective
In the present study, we set out to explore the T helper 17 (TH17) lymphocyte subset, the hallmark of T-cell plasticity, in skin biopsies from patients with erythema nodosum leprosum (ENL) treated with thalidomide.
Method
RNA was extracted from paraffin embedded tissue before and after thalidomide treatment of ENL and RT-PCR was performed.
Results
IL17A, the hallmark of TH17, was consistently seen before and after thalidomide treatment, confirming the TH17 subset to be involved in ENL and potentially up-regulated by thalidomide.
Conclusion
A reduction in CD70, GARP, IDO, IL17B (IL-20) and IL17E(IL-25) coupled with increases in RORγT, ARNT, FoxP3 and IL17C(IL-21) following thalidomide treatment opens the door for the complex understanding of how the immunomodulatory drug-thalidomide can be anti-inflammatory and simultaneously stimulate cell mediated immunity (CMI). We conclude that TH17 is involved in the immunopathogenesis of ENL and that thalidomide suppresses inflammatory components of TH17, while enhancing other components of TH17 potentially involved in CMI.
Introduction
Burnet won the Nobel Prize for the clonal selection theory of immunity in 1960.1 In 1961, Miller published evidence that these lymphocyte “clones” were educated in the thymus.2 In 1962, the British classified leprosy according to the thymus with intact cell mediated immunity labeled as “tuberculoid” and the anergic form as “lepromatous.”3,4 In the 1980’s, the concept of tuberculoid with intact IFNγ and IL-25 and lepromatous with deficient IFNγ, IL-2 and increased IL-4 and IL-106 led to the concept of the TH1–TH2 paradigm based on cytokine profiles.7 This binary concept of the immune system was dominant for 20 years.8 In the year 2000, Oppmann and coworkers discovered through bio-informatics the third submit of IL-12, p19 and designated the p40p19 cytokine as IL-23.9 In 2005, Steinman, discovered the TH17 lymphocyte subset in the autoimmune disorder EAE and provided a major revision of the TH1–TH2 paradigm.10 This discovery has led to multiple review articles and further use of the term T-cell plasticity where TH17 is in ying-yang with suppressor T-cell (Tregs) including hybrid T-cells with both RORγT and FoxP3.11 Surprisingly, these findings may explain the recent reports that have shown Tregs, as measured by CD4CD25-FoxP3 functions, occur throughout the leprosy spectrum, rather than being dominant at the TH2 or anergic lepromatous form of the disease.12,13 Despite the fact that leprosy is the first disease classified according to T-cells, TH17 has not yet been thoroughly studied in the leprosy spectrum. As PMNs are a hallmark of the TH17 subset and type II (ENL) leprosy reactions, we theorized this T-cell subset would be involved in type II (ENL) leprosy reactions.
In 1964, Sheskin prescribed thalidomide as a sedative for a patient with ENL.14 It resulted in spontaneous resolution of the cutaneous lesions, fever and pain related to ENL.14 Haslett et al.15 elucidated the mechanism of action of thalidomide in twenty ENL patients from Nepal who were treated with thalidomide for 21 days. All patients responded to treatment with the majority having complete resolution of cutaneous lesions within seven days.
Despite several studies conducted for over 25 years, thalidomide's mechanism of action in arresting ENL is still unknown despite it being FDA approved. The proven efficacy of thalidomide in the treatment of ENL presents an opportunity to correlate the clinical and immunologic effects of this drug. The dynamic patterns of immunologic changes observed with the effects of thalidomide suggest some new ideas about the possible mechanism of action of the drug against ENL. Results suggest a complex effect of thalidomide that is anti-inflammatory, but also involves a transient boosting of T-cell function. It is hypothesized that thalidomide may actively promote T-cell regulation. In this report, we analyzed skin tissue from seven ENL Nepal patients pre- and post thalidomide treatment and found different relative gene expression patterns for twenty genes in the Th17 pathway.
Materials and Methods
Patient cohort
Our cohort includes seven ENL patients from Nepal treated with thalidomide for 21 days as FFPE skin samples pre- and post treatment with thalidomide (Table 1).
Table 1.
Table 1 of six Nepal ENL patients treated with thalidomide
| ENL HISTOL# |
ENL ID# |
Age (Yr) |
Sex | Duration of Leprosy Treatment (months) |
Ridley-Jopling Classification at Leprosy Diagnosis* |
Bacillary Index at Leprosy Diagnosis¶ |
Duration of Present ENL episode (days) |
Previous Episodes of type II reactions ENL (number) |
Previous Episodes of type I lepra reaction (number) |
|---|---|---|---|---|---|---|---|---|---|
| 482 | 1 | 17 | M | 17 | LL | 4.75 | 24 | 1 | 0 |
| 483 | 2 | 45 | M | 7 | LL | 4.5 | 2 | 2 | 0 |
| 488 | 3 | 32 | M | 5 | BL | 1.25 | 240 | 1 | 0 |
| 478 | 4 | 24 | M | 17 | LL | 4.75 | 8 | 0 | 0 |
| 517 | 5 | 40 | M | 20 | LL | 3.25 | 30 | 1 | 1 |
| 514 | 6 | 37 | M | 10 | LL | 4.25 | 18 | 0 | 0 |
Extraction of RNA and RT-PCR
Three to four 1.0 µ sections of formalin-fixed paraffin-embedded ACD punch biopsy tissues were scraped from glass slides, paraffin removed by addition of 1 ml xylene for 5 min. and tissue precipitated by microcentrifugation for 5 min. The tissue was washed twice with 1 ml 100% ethanol and dried. RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions and re-suspended in 50 µl of DEPC-treated water. RNA was reverse transcribed into ss-cDNA using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions, stored at −80°C and used within two weeks. Primers are listed in Table 2. RNA was equalized to hGAPDH using 3 µl ss-cDNA with 0.5 µl Taq polymerase (Promega, Madison, WI); dissociation at 94°C for 1.0 minute, annealing at 58°C for 1.0 minute and extension at 72°C for 1 minute for 45 cycles with a final 10 minute extension. Nested PCR used 3 µl of the first PCR reaction with annealing at 60°C and a second nested PCR used 3 µl of the first nested PCR reaction with annealing at 62°C. Nested and nested-nested PCR was utilized to detect the expression of the following human genes and primers were derived from published sequences for: hRORγT (16); hCD70 ; hCD27; hIL-17A (IL-17); hIL-17B (IL-20); hIL-17C (IL-21); hIL-17D (IL-27); hIL-17E (IL-25); hIL-17F (IL-24); hPLZF-1; hCTLA4, hAHR, hIL22, hiNOS2, hARNT, hIDO, hGARP, hIL9, hCD46 and hFoxP3 (Table 2). The PCR conditions were: 10 µl ss-cDNA with 0.5 µl Taq polymerase; dissociation at 94°C for 1.0 minute, annealing at 56°C outer for 1.0 minute and extension at 72°C for 1 minute for 45 cycles with a final 10 minute extension. Nested PCR used 3 µl of the first PCR reaction with annealing at 58°C and a second nested PCR used 3 µl of the first nested PCR reaction with annealing at 60°C. Negative reactions (no template DNA) are included in all PCR reactions. The PCR amplicons for hGAPDH (20 µl) were electrophoresed in 2–3% agarose containing ethidium bromide (0.4 µg/ml) and visualized under ultra violet light. The PCR amplicons for remaining 20 genes were electrophoresed on agarose gel or 8–12% PAGE-0.5x TBE followed by staining with ethidium bromide. The gels were photographed with a FB-PDC-34 photo-documentation camera (Fisher Scientific-Springfield, NJ).
Table 2.
Primer sequence and location
| Gene and GenBank accession # |
Location in cDNA | Primer sequence (actual sequence) |
|---|---|---|
|
hGAPDH # M17851 |
outer primers: bp 618-647 bp 997-968 inner primers: bp 681-710 bp 1000-971 inner-inner primers: bp 617-646 bp 936-907 |
5’-CATCCCTGCCTCTACTGGCGCTGCCAAGGC-3’ 5’-AGGCCATGAGGGCCATGAGGTCCACCACCC-3’ 5’-TGTGGGCAAGGTCATCCCTGAGCTG AACGG-3’ 5’-TGTTGCTGTAGCCAAATTCGTTGTCATACC-3’ 5’-GAAGCTCACTGGCATGGCCTTCCGTGTCCC-3’ 5’-AGGAAATGAGCTTGACAAAGTGGTCGTTGA-3’ |
| hRORγT #U16997 |
outer primers: bp 1231-1260 bp 1590-1561 inner primers: bp 1262-1291 bp 1560-1531 inner-inner primers: bp 1293-1322 bp 1526-1495 |
5'-TTCGAGCCTTGGGCTGCAGCGAG CTCATC-3' 5'-GCTGAAGAGCTCCTTGTAGAGTGGAGGGAA-3' 5'-GCTCCATCTTTGACTTCTCCCACTCCCTAA-3' 5'-AGCGGCTTGGACCACGATGGGGTGGAGGTC-3' 5'-TGCCTTGCATTTTCCGAGGA TGAGATTGC-3' 5'-AAGATCTGCAGCCTTTCCACATGC TGGCTACA-3' |
|
hCD70 # BC000725) |
outer primers: bp 301-330 bp 660-631 inner primers: bp 331-360 bp 630-601 inner-inner primers: bp 361-390 bp 600-571 |
5’-CAGGACCCCAGGCTATACTGGCAGGGGGGC-3’ 5’-CACTCCAAAGAAGGTCTCATCAGTGTTTCG-3’ 5’-CCAGCACTGGGCCGCTCCTTCCTGCATGGA-3’ 5’-GGAAGGCAAAAGTGTCCCAGTGAGGTTGGT-3’ 5’-CCAGAGCTGGACAAGGGGCAGCTACGTATC-3’ 5’-GCAGAGTGTGTCCCCTCGGGCCAGGGGCGT-3’ |
|
HCD27 # NM_001242 |
outer primers: bp 541-570 bp 990-961 inner primers: bp 571-600 bp 960-931 inner-inner primers: bp 601-630 bp 930-901 |
5’-GCAGTGCAGGGACAAGGAGTGCACCGAGTG-3’ 5’-GGGGAGCAGGCAGGCTCCGGTTTTCGGTAA-3’ 5’-TGATCCTCTTCCAAACCCTTCGCTGACCGC-3’ 5’-CTTCCCTGGATGGGGATGGAGCTGCCCTCC-3’ 5’-TCGGTCGTCTCAGGCCCTGAGCCCACACCC-3’ 5’-TCCTCCCTGGGGCAGCTGTAACGACAAGGC-3’ |
|
hIL-17A # NM_002190 |
outer primers: bp 361-390 bp 780-750 inner primers: bp 391-420 bp 750-721 inner-inner primers bp 421-450 bp 721-691 |
5’-GTGGACTACCACATGAACTCTGTCCCCATC-3’ 5’-CATAAATAACTTAAAGCCCTTAAAAAAGTA-3’ 5’-CAGCAAGAGATCCTGGTCCTGCGCAGGGAG-3’ 5’-AACTTGAAGCAAATTGGTACTCAGTCAAAC-3’ 5’-CCTCCACACTGCCCCAACTCCTTCCGGCTG-3’ 5’-CTTCCTTCTTGGAAAGAGGGAAAGGTAATT-3’ |
|
hIL-17B # NM_014443 |
outer primers: bp 121-150 bp 590-561 inner primers: bp 391-420 bp 560-531 inner-inner primers: bp181-210 bp 530-501 |
5’-GCCCCAAAAGCAAGAGGAAGGGGCAAGGGC-3’ 5’-GAAGATGCAGGAGCAGCCCACAGCGATGGT-3’ 5’-GGCCTGGGCCCCTGGCCCCTGGCCCTCACC-3’ 5’-CTCCATGACTGCGCGCTGGCGGCAAGGCCC-3’ 5’-AGGTGCCACTGGACCTGGTGTCACGGATGA-3’ 5’-TGTGCGGGGCGGTGGCGGGCAGAGGCGGCG-3’ |
|
hIL-17C # NM_013278 |
outer primers: bp 241-270 bp 640-611 inner primers: bp271-300 bp 610-581 inner-inner primers: bp 301-330 bp 580-551 |
5’-AGCCCTGGTGTCCAGCCTGGAGGCAGCAAG-3’ 5’-CACTGAACGGGGCAGCACGCAGGTGCAGCC-3’ 5’-CCACAGGGGGAGGCACGAGAGGCCCTCAGC-3’ 5’-ACGGGGACGTGGATGAACTCGGTGTGGAA-3’ 5’-TACGACCCAGTGCCCGGTGCTGCGGCCGGA-3’ 5’-GGCAAAGGCCCCAGGTGTGGGGAGCCCCGA-3’ |
|
hIL-17D # NM_138284 |
outer primers: bp 241-270 bp 810-781 inner primers: bp 271-300 bp 780-751 inner-inner primers: bp 301-339 bp 750-721 |
5’-CGCCTGGCGGCCGGCGTGCTCAGTGCCTTC-3’ 5’-CCAGCGGGCGCGTCGTTGGGGCCCAGCAGG-3’ 5’-CACCACACGCTGCAGCTGGGGCCGCGTGAG-3’ 5’-AGCTTGGCGCCCTGTTTGTCGATGCTGGAG-3’ 5’-CAGGCGCGCAACGCGAGCTGCCCGGCAGGG-3’ 5’-TTGATGCTGTCTGCGTCCTTCTCCGGCTCG-3’ |
|
hIL-17E # NM_022789 |
outer primers: bp 421-450 bp 840-810 inner primers: bp 451-480 bp 810-781 inner-inner primers: bp 481-510 bp 780-751 |
5’-CAGCTGCTGCCCCAGCAAAGGGCAGGACAC-3’ 5’-CCCATCACACGGGGCCGCACACACACACAA-3’ 5’-CTCTGAGGAGCTGCTGAGGTGGAGCACTGT-3’ 5’-GCTAAGGAAACACGGTACAGCCTGCGCTCC-3’ 5’-GCCTGTGCCTCCCCTAGAGCCTGCTAGGCC-3’ 5’-AGGCAGTAGCCCTTGTGGGTGCCCTTCTCG-3’ |
|
hIL-17F # BC070124 |
outer primers: bp 81-110 bp 540-511 inner primers: bp 111-140 bp 510-481 inner-inner primers: bp 141-170 bp 480-451 |
5’-AAGACCCTGCATGGCCCAGCCATGGTCAAG-3’ 5’-GGGTGACGCAGGTGCAGCCAACAGTCACCA-3’ 5’-TACTTGCTGCTGTCGATATTGGGGCTTGCC-3’ 5’-GCACCTTCTCCAACTGGAAAGAAACAGAGC-3’ 5’-TTTCTGAGTGAGGCGGCAGCTCGGAAAATC-3’ 5’-AGCCTTGGTGCTTCCTCCGGACGACCAGGG-3’ |
|
HPLZF-1 # BC029812 |
outer primers: bp 1891-1920 bp 2220-2161 inner primers: bp 1921-1950 bp 2160-2131 inner-inner primers: bp 1951-1980 bp 2160-2131 |
5’-GAGTTCTGTGGCAGCTGCTTCCGGGATGAG-3’ 5’-CGGGATCTCCTCGGGCTTGTGGCCCTTCAT-3’ 5’-AGCACACTCAAGAGCCACAAACGCATCCAC-3’ 5’-GTGCTTCTGCATGGAGGAGAGGCTGGGGCA-3’ 5’-ACGGGTGAGAAACCCTACGAGTGCAATGGC-3’ 5’-GTACTCTGTGCAGATGGTGCA CTGGTAGGG-3’ |
|
HCTLA4 # AF414120 |
outer primers: bp 211-240 bp 480-461 inner primers: bp 245-275 bp 460-431 inner-inner primers: bp 281-306 bp 425-396 |
5’-CCTGGCCCTGCACTCTCCTGTTTTTTCTTC-3’ 5’-ATCCCTGTCTTCTGCAAAGCAATGCACGTG-3’ 5’-CATGTAGGTTGCCGCACAGACTTCAGTCAC-3’ 5’-ATGGAATCATCTAGGAAGG TCAACTCATTC-3’ 5’- GCCTGCTGTGGTACTGGCCAGCAGCC -3’ 5’-ACTTGATTTCCACTGGAGGTGCCCGTGCAG-3’ |
|
hFoxP3 # AF277993 |
outer primers: bp 961-990 bp 1440-1471 inner primers: bp 991-1020 bp 1410-1381 inner-inner primers: bp 1021-1050 bp 1380-1351 |
5’-CCCACCTGGCTGGGAAAATGGCACTGACCA-3’ 5’-AGGCTTCATCTGTGGCATCATCCGACAAGG-3’ 5’-GCTCCTGCTGCATCGTAGCTGCTGGCAGCC-3’ 5’-CCCGCACAAAGCACTTGTGCAGACTCAGGT-3’ 5’-CGGTCCACACAGCCCCCTTCTCGCTCTCCA-3’ 5’-TCCGTTTCTTGCGGAACTCCAGCTCATCCA-3’ |
|
hIL22 # NM_020525 |
outer primers: bp 121-150 bp 590-561 inner primers: bp 151-180 bp 560-531 inner-inner primers: bp 181-210 bp 530-501 |
5’-TCTTGGCCCTCTTGGTACAGGGAGGAGCAG-3’ 5’-AATGCAGGCATTTCTCAGAGACATAAACAG-3’ 5’- CTGCGCCCATCAGCTCCCACTGCAGGCTTG-3’ 5’-CAAATCCAGTTCTCCAATTGCTTTGATCTC-3’ 5’- ACAAGTCCAACTTCCAGCAGCCCTATATCA-3’ 5’-TCCACTCTCTCCAAGCTTTTTCACTGAGAC-3’ |
|
hiNOS2 # NM_000625.4 |
outer primers: bp 3121-3150 bp 3720-3691 inner primers: bp 3151-3180 bp 3690-3661 inner-inner primers: bp 3181-3210 bp 3660-3631 |
5’-CCCCAAGACCCAGTGCCCTGCTTTGTGCGG-3’ 5’-CGCTGACATCTCCAGGCTGCTGGGCTGCAC-3’ 5’-AATGCCAGCGGCTTCCACCTCCCCGAGGAT-3’ 5’-CGCCACCCTGTCCTTCTTCGCCTCGTAAGG-3’ 5’-CCCTCCCATCCTTGCATCCTCATCGGGCCT-3’ 5’-AAATTCTGCACCAAAGATATCTTCGTGATA-3’ |
|
hARNT # NM_001668 |
outer primers: bp 2101-2130 bp 2490-2461 inner primers: bp 2131-2160 bp 2460-2431 inner-innr primers: bp 2161-2190 bp 2430-2401 |
5’-CTACCCAGGCTACTGCTAAGACTCGTACTT-3’ 5’-TAGTTAGATCAGGGAATTCTTCATTGTTGT-3’ 5’-CCCAGTTTGGTGTGGGCAGCTTTCAGACTC-3’ 5’-AGCTGTTGCTCTGATCTCCCAGCATGGACA-3’ 5’-CATCCTCCTTCAGCTCCATGTCCCTCCCTG-3’ 5’-GCATCTCCTGGAAGACCTCAGGCTGGCCAG-3’ |
|
hIDO # NM_002164.4 |
outer primers: bp 871-900 bp 1320-1291 inner primers: bp 901-930 bp 1290-1261 inner-inner primers: bp 931-960 bp 1260-1231 |
5’-TGGGAAGACCCAAAGGAGTTTGCAGGGGGC-3’ 5’-TTCCTTCAAAAGGGATTTCTCAGTTGTACT-3’ 5’-AGTGCAGGCCAAAGCAGCGTCTTTCAGTGC-3’ 5’-TCTTACAGTCTTCAGGAAATTCATTAAATC-3’ 5’-TTTGACGTCCTGCTGGGCATCCAGCAGACT-3’ 5’-AGTGCCTCCAGTTCCTTTGGCTTCCAGTTT-3’ |
|
hGARP # Z24680.1 |
outer primers: bp 1591-1620 bp 2070-2041 inner primers: bp 1621-1650 bp 2040-2011 inner-inner primers: bp 1651-1680 bp 2010-1981 |
5’-GGGCAACGGGCTGATGGTCCTGCAGGTGGA-3’ 5’-TGTTGGTTAAACTTCTGCCGGCGGACGCAG-3’ 5’-CCTGCCCTGCTTCATCTGCCTCAAGCGGCT-3’ 5’-CAGCAGGCGGCCAGCGTGGTGCGGAGGATG-3’ 5’-CAATCTTGCCGAGAACCGCCTGAGCCACCT-3’ 5’-GCAGAGACCAGTATGAAGGTGAGGATGATG-3’ |
|
hIL9 # NM_000590 |
outer primers: bp 21-50 bp 440-411 inner primers: bp 51-80 bp 410-381 inner-inner primers: bp 81-110 bp 380-351 |
5’-GCCATGGTCCTTACCTCTGCCCTGCTCCTG-3’ 5’-CTTGCCTCTCATCCCTCTCATCTTTTCTTT-3’ 5’-TGCTCCGTGGCAGGCCAGGGGTGTCCAACC-3’ 5’-CTGGAAAATTTCCAGAAGACTCTTCAGAAA-3’ 5’-TTGGCGGGGATCCTGGACATCAACTTCCTC-3’ 5’-TGTCAGCGCGTTGCCTGCCGTGGTTTGGTT-3’ |
|
hCD46 # NM_002389.4 |
outer primers: bp 691-720 bp 1260-1231 inner primers: bp 721-750 bp 1230-1201 inner-inner primers: bp 751-780 bp 1200--1271 |
5’-ACACACCTTTAGTGAAGTAGAAGTATTTGA-3’ 5’-CAAATTACTGCAACTCCAACAACTATGGCA-3’ 5’-GTATCTTGATGCAGTAACTTATAGTTGTGA-3’ 5’-ATAACAATCACAGCAATGACCCAAACATCC-3’ 5’-TCCTGCACCTGGACCAGATCCATTTTCACT-3’ 5’-AAACTGTCAAGTATTCCTTCCTCAGGTTTA-3’ |
|
hAHR # BC070080.1 |
outer primers: bp 2611-2640 bp 3060-3031 inner primers: bp 2641-2670 bp 3030-3001 inner-inner primers: bp 2671-2700 bp 3000-2971 |
5’-ATGCCTTATACACAGAACTTTATTTCCTGT-3’ 5’-TGTCAAATCAGGAAAAGGTCTGGCTTCTGA-3’ 5’-AATCAGCCTGTATTACCACAACATTCCAAA-3’ 5’-CGGATGATGAAGTGGCTGAAGATGTGTGGT-3’ 5’-TGTACAGAGCTGGACTACCCTATGGGGAGT-3’ 5’-AGTCTGAGTGTTATTTATGTTATTTAATTC-3’ |
Results
Patient cohort
Our cohort was seven ENL patients from Nepal treated with thalidomide for 21 days. Shown in Table 1 is information on six of seven cases.
Molecular studies on the TH17 pathway
Total RNA from FFPE skin tissue from seven patients ENL pre- and post thalidomide treatment (Table 1) were extracted using xylene deparaffination. Total RNA was reverse transcribed by RT-PCR for hGAPDH and genes- hRORγT; hCD70 ; hCD27; hIL-17A ; hIL-17B (IL-20); hIL-17C (IL-21); hIL-17D (IL-27); hIL-17E (IL-25); hIL-17F (IL-24); hPLZF-1; hCTLA4, hAHR, hIL22, hiNOS2, hARNT, hIDO, hGARP, hIL9, hCD46 and hFoxP3 by nested or nested-nested RT-PCR to detect mRNA expression of human TH17. Initially, mRNA was equalized to hGAPDH (Figure 1). Before treatment with thalidomide, we found increased relative expression of hCD70 (3/7 cases), hIL-17B(IL-20) (4/7 cases), hIL-17E(IL-25) (5/7 cases), hIDO (5/7 cases) and hGARP (3/7 cases) while after treatment, they were not detected. Conversely, we found that after treatment there was an increased relative expression of hFoxP3 (4/7 cases), hIL-17C(IL-21) (2/7 cases), hRORγT (2/7 cases) and hARNT (3/7 cases) with no expression before treatment. For genes hCTLA4, hIL-17A(IL-17) and hiNOS2, we found both expression patterns before and after treatment. No expression of hAHR, hIL22, hCD27, hPLZF-1, hIL-17D(IL-27), hIL-17F(IL-24), hIL9 and hCD46 were observed before or after treatment.
Figure 1.
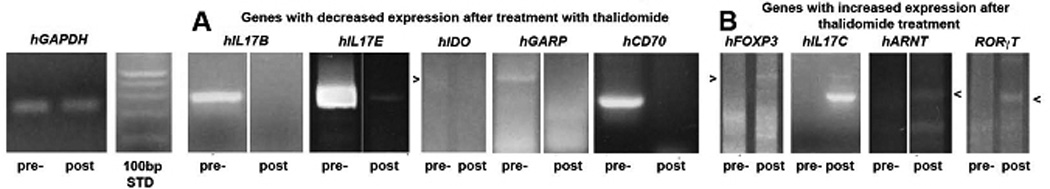
Composite photograph of agarose gel electrophoresis of representative PCR amplicons for genes indicated pre- and post thalidomide treatment for Nepal ENL patients. HGAPDH and 100 bp DNA ladder are in the left three lanes. In A are genes with decreased expression after thalidomide treatment. In B are genes with increased expression after thalidomide treatment.
Discussion
Our results are in keeping with the literature that ENL or type 2 leprosy reaction associated with immune up regulation is ameliorated by an immunomodulatory drug (IMID) thalidomide that is also associated with immune up-regulation. Furthermore, this report extends the original discovery of interleukin-2 and interferon-γ up-regulation of the TH1 T-cell subset by thalidomide16, 17 to the TH17 subset which along with TH1 has been identified in many autoimmune and neoplastic models as the second dominate subset of T-cells involved in cell mediated immunity (CMI). Specifically, IL17A (IL-17), the hallmark cytokine of TH17, was up-regulated both before and after thalidomide treatment. The IL17 pathway is particularly complex as in addition to IL17A(IL-17), there is IL17B-F(IL-24). In this regard IL17B(IL-20) and E(IL-25) were up before thalidomide treatment of ENL, but not detected afterwards. Conversely, IL17C(IL-21) and the TH17 transcription factor-RORγT along with ARNT-a component of the aryl hydrocarbon receptor-implicated in TH17 activation,18 were only detected after thalidomide treatment of ENL. In keeping with the finding reported by Haslett et al.,15 we found an increase in FoxP3-the transcription factor for CD4/CD25+ Tregs, after thalidomide treatment of ENL. We also found CD70, a member of T-cell activation, increased before thalidomide treatment, but not after. These data combined with Haslett’s et al.15 observation of a sustained increase in IL12, suggests the possibility of up-regulation of both TH1 and TH17 immunity during ENL and after thalidomide treatment, yet anti-inflammatory with increased FoxP3 and decreased CD70. While the approach to understanding leprosy type 2 reaction and the effect of thalidomide on the immune system is immunologically interesting, further identification of molecular targets for thalidomide is required. In this regard, indoleamine 2,3-dioxygenase (IDO) needs to be investigated as a potential target of thalidomide as it is up-regulated by INF-γ a hallmark cytokine of ENL and our data show a down-regulation of IDO following thalidomide treatment of ENL. As IDO provides kynurenine19, 20 which down-regulates TH1 and TH17 lymphocytes via the AHR but up-regulates Tregs, a down-regulation of IDO would help explain how thalidomide is immune enhancing yet anti-inflammatory via FoxP3 Tregs. IDO has recently been implicated in the anergy of lepromatous leprosy.21 IDO has also been identified as inhibiting TH17 cell responses by non-hematopoetic cells in a mouse model of M. tb.22 Our data suggest further investigation of IDO as a direct or indirect target of thalidomide and other IMIDs. Park et al.23 have identified the macrophage inducible nitric oxide synthetase (iNos) as a potential target of thalidomide in a mouse macrophage cell line-(RAW264.7). In the present study, iNos2 was found before and after treatment. Recently, Niedbala et al.24 have implicated NO in the regulation of TH17. Lockwood et al.25 have shown iNOS expressed in 78% of 237 leprosy biopsies with reduced levels of non-reactional lepromatous leprosy, indicating this molecule is associated with a well expressed cell-mediated immune response. Furthermore, TGFβ was detected in 94% of the 237 leprosy skin biopsies. TGFβ has traditionally been looked at as a down-regulator of inflammation, but it also implicated as an up-regulator of FoxP3 positive IL17A-producing Th17 cells.26
Pharmacologically thalidomide is predominantly hydrolyzed and renally excreted. However, a portion is catabolized via the liver cytochrome P450 system27 and some of the thalidomides catabolites are likely to be biologically active. Thalidomide treatment of metastatic prostate cancer patients, who have a poorly metabolizing form of CYP2C19, had an inferior outcome.28 Furthermore, AHR inhibition can increase TH17 whereas AHR agonists have been shown to reduce the TH17 transcription factor RORC without affecting T-bet, GATA-3, or FoxP3.29 Further study of thalidomide lenalidomide (Revlimid) and catabolites on the regulation of T-cell subsets via IDO and AHR is indicated. While TNF-α has been suggested as a potential target for thalidomide, Haslett et al.15 were unable to confirm this as important in these patients. As a result, we have emphasized Th-17 transcripts to further our understanding of how thalidomide and other IMIDS function to regulate immunity. It is however likely that additional genes and targets are involved in the complex interplay of small molecular drugs with the human immune system. RNA seq has recently identified a number of new genes involved in the immunopathogenesis of psoriasis30 and offers promise to do the same for understanding the immunopathogenesis of leprosy.
Supplementary Material
Acknowledgements
This research was supported in part by a grant from the CTSI grant-NCRR-NIH 1UL1RR029893.
Funding source-none
Footnotes
Authors have no conflict of interest or disclosures to declare
References
- 1.Burnet FM. The new approach to immunology. N Engl J Med. 1961;5(264):24–34. doi: 10.1056/NEJM196101052640107. [DOI] [PubMed] [Google Scholar]
- 2.Miller JF. Immunological function of the thymus. Lancet. 1961;2:748–749. doi: 10.1016/s0140-6736(61)90693-6. [DOI] [PubMed] [Google Scholar]
- 3.Ridley DS, Jopling WH. A classification of leprosy for research purposes. Lepr Rev. 1962;33:119–128. doi: 10.5935/0305-7518.19620014. [DOI] [PubMed] [Google Scholar]
- 4.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34:255–273. [PubMed] [Google Scholar]
- 5.Nogueira N, Kaplan G, Levy E, et al. Defective gamma interferon production in leprosy. Reversal with antigen and interleukin 2. J Exp Med. 1983;158:2165–2170. doi: 10.1084/jem.158.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamura M, Uyemura K, Deans RJ, et al. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science. 1991;254:277–279. doi: 10.1126/science.254.5029.277. [DOI] [PubMed] [Google Scholar]
- 7.Mosmann TR, Cherwinski H, Bond MW, et al. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 8.Bluestone JA, Mackay CR, O'Shea JJ, Stockinger B. The functional plasticity of T cell subsets. Nat Rev Immunol. 2009;9:811–816. doi: 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 10.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 11.Voo KS, Wang YH, Santori FR, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci. 2009;106:4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massone C, Nunzi E, Ribeiro-Rodrigues R, et al. T regulatory cells and plasmocytoid dentritic cells in hansen disease: a new insight into pathogenesis? Am J Dermatopathol. 2010;32:251–256. doi: 10.1097/DAD.0b013e3181b7fc56. [DOI] [PubMed] [Google Scholar]
- 13.Attia EA, Abdallah M, Saad AA, et al. Circulating CD4+ CD25 high FoxP3+ T cells vary in different clinical forms of leprosy. Int J Dermatol. 2010;49:1152–1158. doi: 10.1111/j.1365-4632.2010.04535.x. [DOI] [PubMed] [Google Scholar]
- 14.Sheskin J. Thalidomide in the treatment of lepra reactions. Clin Pharmacol Ther. 1965;6:303–306. doi: 10.1002/cpt196563303. [DOI] [PubMed] [Google Scholar]
- 15.Haslett PA, Roche P, Butlin CR, et al. Effective treatment of erythema nodosum leprosum with thalidomide is associated with immune stimulation. J Infect Dis. 2005;192:2045–2053. doi: 10.1086/498216. [DOI] [PubMed] [Google Scholar]
- 16.Haslett PA, Corral LG, Albert M, Kaplan G. Thalidomide costimulates primary human T lymphocytes, preferentially inducing proliferation, cytokine production, and cytotoxic responses in the CD8+ subset. J Exp Med. 1998;187:1885–1892. doi: 10.1084/jem.187.11.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon EJ, Sandoval F. Thalidomide increases the synthesis of IL-2 in cultures of human mononuclear cells stimulated with Concanavalin-A, Staphylococcal enterotoxin A, and purified protein derivative. Immunopharmacology. 1995;31:109–116. doi: 10.1016/0162-3109(95)00039-7. [DOI] [PubMed] [Google Scholar]
- 18.Stockinger B, Hirota K, Duarte J, Veldhoen M. External influences on the immune system via activation of the aryl hydrocarbon receptor. Semin Immunol. 2011;23:99–105. doi: 10.1016/j.smim.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen NT, Kimura A, Nakahama T, et al. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezrich JD, Fechner JH, Zhang X, et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Souza Sales J, Lara FA, Amadeu TP, et al. The role of indoleamine 2-3-dioxygenase in lepromatous leprosy immunosuppression. Clin Exp Immunol. 2011;165:251–263. doi: 10.1111/j.1365-2249.2011.04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desvignes L, Ernst JD. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31:974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park E, Levis WR, Greig N, et al. Effect of thalidomide on nitric oxide production in lipopolysaccharide-activated RAW 264.7 cells. J Drugs Dermatol. 2010;9:330–333. [PMC free article] [PubMed] [Google Scholar]
- 24.Niedbala W, Alves-Filho JC, Fukada SY, et al. Regulation of type 17 helper T-cell function by nitric oxide during inflammation. Proc Natl Acad Sci. 2011;108:9220–9225. doi: 10.1073/pnas.1100667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockwood DN, Suneetha L, Sagili KD, et al. Cytokine and protein markers of leprosy reactions in skin and nerves: baseline results for the north Indian INFIR cohort. PLoS Negl Trop Dis. 2011;5:e1327. doi: 10.1371/journal.pntd.0001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovhannisyan Z, Treatman J, Littman DR, Mayer L. Characterization of interleukin-17-producing regulatory T cells in inflamed intestinal mucosa from patients with inflammatory bowel diseases. Gastroenterology. 2011;140:957–965. doi: 10.1053/j.gastro.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ando Y, Fuse E, Figg WD. Thalidomide metabolism by the CYP2C subfamily. Clin Cancer Res. 2002;8:1964–1973. [PubMed] [Google Scholar]
- 28.Ando Y, Price DK, Dahut WL, et al. Pharmacogenetic associations of CYP2C19 genotype with in vivo metabolisms and pharmacological effects of thalidomide. Cancer Biol Ther. 2002;1:669–673. doi: 10.4161/cbt.318. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez JM, Brembilla NC, Sorg O, et al. Activation of the aryl hydrocarbon receptor reveals distinct requirements for IL-22 and IL-17 production by human T helper cells. Eur J Immunol. 2010;40:2450–2459. doi: 10.1002/eji.201040461. [DOI] [PubMed] [Google Scholar]
- 30.Jabbari A, Johnson-Huang LM, Krueger JG. Role of the immune system and immunological circuits in psoriasis. G Ital Dermatol Venereol. 2011;146:17–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


