Abstract
The influence of preexisting immunity to viral vectors is a major issue for the development of viral vectored vaccines. Here, we investigate the effect of preexisting vaccinia virus immunity on the immunogenicity and efficacy of a DNA/MVA SIV vaccine in rhesus macaques using a pathogenic intrarectal SIV251 challenge. Preexisting immunity decreased SIV-specific CD8 and CD4 T cell responses, but preserved the SIV-specific humoral immunity. In addition, preexisting immunity did not diminish the control of a SIV challenge mediated by the DNA/MVA vaccine. The peak and set point viremia was 150- and 17-fold lower, respectively in preimmune animals compared to control animals. The peak and set point viremia correlated directly with colorectal virus at 2 weeks post challenge suggesting that early control of virus replication at the site of viral challenge was critical for viral control. Factors that correlated with early colorectal viral control included (i) the presence of anti-SIV IgA in rectal secretions, (ii) high avidity binding antibody for the native form of Env and (iii) low magnitude of vaccine-elicited SIV-specific CD4 T cells displaying the CCR5 viral co-receptor. The frequency of SIV-specific CD8 T cells in blood and colorectal tissue at 2 weeks post challenge did not correlate with early colorectal viral control. These results suggest that preexisting vaccinia virus immunity may not limit the potential of recombinant MVA vaccines to elicit humoral immunity and highlight the importance of immunodeficiency virus vaccines achieving early control at the mucosal sites of challenge.
INTRODUCTION
Live vector based vaccines have become popular for their ability to induce strong cellular and humoral immunity(1-10). However, preexisting immunity to viral vectors has been a major issue for the development of viral-vectored vaccines. This has been particularly important for vectors such as adenovirus type 5 (Ad5) because of the high prevalence of Ad5-specific immunity in people around the world(11). Similarly, a significant proportion of the US population is preimmune to VV because of vaccination for smallpox. Although routine vaccination with VV to prevent smallpox ceased more than 30 years ago, the US government re-initiated vaccination of certain groups because of perceived bioterrorist threats. Because MVA is an attenuated strain of VV(12), the anti-VV immunity generated by smallpox vaccine may limit the immunogenicity of MVA based vaccines.
Preexisting immunity to Ad5, VV or MVA has been shown to reduce the immunogenicity of the respective recombinant viral vectors in mice(13-16), macaques(17-19) and humans(20, 21). The majority of these studies evaluated the effects on cellular immunity and very little information is available on humoral immunity. In addition, none of these studies evaluated the consequence of this diminished immunogenicity on the efficacy of HIV vaccines using an appropriate challenge model. Furthermore, the results of a recent human trial for an Ad5 based vaccine revealed a higher rate of HIV infection in uncircumcised males with preexisting Ad5 immunity(22, 23). These results showed preexisting immunity to the vaccine vector affecting the efficacy of an HIV vaccine. Thus it is important to study the effect of preexisting anti-vector immunity not only on the immunogenicity but also on the efficacy of a candidate HIV vaccine.
DNA prime and live vector boost vaccines have become popular for their ability to elicit high levels of vaccine-specific cellular and humoral immunity (2, 17, 24-32). Our previous preclinical studies in macaques demonstrated that DNA priming and recombinant modified vaccinia Ankara (rMVA) boosting elicited high frequencies of virus-specific CD4 and CD8 T cells and controlled a pathogenic SHIV 89.6P challenge (2, 3, 33, 34). The prototype HIV-1 clade B version of this DNA/MVA vaccine (35) has successfully completed phase I safety testing and entered phase II trials in humans in US. The preexisting anti-VV immunity generated by the smallpox vaccine may limit the boosting ability of rMVA, and hence the efficacy of DNA/MVA vaccines. Here, we evaluated the effect of preexisting VV immunity on the immunogenicity and efficacy of a DNA/MVA SIV vaccine in rhesus macaques using a high dose pathogenic intrarectal SIV251 challenge. Our results demonstrate that preexisting immunity diminishes cellular but not humoral immunity. They also demonstrate that diminished cellular immunity does not reduce the efficacy of the DNA/MVA vaccine and suggest a role for non-neutralizing anti-viral antibody in viral control.
MATERIALS AND METHODS
Immunizations and challenge
Young adult Indian rhesus macaques from the Yerkes breeding colony were cared for under guidelines established by the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals” using protocols approved by the Emory University Institutional Animal Care and Use Committee. Macaques were typed for the Mamu-A*01, Mamu B08 and Mamu B17 alleles as described before(36-38). Macaques were randomized into 3 trial groups of eight animals each based on weight and A*01 status. There were four Mamu A*01 macaques in each group. Trial groups were randomized into three inoculation and sampling groups. Of the 24 macaques, 16 were vaccinated with a DNA/MVA SIV vaccine and 8 were unvaccinated. The DNA and recombinant MVA (rMVA) immunizations were delivered intramuscularly in PBS using a hypodermic needle in the outer thigh. The DNA immunogen expressed SIV239 Gag-Pol, Env, Tat and Rev. The DNA immunogen was constructed by replacing the EcoRI-NheI fragment of SHIV DNA construct(39) containing HIV-1 89.6 Tat, Rev and Env genes with an EcoRI-NheI fragment containing SIV Tat, Rev and Env. Two MVA recombinants, one expressing SIV239 Gag-Pol(40) and the other expressing SIV239 Env(41) were premixed and used for immunizations. Two DNA inoculations were given on weeks 0 and 8, and two rMVA boosters were given on weeks 16 and 24. The DNA was delivered at 1.2 mg/dose and the rMVA was delivered at 1×108 pfu/dose. Of the 16 vaccinated animals, 8 received standard dose of Dryvax (~105 pfu) by percutaneous route 17 months prior to first DNA inoculation. At 9 months after the final rMVA booster, animals received an intrarectal challenge with SIVmac251 by using a pediatric feeding tube 15 to 20 cm into the rectum. Dr. Nancy Miller at NIH provided the challenge stock and 100μl of the stock diluted in 1 ml RPMI was used per animal. 100% of animals can be infected under these conditions.
Collection and processing of rectal secretions, biopsies and blood
Rectal secretions were collected with and eluted from Weck-Cel sponges as previously described(39, 42). Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood according to the standard procedures as described before(33). Lymphocytes from pinch biopsies from the rectum were obtained as described before(43). Briefly, 10 to 20 pinch biopsies were collected in complete RPMI and washed two times with ice-cold Hank’s buffered salt solution. Biopsies were digested with 200 units/ml of collagenase IV (Worthington, Lake wood, NJ) and DNase I (Roche, Indianapolis, IN), passed through decreasing sizes of needles (16-, 18-, and 20-gauge, five to six times with each needle), and filtered through a 100μm filter. Cells were washed twice with RPMI and resuspended in complete RPMI for analysis.
T cell responses
Intracellular cytokine production was assessed as previously described with a few modifications(2). Briefly, two million PBMCs were stimulated in 200μl of RPMI with 10% FBS in a 5 ml polypropylene tube. For measuring vaccinia-specific responses, approximately 5×106 pfu (multiplicity of infection of ~2.5) of vaccinia virus strain WR was added in a volume of 100 μL. SIV-specific stimulations were conducted using a single pool of 125 SIV239 Gag peptides, a single pool of 221 SIV239 Env peptides and 2 pools (132 peptides/pool) of SIV239 Pol peptides (NIH AIDS Research and Reference Reagent Program). All peptides were 15-mers overlapping by 11. Staphylococcal enterotoxin B (SEB) was used as a positive control at 1 μg/ml. Stimulations were performed in presence of anti-CD28 and anti-CD49d antibodies (1 μg/ml; BD Pharmingen, San Diego, CA). For all stimuations, cells were incubated at 37°C in the presence of 5% CO2 for 6 hours except for stimulations using vaccinia virus, where stimulations were performed for 10-12 hours. Brefeldin A (10 μg/ml) was added for the last 4 hours of incubation. At the end of stimulation, cells were washed once with phosphate-buffered saline (PBS) containing 2% FBS, surface stained with anti-human CCR5-PE (clone 3A9, BD Pharmingen), anti human CCR7-FITC (clone 150503, R&D Systems, Minneapolis, MN USA), anti-human CD4-PerCP (clone L200; BD Pharmingen, San Diego, CA) and anti-human CD8-Amcyan (clone SK1; BD Biosciences, San Jose, CA), fixed with cytofix/cytoperm (BD Pharmingen, San Diego, CA) and permeabilized with 1× permwash (BD Pharmingen, San Diego, CA). Cells were then stained using a cocktail of antibodies containing anti-human CD3-Pacific Blue (clone SP34-2, BD Pharmingen), anti-human IFN-γ Alexa 700 (clone B27, BD Pharmingen), anti-human IL-2-APC (clone MQ1-17H12, BD Pharmingen) and anti-human TNF-α-PE-Cy7 (clone Mab11, eBiosciences), washed twice with Permwash, once with 2% FBS in PBS and resuspended in 1% formalin in PBS. Approximately 500,000 lymphocytes were acquired on the LSRII (BD Immunocytometry systems) and analyzed using FlowJo software (Treestar, Inc., San Carlos, California). Lymphocytes were identified based on their scatter pattern, and CD3+, CD8-, CD4+ cells were considered as CD4 T cells and CD3+CD8+CD4-cells were considered as CD8 T cells. These CD4 or CD8 T cells were then gated for cytokine positive cells.
T cells were subjected to tetramer staining and typing for the presence of CD4 and CD8 T cells. This was done using a cocktail of the following antibodies and Gag-CM9 tetramer conjugated to APC: anti-human CD3-Alexa Fluor 700 (clone SP34-2, BD Biosciences PharMingen), anti-human CD4-Percp (clone L200), anti-human CD8-Amcyan (clone SK1; BD Biosciences), anti-human CD28-PE-Cy7 (clone CD28.2, Beckman Coulter) and anti-human CD95-Pacific blue (clone DX2, Invitrogen Corp.). The levels of CD4 T cells in intestinal biopsies are presented as a % of total CD3+ T cells.
CFSE dilution assays were performed as described previously(43). Briefly, PBMC were pre-stained with CFSE and approximately 1×106 cells were stimulated in 48 well plates in a volume of 600μl in RPMI containing 10% human serum at 37°C under 5% CO2 for six days. Cells were stimulated with pooled peptides spanning the entire SIV Gag protein (single pool of 125 peptides, Cat# 6204, NIH AIDS Research and Reference Reagent Program) at a concentration of 1.0 μg/ml of each peptide. Unstimulated cells served as negative controls. At the end of six days in culture, the cells were stained intracellularly using antibodies specific for Ki-67 (clone B56), CD8 (clone SK1) and CD3 (clone SP34-2), and acquired on a FACS Calibur and analyzed using FlowJo software (Treestar, Inc., San Carlos, California).
Measurement of binding antibody responses
SIV Env-specific binding antibodies were measured with ELISA using tissue culture produced SIV ENV, captured on a concanavalin A (con A) coated plate as described previously(39). Briefly, ELISA plates (Costar, Corning Life Sciences) were coated with ConA (25 μg per ml) overnight at 4 °C. Plates were washed and incubated with 100μl of undiluted VLP supernatant (generated by transient transfection of 293T cells with the above described SIV239 DNA vaccine expressing Gag, Pol and Env) for one hour. Plates washed and blocked for 1 hour (PBS-tween with 4% whey and 5% dry milk). Test sera were added to duplicate wells in serial 3-fold dilutions and incubated for 1 hour. Plates were then washed and bound Ab was detected using peroxidase conjugated anti-monkey IgG (Accurate Chemical and Scientific Corp) and TMB substrate (KPL). Reactions were stopped with 100 μl of 2 N H2SO4. Each plate included a standard curve generated using goat anti-monkey IgG and rhesus IgG (both from Accurate Chemicals) as previously(39). Standard curves were fitted and sample concentrations interpolated as μg of Ab per ml of serum using SOFTmax 2.3 software (Molecular Devices). The concentrations of IgG are relative to our standard curve, not absolute values.
A NaSCN displacement ELISA assay modeled after that described by Vermont was used for determining avidity(44). This assay was conducted using parallel titrations of test sera in our standard ELISA assay. Following the binding of the test sera, the parallel titrations were treated for 15 minutes at room temperature with PBS or 1.5M NaSCN (prepared fresh in PBS). Then, the relative levels of bound Ab were determined using the standard ELISA procedure (see above). The avidity index was calculated by dividing the dilution of the serum that gave an OD of 0.5 with NaSCN treatment by the dilution of the serum that gave an OD of 0.5 without NaSCN treatment and multiplying by 100. Each assay included one plate with a standard serum with known avidity. Inter assay variation in the avidity index for the standard serum was ± 3 for an index of 27.
Measurements for total IgA, anti-SIV env IgA, or anti-SIV gag,pol IgA or IgG were done by ELISA using microtiter plates coated respectively with 100 μl of 0.5 μg/ml goat anti-monkey IgA (Rockland, Gilbertsville, PA), 1 μg/ml SIVmac251 rgp130 (ImmunoDiagnostics, Woburn, MA) or 1/400 SIVmac251 viral lysate (Advanced Biotechnologies Inc., Columbia, MD), which lacks detectable envelope protein at this dilution. These ELISAs and the serum standards have been described previously(39). Plates were developed by consecutive treatments with biotinylated goat anti-monkey IgA (Alpha Diagnostics, San Antonio, TX) or –human IgG (SouthernBiotech, Birmingham, AL), avidin-peroxidase, TMB, and 2N H2SO4. For rectal secretions, the concentration of anti-env or -gag,pol IgA was divided by the total IgA concentration to obtain the specific activity. A secretion was considered IgA antibody positive if the env or gag,pol specific activity was greater than or equal to 0.145 or 0.224, respectively. These cut-offs represent the mean specific activity + 3 SD previously established for rectal secretions from naïve macaques.
Measurement of neutralizing antibodies
SIV-specific neutralization was measured as a function of reductions in luciferase reporter gene expression after a single round of infection in TZM-bl cells as described(45). TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu.
Vaccinia virus neutralization assays were performed as described previously(46). Briefly, two-fold serial dilutions of sera were incubated with VV-NP-S-EGFP for 1 h at 37°C. HeLa S3 cells were added and incubated overnight in the presence of cytosine arabinoside. Fluorescent cells were enumerated with a FACScalibur flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star, Inc.). IC50 values were determined with PRISM software (Graph Pad).
Quantitation of SIV RNA Plasma Load
The SIV copy number was determined using a quantitative real-time PCR as previously described(2). All specimens were extracted and amplified in duplicates, with the mean results reported. For viral load determinations in gut, total RNA was extracted from about one million cells obtained from gut biopsies and used for quantitative real-time PCR analyses.
Statistical Analysis
Wilcoxon–Mann–Whitney test was used for comparisons of T cell responses, Ab responses and viral RNA levels between Dryvax-immune and Dryvax-naïve groups. This method was used because data did not meet with parametric assumptions. P values were given before correcting for any multiple comparisons. Pearson’s product moment correlation method was used for correlation analysis when data met with parametric assumptions. Spearman’s rank correlation method was used for non-parametric data correlations. This was indicated with ‘s’ as a subscript for ‘r’ values on the graphs. A two-sided p<0.05 was considered significant. Statistical analyses were performed using TIBCO Spotfire S-PLUS 8.1.
RESULTS
We vaccinated two groups of eight macaques with our DNA/MVA SIV239 vaccine that expresses Gag, Pol and Env, and produces non-infectious virus-like particles. The vaccination regimen consisted of intramuscular priming with DNA at weeks 0 and 8, and intramuscular boosting with MVA at weeks 16 and 24. One of the two vaccinated groups received a single inoculation of Dryvax smallpox vaccine 17 months before the first DNA prime (Dryvax-immune group). Another eight unvaccinated macaques served as a control group. In each group, 4 macaques expressed the Mamu A*01 histocompatability molecule that allowed us to use MHC I tetrameric complexes to follow CD8 T cell responses to the immunodominant Gag epitope CM9(47). Macaques RLk7 and RGd8 of the Dryvax-naïve group were positive for Mamu B08 and the macaque RNv9 of the control group was positive for Mamu B17. All macaques were challenged intrarectally with the highly pathogenic uncloned SIV251 at 9 months after the final MVA boost. We used uncloned SIV251 rather than SIV239 because the former is a quasi species.
Preexisting vaccinia virus immunity diminishes SIV-specific T cell responses
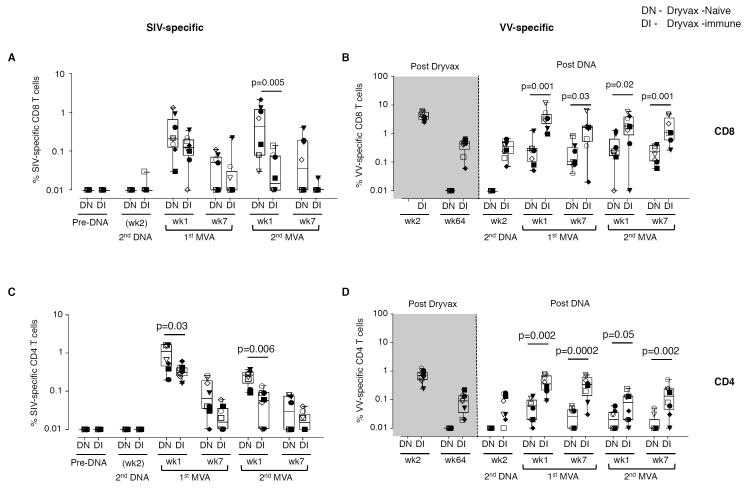
As anticipated, prior immunization with Dryvax diminished the magnitude of SIV -specific CD8 and CD4 T cell responses elicited by the DNA/MVA vaccine (Fig. 1A, 1C). We measured the magnitude of SIV Gag, Pol and Env-specific T cell responses in an intracellular cytokine staining assay following the stimulation of PBMC with peptide pools and. Consistent with our previous reports, SIV-specific immune responses were not detected until after the MVA boost(2, 3, 48-50). At 1 week following the 1st MVA boost, SIV-specific CD8 T cells ranged from 0.03%-1.3% (geometric mean, GM of 0.24%) of total CD8 T cells in the Dryvax-naïve group (Fig. 1 A). Responses were equally directed against Gag and Env. Responses against Pol were observed only in some animals. The SIV-specific responses were further boosted at 1 week following the 2nd MVA boost to a GM of 0.3% of total CD8 T cells. In contrast, in the Dryvax-immune group, SIV-specific CD8 T cells were boosted by the 1st but not the 2nd MVA and were 2.4-fold lower (GM of 0.1%)(p=ns) and 12-fold lower (GM of 0.03%)(p=0.005) than in the Dryvax-naïve group at 1 week following the 1st and 2nd MVA boosts, respectively (Fig. 1A). This was true for responses directed against Gag or Env (data not shown). Similar patterns were observed for the magnitude of Gag-CM9 tetramer positive cells in Mamu A*01 positive animals (Fig. S1A). CFSE proliferation assays on PBMC prior to challenge also revealed significantly lower Gag-specific CD8 T cell responses in the Dryvax-immune than Dryvax-naïve group (Fig. S1B).
Figure 1. Pre-existing immunity to vaccinia limits the MVA insert-specific cellular immunity elicited by a DNA/MVA SIV vaccine.
(A) SIV (Gag, Env and Pol)-specific IFNγ producing CD8 T cell responses. (B) VV-specific IFNγ producing CD8 T cell responses. (C) SIV (Gag, Env and Pol)-specific IFNγ producing CD4 T cell responses. (D) VV-specific IFNγ producing CD4 T cell responses. Closed symbols represent Mamu A*01 positive animals and open symbols represent Mamu A*01 negative animals. Boxes represent medians with 25th and 75th percentiles for the group. Key to animal names is presented in Figure 3B. DN, Dryvax-naïve (n=8); DI, Dryvax-immune (n=8).
The magnitude of the SIV-specific CD4 response was also lower in the Dryvax-immune than the Dryvax-naïve group (Fig. 1C). In the Dryvax-naïve group, the SIV-specific CD4 T cell responses ranged from 0.2%-1.6% (GM of 0.8%) of total CD4 T cells at 1 week following the 1st MVA boost (Fig. 1C) and underwent only a minimal boost following the 2nd MVA immunization. In the Dryvax-immune group, these responses were 2.5-fold lower (GM of 0.3%)(p=0.03) and 6-fold lower (GM of 0.04%)(p=0.006) at 1 week following the 1st and 2nd MVA boosts, respectively. CFSE proliferation assays on PBMC prior to challenge also revealed lower Gag-specific CD4 T cell responses in the Dryvax-immune than Dryvax-naïve group (Fig. S1B). The magnitude of CD4 T cells that co-produced IFNγ, TNFα and IL-2 was also higher in the Dryvax-naive than Dryvax-immune animals (Fig. S2 B). However, the proportion of IFNγ, TNFα and IL-2 co-producing cells as a percent of total cytokine positive cells was similar between the two groups (Fig. S2 C). A similar pattern was observed for SIV-specific CD8 T cell responses (data not shown).
In contrast to the SIV-specific T cell responses, VV-specific T cell responses were higher in the Dryvax-immune than Dryvax-naïve animals. Vaccination with Dryvax elicited a robust VV-specific cellular immune response (Fig. 1, B and D). Following the MVA boosts, both VV-specific CD8 and CD4 T cell responses had about 10-fold higher peaks in the Dryvax-immune than in the Dryvax-naive animals. To understand the influence of preexisting VV immunity on SIV-specific CD8 T cell response, we performed correlations between the magnitude of VV-specific CD8 T cells or VV-specific neutralizing Ab after the 1st MVA boost and SIV-specific CD8 T cells after the 2nd MVA boost. We chose to look at the effect of preexisting VV immunity at the time of 2nd MVA boost because this allowed us to look at all animals (Dryvax-immune and Dryvax-naïve) at a time when the maximum effect of preexisting immunity would be observed. VV-specific neutralizing Ab as well as VV-specific CD8 T cells prior to the 2nd MVA boost showed inverse correlations with SIV-specific CD8 T cells post the 2nd MVA boost suggesting that both of these responses contributed to the diminished immunogenicity of MVA/SIV vaccine (Fig. S3).
Preexisting vaccinia virus immunity does not diminish SIV-specific humoral immunity
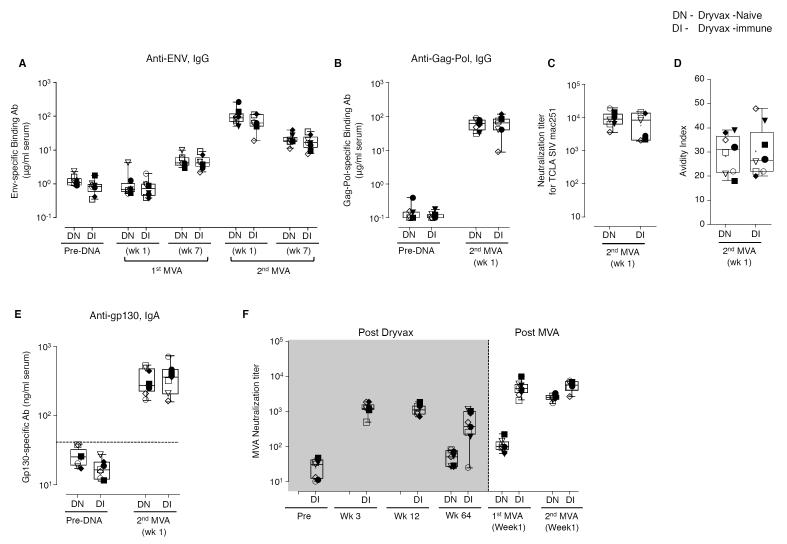
In contrast to the SIV-specific T cell responses, the preexisting VV immunity did not diminish SIV-specific Ab responses (Fig. 2). To evaluate the effect of preexisting VV immunity on the SIV-specific humoral immunity, we measured the titers of binding Ab specific to SIV Env and Gag-Pol and neutralizing Ab to SIV251 in serum. In addition, we determined the avidity index of anti-Env binding Ab in serum. Consistent with our previous reports, SIV Env-specific Ab responses were not detected until after the MVA boost(2, 3, 48-50). At 7 weeks following the 1st MVA boost, the titers of SIV Env-specific binding Ab ranged from 3-10 μg/ml (mean of 5 μg/ml) of serum in the Dryvax-naïve group (Fig. 2A). These were further boosted to a mean of 108 μg/ml at 1 week following the 2nd MVA boost. Throughout the period of vaccination, similar titers of Env-specific binding Ab were found in the Dryvax-immune and Dryvax-naïve groups. Consistent with the titers of Env-specific binding Ab, at the peak vaccine response (1 week post 2nd MVA boost), the titers of Gag-Pol-specific binding Ab (Fig. 2B), neutralizing Ab against T-cell line adapted SIV251 (Fig. 2C), and the avidity of Env-specific binding Ab (Fig. 2D) were also similar in the two vaccine groups. Neither group had neutralizing activity for non-T cell line adapted SIV251 (data not shown).
Figure 2. Pre-existing immunity to vaccinia does not limit the MVA insert-specific humoral immunity elicited by a DNA/MVA SIV vaccine.
(A) SIV Env-specific IgG in serum. (B) SIV Gag-Pol-specific IgG in serum. (C) TCLA SIV-specific neutralizing Ab titer in serum. (D) Avidity index of SIV Env-specific IgG in serum. (E) SIV Env-specific IgA in serum. The horizontal dashed line indicated the limit of background. (F) VV-specific neutralizing Ab titer in serum. Boxes represent medians with 25th and 75th percentiles for the group. Key to animal names is presented in Figure 3B. DN, Dryvax-naïve (n=8); DI, Dryvax-immune (n=8).
We next investigated the effect of preexisting immunity on SIV-specific IgA responses in serum and rectal secretions following vaccination. Measurements in serum were performed at 1 week and 9 months (six weeks prior to challenge) following the 2nd MVA boost. Measurements in rectal secretions were performed prior to challenge. At 1 week following the 2nd MVA boost, all animals had Env-specific serum IgA (Fig. 2E) and one animal in each group had significant levels of Gag-Pol-specific IgA (data not shown). These Env responses were similar between the two vaccine groups demonstrating that preexisting VV immunity did not diminish the SIV-specific IgA response. However, the serum IgA response was transient, and prior to challenge, all animals were negative for Env-specific serum IgA (data not shown). In contrast, six weeks prior to challenge, SIV Gag-Pol (Table I), but not Env (data not shown) – specific IgA was present in rectal secretions of 4 of the 8 Dryvax-immune animals. This IgA was found in only 1 of the 8 Dryvax-naïve animals and none of the unvaccinated controls suggesting that preexisting VV immunity had promoted the generation of long lasting SIV-specific rectal IgA.
Table I.
The magnitude of SIV Gag-Pol specific IgA in rectal secretions
| ng of SIV Gag-pol- specific IgA per μg of total IgA* |
||
|---|---|---|
| Macaque | 6 weeks prior to challenge |
2 weeks after challenge |
| Dryvax-Naive | ||
| ROW7 | 0.166 | 0.211 |
| RLK7 | 0.048 | 0.226 |
| REP7 | 0.084 | 0.050 |
| RNI7 | 1.257 | 0.694 |
| RHS7 | 0.079 | 0.733 |
| RGD8 | 0.069 | 0.103 |
| RIN8 | 0.134 | 0.198 |
| RTM7 | 0.081 | 0.756 |
| Dryvax-Immune | ||
| RGR8 | 0.029 | 0.074 |
| RVP8 | 0.120 | 0.119 |
| RBS8 | 0.372 | 0.511 |
| RLR8 | 0.052 | 0.501 |
| RLS8 | 0.948 | 1.629 |
| ROB8 | 0.064 | 0.198 |
| RNK8 | 0.041 | 0.290 |
| RCK8 | 0.376 | 1.199 |
| Controls | ||
| RPW9 | 0.058 | 0.045 |
| RAC10 | 0.090 | 0.069 |
| RQJ9 | 0.054 | 0.037 |
| RRK10 | 0.062 | 0.057 |
| RBY9 | 0.040 | 0.338 |
| RYD9 | 0.015 | 0.024 |
| RNV9 | 0.031 | 0.085 |
| RFP9 | 0.032 | 0.178 |
-Positive responses (>0.224) are highlighted in bold.
As expected, MVA-specific neutralizing Ab responses were higher in the Dryvax-immune than Dryvax-naïve animals (Fig. 2F). These responses were 100-fold higher in the Dryvax-immune than in the Dryvax-naïve animals following the 1st MVA boost and about 2-fold higher following the second MVA boost. Analyses for VV-specific neutralizing Ab activity showed a similar pattern (data not shown).
Preexisting VV immunity does not diminish the control of a pathogenic intrarectal SIV challenge
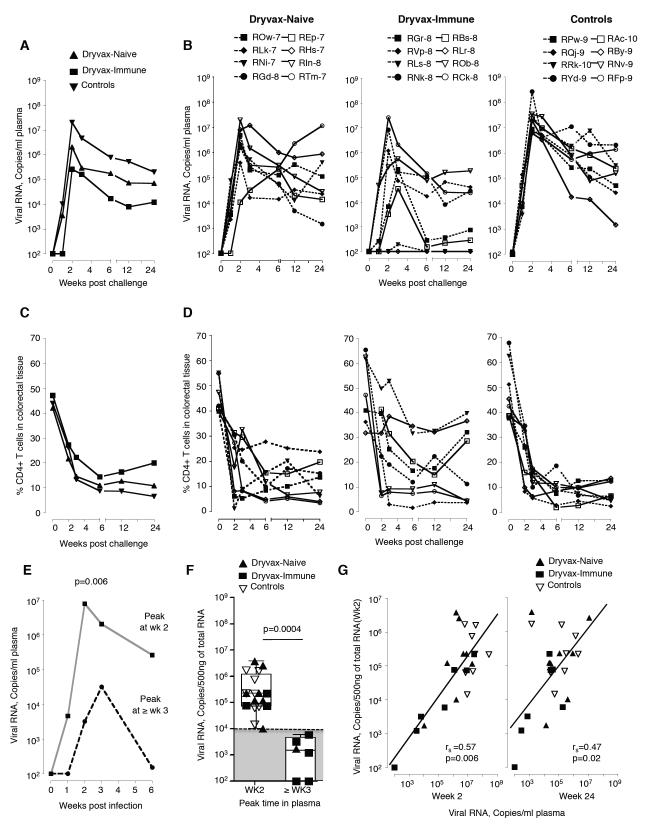
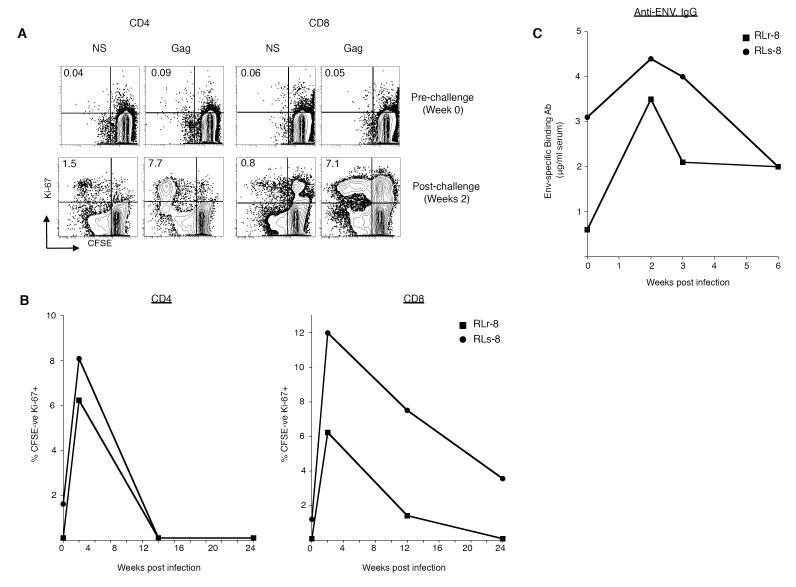
Despite their low SIV-specific T cell responses, the Dryvax-immune macaques exhibited the best control of an intrarectal SIV challenge (Fig. 3 and Supplementary Fig. 5). Following SIV challenge, viremia was detected in all animals except RLr8 of the Dryvax-immune group (Fig. 3A). In RLs8, a low level of viremia was detected, but only at 2 weeks post challenge (Fig. 3A). We observed a 5-60-fold increase in the frequency of proliferating (Ki-67+, CFSElo) Gag-specific CD4 and CD8 T cells at 2 weeks post challenge in RLr8 and RLs8 suggesting both animals had been infected (Fig. 4A and 4B). In addition, the titer of serum anti-Env binding Ab increased 3-fold in RLr8 and 1.5-fold in RLs8 at 2 weeks post challenge (Fig. 4C).
Figure 3. Preexisting VV immunity does not diminish the control of a pathogenic intrarectal SIV challenge.
Temporal viremia (A) median for the group and (B) for individual animals. Frequency of CD4 T cells expressed as a percent of total T cells in the colorectal tissue (C) median for group and (D) for individual animals. Dotted lines indicate Mamu A*01 positive animals and solid lines indicate Mamu A*01 negative animals. (E) Median temporal levels of plasma viral RNA in animals that peaked at week 2 and > week 3. (F) Comparison of colorectal virus at 2 weeks post challenge by time of peak in plasma. Key to animal names is presented in Figure 3B. Boxes represent medians with 25th and 75th percentiles for the group. (G) Correlation between levels of viral RNA in the rectum at week 2 and plasma at weeks 2 and 24. Dryvax-naïve animals are shown in closed triangles (n= 8), Dryvax-immune in closed squares (n= 8) and controls in open triangles (n= 8). The sensitivity of viral load assay was 80 copies of RNA/ml, and animals with levels of virus below 80 were scored at 100. rs, Spearman’s rank correlation.
Figure 4. Evidence for ‘take’ of infection in RLr8 and RLs8.
(A) Magnitude of in vitro proliferating (Ki67+, CFSElo) SIV-specific CD4 and CD8 T cells following SIV infection. (A) Representative FACS plots are shown demonstrating the frequency of Gag-specific CD4 or CD8 T cells following in vitro stimulation with a SIV Gag peptide pool for 6 days. Cells were gated on either CD3+ CD4+ or CD3+CD8 + T cells respectively. Numbers on the graphs represent the frequency of proliferating CFSElo Ki67+ T cells as a percent of total CD4 or CD8+ T cells. (B) Magnitude of proliferating SIV-Gag-specific T cells from multiple time points are plotted for CD4 T cells on the left and CD8 T cells on the right. (C) Magnitude of SIV Env-specific binding Ab.
At 2 weeks post challenge, plasma viral RNA in the control animals ranged from 6×106 −2.6×108 copies per ml with median copies of 2.0×107/ml. At this time, the median levels of plasma viral RNA were 10 times lower in the Dryvax-naïve animals (p=0.005) and 150 times lower in the Dryvax-immune animals (p=0.01) than in the unvaccinated control animals (Fig. 3A and 3B). The median levels of plasma viral RNA in Dryvax-immune animals were 15 times lower than in the Dryvax-naïve animals (Fig. 3A and 3B). Viral control during set point was also lower in the Dryvax-immune than control animals. At 24 weeks, the Dryvax-immune animals had levels of virus that were 6 times lower than in the Dryvax-naïve animals (p=0.03) and 17 times lower than in the unvaccinated control animals (p=0.02)(Fig. 3, A and B). At this time, the median levels of plasma viral RNA were not significantly lower in the Dryvax-naïve animals than in the control animals (Fig. 3A and 3B).
We further analyzed the viral control based on the Mamu A*01 status of the animals (Fig. S4). The viral control in the Dryvax-immune animals was not restricted to Mamu A*01 positive animals, whereas in the Dryvax-naïve animals it was better in Mamu A*01 positive than Mamu A*01 negative animals (Fig. S4). In the Mamu A*01 positive animals, the viremia at week 2 post infection was 20 times lower in the Dryvax-naïve animals (p=0.01) and 54 times lower in the Dryvax-immune animals (p=0.02) than in the unvaccinated control animals (Fig. S4A and S4B). In the non-Mamu A*01 animals, the viremia at week 2 post infection was not significantly lower in the vaccinated animals than in the unvaccinated animals. However, at 24 weeks post infection, it was 21 times lower in the Dryvax-immune animals (p=0.04) but not in the Dryvax-naïve animals than in the unvaccinated controls (Fig. S4C and S4D). Furthermore, none of the Dryvax-immune animals were positive for Mamu B08 and B17 indicating that the enhanced control was not due to expression of known protective Mamu class I alleles.
Consistent with the lower levels of virus, depletion of colorectal CD4 T cells was slower and less severe in the Dryvax-immune than in the Dryvax-naïve and control animals (Fig. 3, C and D). In the majority of the Dryvax-immune animals, the nadir of CD4 T cells did not occur until 6 weeks post challenge, whereas in the majority of animals in the other two groups the nadir of CD4 T cells had occurred by 3 weeks post challenge (Fig. 3, C and D). Furthermore, in 4 out of the 8 Dryvax-immune animals, the colorectal CD4 T cells were rebounding by 24 weeks. At 24 weeks post challenge, the frequencies of central memory CD4 T cells in the blood (p=0.03) were higher in the vaccinated than the control animals (Fig. S5 A). The preservation of central memory T cells is a predictor for better survival in SIV-infected macaques (51).
Enhanced viral control correlates with lower colorectal virus early post challenge
A closer look at the kinetics of viremia during the first 3 weeks of infection revealed slow expansion of viremia in 5 Dryvax-immune and one Dryvax-naïve animal (Fig. 3, A, and B). In these six animals, virus reached peak levels at > 3 weeks, whereas in the remaining vaccinated and control animals, virus reached peak levels at 2 weeks post challenge (Fig. 3B). The slower expansion of viremia resulted in lower peak and set point viremia. Animals in which virus peaked at > 3 weeks had a 243-fold lower median peak viremia and a 159-fold lower median set point viremia (week 24)(Fig. 3E).
Impressively, the levels of colorectal virus at 2 weeks post challenge influenced the kinetics and magnitude of viremia. The median level of colorectal virus was 150-fold lower in animals in which peak viremia was delayed to > 3 weeks (Fig. 3F). All animals in which viremia peaked at week 2 had levels of colorectal virus above 104 copies/500ng of total tissue RNA, whereas all animals in which viremia peaked at > week 3 had colorectal virus below this level. The levels of colorectal virus in 5 of the Dryvax-immune and one of the Dryvax-naïve animals were below this level, whereas in all of the remaining vaccinated and control animals colorectal virus was above this level (Fig. 3F). The median level of colorectal virus in Dryvax-immune animals was 39 fold and 49 fold lower than in Dryvax-naïve (p=0.02) and control (p=0.02) animals, respectively. Furthermore, the levels of colorectal virus showed a strong direct correlation with peak (week 2; p=0.006) and set point (week 24; p=0.02) viremia (Fig. 3G). In addition, an inverse correlation was observed between the levels of colorectal virus and the frequencies of total colorectal CD4 T cells at weeks 3 (p=0.02) and 24 (p=0.05) post challenge (Fig. S5, B and C). These results demonstrate that the enhanced control of viremia correlated with lower levels of virus at the site of challenge early post infection.
No clear protective association between SIV-specific CD8 T cells and colorectal virus early following infection
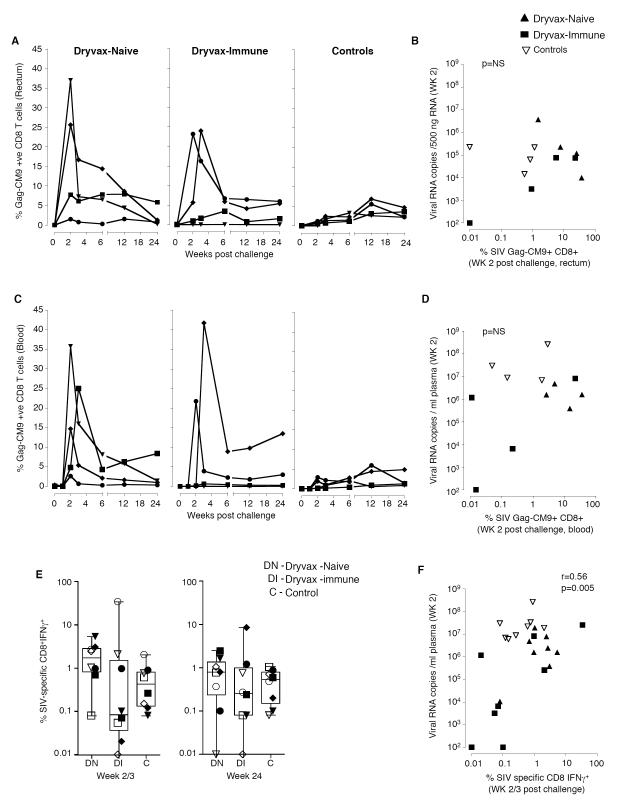
The frequencies of virus-specific CD8 T cells in colorectal tissue and blood at 2 weeks post challenge did not correlate with reduced levels of colorectal virus (Fig. 5). To understand the role of SIV-specific CD8 T cells in viral control, we measured the SIV Gag and Env-specific IFNγ producing CD8 T cells in blood at acute (week 2 or 3) and set point (weeks 24) phases following challenge. In addition, we measured the frequency of Gag CM9-specific CD8 T cells in blood and colorectal tissue. However, the tetramer analyses were restricted to Mamu A*01 positive animals. At 2 weeks following challenge, expansion of Gag-CM9-specific T cells was higher in colorectal tissue (Fig. 5A) and blood (Fig. 5C) of vaccinated than unvaccinated animals. In general, in vaccinated animals, expansion was higher in animals with higher levels of virus suggesting that higher expansion was in response to higher levels of antigen (Fig. 5B and 5D). Analyses for SIV-specific IFN-γ producing CD8 T cells in blood revealed similar patterns (Fig. 5, E and F). Correlations between SIV-specific CD8 T cells at 1 week following the 2nd MVA boost and viral load at 2 weeks following challenge in colorectal tissue or plasma also revealed a direct correlation (data not shown). Consistent with the variability in viremia in the Dryvax-immune group (Fig. 3B), the frequencies of SIV-specific CD8 T cells post challenge showed the most variability in this group (Fig. 5A, 5C and 5E). In some of the Dryvax-immune animals, high levels of expansion were observed despite these animals having low levels of CD8 responses following vaccination (Fig. 5E).
Figure 5. No clear association between the SIV-specific CD8 T cell response and viremia in colorectal tissue early following infection.
(A) Temporal SIV Gag-CM9 specific CD8 T cell responses in the colorectal tissue of Mamu A*01 positive animals. (B) Correlation between tetramer-specific CD8 T cells and levels of virus in the colorectal tissue. (C) Temporal SIV Gag-CM9 specific CD8 T cell responses in blood of Mamu A*01 positive animals. (D) Correlation between tetramer-specific CD8 T cells and levels of virus in blood. (E) SIV (Gag and Env)-specific IFNγ producing CD8 T cells in blood. Boxes represent medians with 25th and 75th percentiles for the group. (F) Correlation between SIV-specific CD8 T cells and levels of virus in blood. Key to animal names is presented in Figure 3B. DN, Dryvax-naïve; DI, Dryvax-immune; C, Controls.
Non-CD8 T cell correlates for reduced levels of colorectal virus
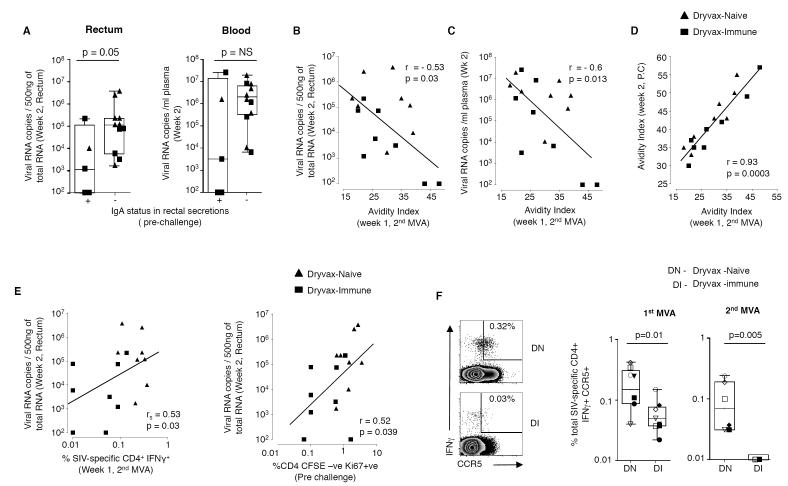
We next investigated the relationship between vaccine-elicited antibody responses, CD4 T cell responses and colorectal virus (Fig. 6). These analyses suggested significant roles for SIV-specific IgA in rectal secretions, the avidity of serum binding Ab to the native form of Env and the magnitude of vaccine-elicited SIV-specific CD4 T cells in the early control of colorectal virus. Animals that were positive for anti-viral IgA in rectal secretions prior to challenge had 96-fold lower levels of virus in colorectal tissue at 2 weeks post challenge than animals that were negative for IgA (Fig. 6A). At 2 weeks after challenge, the SIV-specific IgA responses were present in rectal secretions of all animals that were positive prior to challenge, except for RLr8 that had levels of virus below detection (Table I). At this time, 3 additional animals in the Dryvax-naïve group, 1 additional animal in the Dryvax-immune group and the only spontaneous controller in the unvaccinated control group (RBy9) had developed SIV-specific IgA responses in rectal secretions.
Figure 6. Non-CD8 T cell mediated correlates for enhanced colorectal viral control.
(A) Comparison of levels of virus in the colorectal tissue and blood based on IgA positivity in rectal secretions prior to challenge. Correlation between avidity index following vaccination and viral load (B) in the rectum and (C) blood. (D) Correlation between avidity indices following vaccination and challenge. (E) Correlation between SIV-specific IFNγ secreting CD4 T cells at 1 week after the 2nd MVA boost or SIV Gag-specific proliferating CD4 T cells 4 weeks prior to SIV challenge and levels of colorectal virus at 2 weeks post challenge. (F) Frequency of SIV Gag-specific CCR5+ CD4 T cells. On the left, representative FACS plots. Cells were gated on CD3+ and CD4+. Numbers on the graphs represent the frequency of CCR5 positive cells as a percent of total CD4 T cells. On the right, summary of CCR5+ CD4 T cells following 1st and 2nd MVA boosts. Boxes represent medians with 25th and 75th percentiles for the group. Key to animal names is presented in Figure 3B. DN, Dryvax-naïve; DI, Dryvax-immune. rs, Spearman’s rank correlation.
The avidity of vaccine-elicited antibody for the native form of Env also showed an inverse correlation with viral RNA levels at 2 weeks following challenge both in colorectal tissue (p=0.03) and blood (p=0.013) (Fig. 6B and 6C). As expected, the avidity indices were higher post challenge than post vaccination. A direct correlation was observed for avidity indices post vaccination and post challenge suggesting that animals that had higher avidity antibody post vaccination also had higher avidity antibody post challenge (Fig. 6D). In contrast to the avidity indices where there was a correlation with reduced viremia, the titers of anti-Env binding Ab or neutralizing Ab against T-cell line adapted SIV251 showed direct correlations with plasma viral load suggesting the expansion of these responses to higher levels of viral antigen (p<0.01; data not shown). None of the vaccinated and control animals developed neutralizing activity for non-T cell line adapted SIV251 until 12 weeks post challenge (data not shown).
The frequency of IFNγ secreting virus-specific CD4 T cells at one week following the 2nd MVA boost showed a direct correlation with levels of viral RNA in colorectal tissue at 2 weeks following challenge (Fig. 6E)(p=0.03). A direct correlation was also observed between the frequency of Gag-specific proliferating CD4 T cells at 4 weeks prior to challenge and colorectal virus at 2 weeks following challenge (Fig. 6E)(p=0.04). Furthermore, the frequencies of virus-specific CD4 T cells displaying the CCR5 viral co-receptor were 3-fold higher (p=0.01) after the 1st MVA boost and 7-fold higher (p=0.005) after the 2nd MVA boost in the Dryvax-naive than Dryvax-immune animals (Fig. 6F) demonstrating that preexisting VV immunity had diminished the elicitation of CCR5+ CD4 T cells.
DISCUSSION
Our study evaluating the effect of preexisting VV immunity on cellular and humoral immunity elicited by a DNA/MVA SIV vaccine in rhesus macaques demonstrates that preexisting immunity reduces the magnitude of SIV-specific cellular but not humoral immunity. This was true for both IgG as well as IgA responses in serum. To our knowledge this is the first report demonstrating that preexisting poxvirus immunity does not influence the elicitation of humoral immunity following an intramuscular immunization with a poxvirus vector. These results strongly suggest that preexisting VV immunity may not be a limitation for the induction of antibody responses by recombinant MVA vaccines.
The mechanisms that contributed to the inability of preexisting VV immunity to diminish MVA insert-specific humoral immunity are not clear and occurred despite the presence of high titers of MVA-specific neutralizing Ab and T cell responses at the time of MVA boosts. Previous studies have shown that a relatively small amount of antigen is needed for inducing a strong humoral immune response if the antigen is presented in the form of immune complexes (52-54). We speculate that immune complexes formed between preexisting SIV-specific Ab prior to the boost and low levels of SIV antigen present after the boost contributed to the observed strong boost of SIV-specific humoral immunity in Dryvax-immune animals. If this is true, similar mechanisms may be applicable for other viral vectors.
A critical finding of our study is that preexisting VV immunity does not diminish the ability of a DNA/MVA SIV vaccine to control a pathogenic intrarectal SIV challenge despite reducing the magnitude of SIV-specific cellular immunity. In fact, 4 out of the 8 Dryvax-immune animals controlled the virus below 1000 copies/ml of plasma. This control was not due to the presence of known protective Mamu class I alleles. However, we did observe strong anti-viral CD8 T cell responses post challenge in the Dryvax-immune animals despite the low frequencies of these cells post vaccination. These results suggest that pre-existing immunity could have influenced the functional quality of SIV-specific CD8 T cells elicited by the DNA/MVA vaccine such that they possess enhanced expansion potential.
In our study the level of virus replication in colorectal tissue at 2 weeks post challenge correlated with the tempo of infection in blood. Levels of colorectal virus below 104 copies/500ng of tissue RNA were associated with viremia that peaked at > 3 weeks, whereas levels greater than this were associated with viremia that peaked at 2 weeks. Impressively, the magnitude of peak viremia was 248-fold lower in animals in which viremia peaked at > 3 weeks than 2 weeks. These results demonstrate that early control of virus replication at the colorectal site of challenge correlates with protection and highlight the importance of immunodeficiency virus vaccines achieving early control at mucosal sites of challenge.
Our results suggest that immune mechanisms that block virus infection and reduce the frequency of target cells in colorectal tissue limit virus replication at the site of infection. We identified three factors; anti-viral IgA in prechallenge rectal secretions, the avidity of anti-Env IgG and the level of vaccine-elicited CD4 T cells bearing the CCR5 viral co-receptor that influenced the magnitude of colorectal virus early following challenge. The presence prechallenge of anti-viral IgA in rectal secretions was associated with enhanced control of colorectal virus. The anti-viral IgA in rectal secretions in our assays was directed against Gag-Pol and not Env. Anti-Gag-Pol IgA has been shown to neutralize HIV-1 replication inside epithelial cells (55). More importantly, the anti-Gag-Pol activity may have served as an indicator of protective functions that we do not understand. Consistent with our previous studies with SHIV challenges (39, 56), the avidity of anti-Env IgG for the native form of the SIV Env correlated inversely with the levels of viral RNA in colorectal tissue at 2 weeks post SIV challenge. In contrast to the avidity indices, the titers of anti-Env binding Ab did not correlate with viral control (data not shown). These results suggest that high avidity anti-viral IgG as well as anti-viral IgA contribute to colorectal viral control and highlight the importance of immunodeficiency virus vaccines eliciting these responses. We are yet to study the mechanisms by which high avidity binding Ab and rectal IgA contibute to viral control. However, a recent study demonstrated that higher avidity binding Ab correlates with ADCC and ADCVI activities that in turn correlate with enhanced control of a SHIV challenge (57). In the same study, a direct correlation was also observed between the magnitude of HIV env-specific IgA in rectal secretions and inhibition of transcytosis activity of HIV in vitro. Thus we speculate that high avidity Ab in our study may be working through ADCC and ADCVI activities, and the rectal IgA through inhibition of transcytosis activity.
In our study, low levels of vaccine-elicited CD4 T cells showed a moderate correlation with enhanced control of colorectal virus. While the role of vaccine-elicited CD4 T cells in the initiation of HIV/SIV infection during a mucosal challenge remains to be delineated, there is growing evidence suggesting that these cells preferentially support virus replication (58, 59). Mattapallil et. al., demonstrated a preferential infection of virus-specific CD4 T cells in the blood of vaccinated but not unvaccinated animals at day 7 following SIV challenge (60). In the future, it is important to measure the frequency of virus-specific CD4 T cells in the colorectal tissue at the time and early after challenge. However, a recent study demonstrated the rapid expression of MIP-1β (ligand for CCR5) and massive recruitment of CD4 T cells to the site of infection as early as day 4 following a mucosal challenge (61) suggesting that CCR5+ SIV-specific CD4 T cells from blood and other tissues may be recruited to the mucosal site of infection soon after challenge.
Two of the Dryvax-immune animals showed minimal to undetectable levels of virus infection. We think that this protection was due to vaccine-elicited immunity. Prior to challenge, these animals were the two animals in the Dryvax-immune group with the highest specific activities of anti-viral IgA and the highest avidities of anti-Env Ab. These animals also had low levels of vaccine-induced CD4 T cells. One of the two (RLs8) showed definitive signs of infection including a low level of viremia at 3 weeks post challenge and a transient depletion of colorectal CD4 T cells and anamnestic anti-viral IgG and IgA responses post challenge. The second (RLr8) showed a transient 3-fold expansion of anamnestic anti-viral IgG in serum at 2 weeks post challenge but no detectable levels of viremia. Both animals showed strong anamnestic Gag-specific CD8 and CD4 T cell responses at 2 weeks post challenge. We consider this as potential sterilizing immunity mediated by a combination of high avidity binding Ab, rectal IgA and the presence of low levels of anti-viral CD4 T cell targets for infection.
In conclusion, our results show that preexisting VV immunity does not diminish the control of a pathogenic intrarectal immunodeficiency virus challenge by a DNA/MVA vaccine despite it reducing anti-viral cellular but not humoral responses. Our results show that immune mechanisms mediating early viral control in colorectal tissue enhance control of both acute and chronic phases of immunodeficiency virus infections and highlight the critical need for controlling virus replication at the site of viral challenge. And finally, our results show two vaccine elicited Ab responses, anti-viral mucosal IgA and high avidity anti-Env IgG, contributing to early mucosal viral control.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. J. Altman for provision of Gag CM9 tetramer, Mr. J. Americo for preparation of rMVA vaccines and for MVA neutralization assays and Mrs. Helen Drake-Perrow for outstanding administrative support. We thank Dr. F. Villinger for critical reading of the manuscript. We are thankful to Dr. D. Watkins and Wisconsin National Primate Research Center Genotyping Service for help with Mamu B08 and B17 typing of animals. We are also thankful to the Yerkes Division of Research Resources for the consistently excellent pathology support. We thank Emory CFAR virology core for viral load assays. Also, we thank the NIH AIDS Research and Reference Reagent Program for the provision of peptides.
Grant Support: This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants R01 AI057029 and R01 AI071852 to RRA; Yerkes National Primate Research Center base grant, P51 RR00165; Emory CFAR grant P30 AI050409; and R24 RR16038 to David I. Watkins.
REFERENCES
- 1.Shiver JW, Fu T, Chen L, Casimiro D, Davies ME, Evans RK, Zhang Z-Q, Simon A, Trigona WL, Dubey S, Huang L, Harris VA, Long RS, Xiaoping L, Handt L, Schleif WA, Zhu L, Freed DC, Persaud NV, Guan L, Punt K, Tang A, Chen M, Wilson KA, Collins KB, Heidecker GJ, Perry HC, Joyce JG, Grimm KM, Cook JC, Keller PM, Kresock DS, Mach H, Troutman RD, Iospi LA, Williams DM, Xu Z, Bohannon KE, Volkin DB, Montefiori D, Miura A, Krivulka GR, Lifton MA, Kuroda MJ, Schmitz JE, Letvin NL, Caulfield MJ, Bett AJ, Youil R, Kaslow DC, Emini EA. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency virus immunity. Nature. 2002;415:331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 2.Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans S, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma H-L, Grimm BD, Hulsey ML, Miller J, McClure HM, McNicholl JM, Moss B, Robinson HL. Control of a Mucosal Challenge and Prevention of AIDS by a Multiprotein DNA/MVA Vaccine. Science. 2001;292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 3.Amara RR, Villinger F, Staprans S, Altman JD, Montefiori D, Kozyr NL, Xu Y, Wyatt L, Earl PL, Herndon JG, McClure HM, Moss B, Robinson HL. Different patterns of immune responses but similar control of a mucosal immunodeficiency virus challenge by MVA and DNA/MVA vaccines. J Virol. 2002;76:7625–7631. doi: 10.1128/JVI.76.15.7625-7631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, O’Brien KL, Lynch DM, Simmons NL, Porte A, Riggs A. M. La, Abbink P, Coffey RT, Grandpre LE, Seaman MS, Landucci G, Forthal DN, Montefiori DC, Carville A, Mansfield KG, Havenga MJ, Pau MG, Goudsmit J, Barouch DH. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts DM, Nanda A, Havenga MJ, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner AR, Swanson PE, Gorgone DA, Lifton MA, Lemckert AA, Holterman L, Chen B, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 6.Patterson LJ, Malkevitch N, Venzon D, Pinczewski J, Gomez-Roman VR, Wang L, Kalyanaraman VS, Markham PD, Robey FA, Robert-Guroff M. Protection against mucosal simian immunodeficiency virus SIV(mac251) challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J Virol. 2004;78:2212–2221. doi: 10.1128/JVI.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson J, Chougnet C, Robert-Guroff M, Montefiori D, Markham P, Shearer G, Gallo RC, Cranage M, Paoletti E, Limbach K, Venzon D, Tartaglia J, Franchini G. Recombinant vaccine-induced protection against the highly pathogenic simian immunodeficiency virus SIV(mac251): dependence on route of challenge exposure. Journal of Virology. 1998;72:4170–4182. doi: 10.1128/jvi.72.5.4170-4182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, Tryniszewska E, Lewis MG, VanCott TC, Hirsch V, Woodward R, Gibson A, Grace M, Dobratz E, Markham PD, Hel Z, Nacsa J, Klein M, Tartaglia J, Franchini G. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J Virol. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaur A, Sanford HB, Garry D, Lang S, Klumpp SA, Watanabe D, Bronson RT, Lifson JD, Rosati M, Pavlakis GN, Felber BK, Knipe DM, Desrosiers RC. Ability of herpes simplex virus vectors to boost immune responses to DNA vectors and to protect against challenge by simian immunodeficiency virus. Virology. 2007;357:199–214. doi: 10.1016/j.virol.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Publicover J, Ramsburg E, Rose JK. A Single-Cycle Vaccine Vector Based on Vesicular Stomatitis Virus Can Induce Immune Responses Comparable to Those Generated by a Replication-Competent Vector. J. Virol. 2005;79:13231–13238. doi: 10.1128/JVI.79.21.13231-13238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nwanegbo E, Vardas E, Gao W, Whittle H, Sun H, Rowe D, Robbins PD, Gambotto A. Prevalence of Neutralizing Antibodies to Adenoviral Serotypes 5 and 35 in the Adult Populations of The Gambia, South Africa, and the United States 10.1128/CDLI.11.2.351-357.2004. Clin. Diagn. Lab. Immunol. 2004;11:351–357. doi: 10.1128/CDLI.11.2.351-357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayr A, Hochstein-Mintzel, Stickl H. Abstammung, Eigenschaften und Verwendung des attenuierten Vaccinia-stammes MVA. Infection. 1975;3:6–14. [Google Scholar]
- 13.Rooney JF, Wohlenberg C, Cremer KJ, Moss B, Notkins AL. Immunization with a vaccinia virus recombinant expressing herpes simplex virus type 1 glycoprotein D: long-term protection and effect of revaccination. J Virol. 1988;62:1530–1534. doi: 10.1128/jvi.62.5.1530-1534.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z.-y., Wyatt LS, Kong W.-p., Moodie Z, Moss B, Nabel GJ. Overcoming Immunity to a Viral Vaccine by DNA Priming before Vector Boosting. J. Virol. 2003;77:799–803. doi: 10.1128/JVI.77.1.799-803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belyakov IM, Moss B, Strober W, Berzofsky JA. Mucosal vaccination overcomes the barrier to recombinant vaccinia immunization caused by preexisting poxvirus immunity. PNAS. 1999;96:4512–4517. doi: 10.1073/pnas.96.8.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sumida SM, Truitt DM, Kishko MG, Arthur JC, Jackson SS, Gorgone DA, Lifton MA, Koudstaal W, Pau MG, Kostense S, Havenga MJ, Goudsmit J, Letvin NL, Barouch DH. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J Virol. 2004;78:2666–2673. doi: 10.1128/JVI.78.6.2666-2673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casimiro DR, Chen L, Fu T-M, Evans RK, Caulfield MJ, Davies M-E, Tang A, Chen M, Huang L, Harris V, Freed DC, Wilson KA, Dubey S, Zhu D-M, Nawrocki D, Mach H, Troutman R, Isopi L, Williams D, Hurni W, Xu Z, Smith JG, Wang S, Liu X, Guan L, Long R, Trigona W, Heidecker GJ, Perry HC, Persaud N, Toner TJ, Su Q, Liang X, Youil R, Chastain M, Bett AJ, Volkin DB, Emini EA, Shiver JW. Comparative Immunogenicity in Rhesus Monkeys of DNA Plasmid, Recombinant Vaccinia Virus, and Replication-Defective Adenovirus Vectors Expressing a Human Immunodeficiency Virus Type 1 gag Gene 10.1128/JVI.77.11.6305-6313.2003. J. Virol. 2003;77:6305–6313. doi: 10.1128/JVI.77.11.6305-6313.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts DM, Nanda A, Havenga MJE, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner AR, Swanson PE, Gorgone DA, Lifton MA, Lemckert AAC, Holterman L, Chen B, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. 2006;441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 19.McCoy K, Tatsis N, Korioth-Schmitz B, Lasaro MO, Hensley SE, Lin S-W, Li Y, Giles-Davis W, Cun A, Zhou D, Xiang Z, Letvin NL, Ertl HCJ. Effect of Preexisting Immunity to Adenovirus Human Serotype 5 Antigens on the Immune Responses of Nonhuman Primates to Vaccine Regimens Based on Human- or Chimpanzee-Derived Adenovirus Vectors10.1128/JVI.02497-06. J. Virol. 2007;81:6594–6604. doi: 10.1128/JVI.02497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiver JW, Emini EA. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annu Rev Med. 2004;55:355–372. doi: 10.1146/annurev.med.55.091902.104344. [DOI] [PubMed] [Google Scholar]
- 21.Cooney EL, Collier AC, Greenberg PD, Coombs RW, Zarling J, Arditti DE, Hoffman MC, Hu SL, Corey L. Safety of and immunological response to a recombinant vaccinia virus vaccine expressing HIV envelope glycoprotein [see comments] Lancet. 1991;337:567–572. doi: 10.1016/0140-6736(91)91636-9. [DOI] [PubMed] [Google Scholar]
- 22.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantaleo G. HIV-1 T-cell vaccines: evaluating the next step. The Lancet infectious diseases. 2008;8:82–83. doi: 10.1016/S1473-3099(07)70266-9. [DOI] [PubMed] [Google Scholar]
- 24.Robinson HL, Amara RR. T cell vaccines for microbial infections. Nat Med. 2005;11:S25–32. doi: 10.1038/nm1212. [DOI] [PubMed] [Google Scholar]
- 25.Hanke T, Samuel RV, Blanchard TJ, Neumann VC, Allen TM, Boyson JE, Sharpe SA, Cook N, Smith GL, Watkins DI, Cranage MP, McMichael AJ. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J Virol. 1999;73:7524–7532. doi: 10.1128/jvi.73.9.7524-7532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hel Z, Nacsa J, Tryniszewska E, Tsai WP, Parks RW, Montefiori DC, Felber BK, Tartaglia J, Pavlakis GN, Franchini G. Containment of simian immunodeficiency virus infection in vaccinated macaques: correlation with the magnitude of virus-specific pre- and postchallenge CD4+ and CD8+ T cell responses. J Immunol. 2002;169:4778–4787. doi: 10.4049/jimmunol.169.9.4778. [DOI] [PubMed] [Google Scholar]
- 27.Horton H, Vogel TU, Carter DK, Vielhuber K, Fuller DH, Shipley T, Fuller JT, Kunstman KJ, Sutter G, Montefiori DC, Erfle V, Desrosiers RC, Wilson N, Picker LJ, Wolinsky SM, Wang C, Allison DB, Watkins DI. Immunization of Rhesus Macaques with a DNA Prime/Modified Vaccinia Virus Ankara Boost Regimen Induces Broad Simian Immunodeficiency Virus (SIV)-Specific T-Cell Responses and Reduces Initial Viral Replication but Does Not Prevent Disease Progression following Challenge with Pathogenic SIVmac239. J. Virol. 2002;76:7187–7202. doi: 10.1128/JVI.76.14.7187-7202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dale CJ, De Rose R, Stratov I, Chea S, Montefiori DC, Thomson S, Ramshaw IA, Coupar BE, Boyle DB, Law M, Kent SJ. Efficacy of DNA and fowlpox virus priming/boosting vaccines for simian/human immunodeficiency virus. J Virol. 2004;78:13819–13828. doi: 10.1128/JVI.78.24.13819-13828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, Davies ME, McDermott AB, O’Connor DH, Fridman A, Bagchi A, Tussey LG, Bett AJ, Finnefrock AC, Fu TM, Tang A, Wilson KA, Chen M, Perry HC, Heidecker GJ, Freed DC, Carella A, Punt KS, Sykes KJ, Huang L, Ausensi VI, Bachinsky M, Sadasivan-Nair U, Watkins DI, Emini EA, Shiver JW. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol. 2005;79:15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goonetilleke N, Moore S, Dally L, Winstone N, Cebere I, Mahmoud A, Pinheiro S, Gillespie G, Brown D, Loach V, Roberts J, Guimaraes-Walker A, Hayes P, Loughran K, Smith C, De Bont J, Verlinde C, Vooijs D, Schmidt C, Boaz M, Gilmour J, Fast P, Dorrell L, Hanke T, McMichael AJ. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J Virol. 2006;80:4717–4728. doi: 10.1128/JVI.80.10.4717-4728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson NA, Reed J, Napoe GS, Piaskowski S, Szymanski A, Furlott J, Gonzalez EJ, Yant LJ, Maness NJ, May GE, Soma T, Reynolds MR, Rakasz E, Rudersdorf R, McDermott AB, O’Connor DH, Friedrich TC, Allison DB, Patki A, Picker LJ, Burton DR, Lin J, Huang L, Patel D, Heindecker G, Fan J, Citron M, Horton M, Wang F, Liang X, Shiver JW, Casimiro DR, Watkins DI. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J Virol. 2006;80:5875–5885. doi: 10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harari A, Bart PA, Stohr W, Tapia G, Garcia M, Medjitna-Rais E, Burnet S, Cellerai C, Erlwein O, Barber T, Moog C, Liljestrom P, Wagner R, Wolf H, Kraehenbuhl JP, Esteban M, Heeney J, Frachette MJ, Tartaglia J, McCormack S, Babiker A, Weber J, Pantaleo G. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med. 2008;205:63–77. doi: 10.1084/jem.20071331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amara RR, Smith JM, Staprans S, Montefiori D, Villinger F, Altman JD, O’Neil SP, Kozyr NL, Xu Y, Wyatt L, Earl PL, Herndon JG, McNicholl JM, McClure HM, Moss B, Robinson HL. Critical role for Env as well as Gag-Pol for the control of a pathogenic SHIV challenge by a DNA/rMVA Vaccine. J Virol. 2002;76:6138–6146. doi: 10.1128/JVI.76.12.6138-6146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadagopal S, Amara RR, Montefiori DC, Wyatt LS, Staprans SI, Kozyr NL, McClure HM, Moss B, Robinson HL. Signature for Long-Term Vaccine-Mediated Control of a Simian and Human Immunodeficiency Virus 89.6P Challenge: Stable Low-Breadth and Low-Frequency T-Cell Response Capable of Coproducing Gamma Interferon and Interleukin-2. J. Virol. 2005;79:3243–3253. doi: 10.1128/JVI.79.6.3243-3253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson HL, Sharma S, Zhao J, Kannanganat S, Lai L, Chennareddi L, Yu T, Montefiori DC, Amara RR, Wyatt LS, Moss B. Immunogenicity in macaques of the clinical product for a clade B DNA/MVA HIV vaccine: elicitation of IFN-gamma, IL-2, and TNF-alpha coproducing CD4 and CD8 T cells. AIDS Res Hum Retroviruses. 2007;23:1555–1562. doi: 10.1089/aid.2007.0165. [DOI] [PubMed] [Google Scholar]
- 36.Knapp LA, Lehmann E, Piekarczyk MS, Urvater JA, Watkins DI. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens. 1997;50:657–661. doi: 10.1111/j.1399-0039.1997.tb02927.x. [DOI] [PubMed] [Google Scholar]
- 37.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O’Connor DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai L, Vodros D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, Chennareddi L, Yu T, Kannanganat S, Ofielu L, Villinger F, Wyatt LS, Moss B, Amara RR, Robinson HL. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology. 2007;369:153–167. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Earl PL, Wyatt LS, Montefiori DC, Bilska M, Woodward R, Markham PD, Malley JD, Vogel TU, Allen TM, Watkins DI, Miller N, Moss B. Comparison of vaccine strategies using recombinant env-gag-pol MVA with or without an oligomeric Env protein boost in the SHIV rhesus macaque model. Virology. 2002;294:270–281. doi: 10.1006/viro.2001.1345. [DOI] [PubMed] [Google Scholar]
- 41.Van Rompay KK, Abel K, Lawson JR, Singh RP, Schmidt KA, Evans T, Earl P, Harvey D, Franchini G, Tartaglia J, Montefiori D, Hattangadi S, Moss B, Marthas ML. Attenuated poxvirus-based simian immunodeficiency virus (SIV) vaccines given in infancy partially protect infant and juvenile macaques against repeated oral challenge with virulent SIV. J Acquir Immune Defic Syndr. 2005;38:124–134. doi: 10.1097/00126334-200502010-00002. [DOI] [PubMed] [Google Scholar]
- 42.Kozlowski PA, Lynch RM, Patterson RR, Cu-Uvin S, Flanigan TP, Neutra MR. Modified wick method using Weck-Cel sponges for collection of human rectal secretions and analysis of mucosal HIV antibody. J Acquir Immune Defic Syndr. 2000;24:297–309. doi: 10.1097/00126334-200008010-00001. [DOI] [PubMed] [Google Scholar]
- 43.Velu V, Kannanganat S, Ibegbu C, Chennareddi L, Villinger F, Freeman GJ, Ahmed R, Amara RR. Elevated expression levels of inhibitory receptor programmed death 1 on simian immunodeficiency virus-specific CD8 T cells during chronic infection but not after vaccination. J Virol. 2007;81:5819–5828. doi: 10.1128/JVI.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermont CL, van Dijken HH, van Limpt CJ, de Groot R, van Alphen L, van Den Dobbelsteen GP. Antibody avidity and immunoglobulin G isotype distribution following immunization with a monovalent meningococcal B outer membrane vesicle vaccine. Infect Immun. 2002;70:584–590. doi: 10.1128/IAI.70.2.584-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Earl PL, Americo JL, Moss B. Development and use of a vaccinia virus neutralization assay based on flow cytometric detection of green fluorescent protein. J Virol. 2003;77:10684–10688. doi: 10.1128/JVI.77.19.10684-10688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, DeMars R, Pauza CD, Johnson RP, Sette A, Watkins DI. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. Journal of Immunology. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 48.Smith JM, Amara RR, McClure HM, Patel M, Sharma S, Yi H, Chennareddi L, Herndon JG, Butera ST, Heneine W, Ellenberger DL, Parekh B, Earl PL, Wyatt LS, Moss B, Robinson HL. Multiprotein HIV type 1 clade B DNA/MVA vaccine: construction, safety, and immunogenicity in Macaques. AIDS Res Hum Retroviruses. 2004;20:654–665. doi: 10.1089/0889222041217419. [DOI] [PubMed] [Google Scholar]
- 49.Robinson HL, Montefiori DC, Villinger F, Robinson JE, Sharma S, Wyatt LS, Earl PL, McClure HM, Moss B, Amara RR. Studies on GM-CSF DNA as an adjuvant for neutralizing Ab elicited by a DNA/MVA immunodeficiency virus vaccine. Virology. 2006;352:285–294. doi: 10.1016/j.virol.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 50.Kwissa M, Amara RR, Robinson HL, Moss B, Alkan S, Jabbar A, Villinger F, Pulendran B. Adjuvanting a DNA vaccine with a TLR9 ligand plus Flt3 ligand results in enhanced cellular immunity against the simian immunodeficiency virus. J Exp Med. 2007;204:2733–2746. doi: 10.1084/jem.20071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letvin NL, Mascola JR, Sun Y, Gorgone DA, Buzby AP, Xu L, Yang ZY, Chakrabarti B, Rao SS, Schmitz JE, Montefiori DC, Barker BR, Bookstein FL, Nabel GJ. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkl A, Klaus GG. The generation of memory cells. IV. Immunization with antigen-antibody complexes accelerates the development of B-memory cells, the formation of germinal centres and the maturation of antibody affinity in the secondary response. Immunology. 1981;43:371–378. [PMC free article] [PubMed] [Google Scholar]
- 53.Visciano ML, Tuen M, Gorny MK, Hioe CE. In vivo alteration of humoral responses to HIV-1 envelope glycoprotein gp120 by antibodies to the CD4-binding site of gp120. Virology. 2008;372:409–420. doi: 10.1016/j.virol.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanke T, Botting C, Green EA, Szawlowski PW, Rud E, Randall RE. Expression and purification of nonglycosylated SIV proteins, and their use in induction and detection of SIV-specific immune responses. AIDS Res Hum Retroviruses. 1994;10:665–674. doi: 10.1089/aid.1994.10.665. [DOI] [PubMed] [Google Scholar]
- 55.Wright A, Yan H, Lamm ME, Huang YT. Immunoglobulin A antibodies against internal HIV-1 proteins neutralize HIV-1 replication inside epithelial cells. Virology. 2006;356:165–170. doi: 10.1016/j.virol.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J, Lai L, Amara RR, Montefiori DC, Villinger F, Chennareddi L, Wyatt LS, Moss B, Robinson HL. Preclinical studies of human immunodeficiency virus/AIDS vaccines: inverse correlation between avidity of anti-Env antibodies and peak postchallenge viremia. J Virol. 2009;83:4102–4111. doi: 10.1128/JVI.02173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao P, Zhao J, Patterson LJ, Brocca-Cofano E, Venzon D, Kozlowski PA, Hidajat R, Demberg T, Robert-Guroff M. Multiple Vaccine-Elicited Nonneutralizing Antienvelope Antibody Activities Contribute to Protective Efficacy by Reducing both Acute and Chronic Viremia following Simian/Human Immunodeficiency Virus SHIV89.6P Challenge in Rhesus Macaques. J. Virol. 84:7161–7173. doi: 10.1128/JVI.00410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 59.Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4(+) T cells depletes gut lamina propria CD4(+) T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 60.Mattapallil JJ, Douek DC, Buckler-White A, Montefiori D, Letvin NL, Nabel GJ, Roederer M. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge 10.1084/jem.20060657. J. Exp. Med. 2006;203:1533–1541. doi: 10.1084/jem.20060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, Reilly CS, Peterson ML, Schultz-Darken N, Brunner KG, Nephew KR, Pambuccian S, Lifson JD, Carlis JV, Haase AT. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.