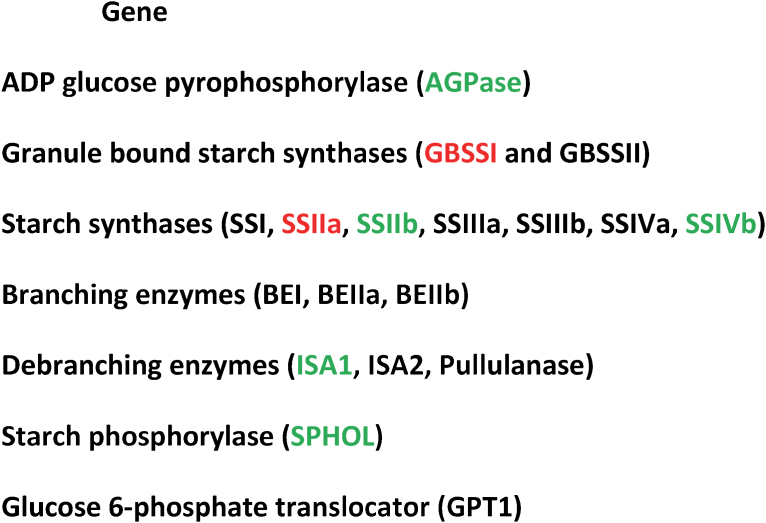
Abstract
Starch is a major component of human diets. The relative contribution of variation in the genes of starch biosynthesis to the nutritional and functional properties of the rice was evaluated in a rice breeding population. Sequencing 18 genes involved in starch synthesis in a population of 233 rice breeding lines discovered 66 functional SNPs in exonic regions. Five genes, AGPS2b, Isoamylase1, SPHOL, SSIIb and SSIVb showed no polymorphism. Association analysis found 31 of the SNP were associated with differences in pasting and cooking quality properties of the rice lines. Two genes appear to be the major loci controlling traits under human selection in rice, GBSSI (waxy gene) and SSIIa. GBSSI influenced amylose content and retrogradation. Other genes contributing to retrogradation were GPT1, SSI, BEI and SSIIIa. SSIIa explained much of the variation in cooking characteristics. Other genes had relatively small effects.
Rice is a major human food composed largely of starch. Starch properties determine the key functional properties of rice such as cooking temperature and influence human health through its contribution to the glycemic index and levels of resistant starch. The incomplete digestion-absorption of resistant starch in the small intestine leads to non-digestible starch fractions with physiological functions similar to dietary fibre with significant beneficial impacts1.
Retrogradation describes the hardening of cooked starch after cooling due to re-crystallization of gelatinized starch components during storage2. It is believed there is a significant correlation between the tendency of any one starch sample to retrograde and its levels of resistant starch. Hence, in this study the term retrograded-resistant rice starch is used. Assessment of the in vivo digestion and structural features of maize, bean and potato flake high amylose resistant-retrograded starch in the ileal contents of four human populations found resistant starch consisted mainly of retrograded amylose with degree of polymerization of approximately 35 glucose units and a melting temperature of 150°C3. Pea, maize, wheat, and potato retrograded amylose are highly resistant to amylolysis and digestibility4. Factors other than amylose content which may have a direct or indirect influence on the rate of starch retrogradation, firmness and resilience of rice starch after cooking are protein and lipid contents5.
High-amylose rice cultivars usually have more resistant starch (RS) and lower estimated glycemic index (EGS), suggesting highly-retrograded cooked rice cultivars tend to a reduction of hydrolysis index (HI) and glycemic index (GI)6. Conversely, starch of low-amylose rices, which have higher HI, are more quickly hydrolysed than intermediate and high-amylose rice (high HI)6,7. Characteristics of high amylose rice cultivars are normally determined by RVA (Rapid Visco Analysis) which are described by parameters such as peak viscosity (PKV), hot paste viscosity (HPV) and cool paste viscosity (CPV).
Seven starch synthesis enzyme classes have been defined, including ADP-glucose pyrophosphorylase (AGPase), granule bound starch synthase (GBSS), starch synthase (SS), branching enzyme (BE), debranching enzyme (DBE), starch phosphorylase (PHO) and glucose 6-phosphate translocator (GPT). These genes/enzymes contribute directly or indirectly to the production of starch granules.
The link between natural variation in particular starch synthesis genes and starch properties is well established in some cases. GBSSI (waxy gene) is primarily responsible for the synthesis of linear chains of glucose molecules found in amylose is the most well characterised cereal grain starch synthesis enzyme. A number of SNP in the rice waxy gene, at the intron/exon 1 junction site, exon 6 and exon 10, impact starch quality8,9,10 by effecting amylose content. The gene encoding starch synthase IIa (SSIIa), alk, is exclusively expressed in the rice endosperm and has been extensively studied in the context of its effect on cooking quality and starch texture11,12. Two SNPs within exon 8, [A/G] and [GC/TT] are significantly associated with rice alkali disintegration and eating quality and starch gelatinisation temperature (GT)13.
More recently, Yan et al. (2010) analysed the association of 17 starch synthesis genes with RVA profile parameters in a collection of 118 glutinous rice accessions using 43 gene-specific molecular markers. They found 10 of 17 starch-related genes have an impact on rapid visco analyzer (RVA) profile parameters. The association analysis revealed pullulanase plays a dominant role in control of PKV, HPV, CPV, breakdown viscosity (BDV), peak time (PKT), and pasting temperature (PT) in glutinous rice. Nine other starch genes had a minor impact on only a few RVA profile parameters. However, RVA parameters such as starch paste viscosity and other starch quality traits may be controlled by a complex genetic system involving many starch-related genes14.
Many induced mutations that have been studied15 result in loss of function or drastically alter starch biosynthesis resulting in poor yielding rice. These studies are useful in understanding the biochemical function of enzymes in starch biosynthesis. However, because the rice varieties are not viable as crop plants these mutants are not directly relevant to rice improvement or the understanding of human selection during domestication that involves more subtle selection for mutants that do not impact adversely on productivity. In this study we have worked with material within a rice breeding program to explore the diversity that is available in the domesticated genepool. With the exception of GBSSI and SSIIa, most studies of the molecular basis of starch synthesis have focused on comparison of gene-deficient mutants15 rather than analysis of allelic diversity, perhaps in part due to a lack of high-throughput technologies to discover and analyse new variants in diverse populations. Single nucleotide polymorphisms (SNP) are the most abundant type of genetic variation found within all species and many important plant traits and human diseases are attributed to these sequence variations16. Identifying SNP and associating them with grain starch quality advances our understanding of the starch biosynthesis pathway and highlights ways to improve crops that are higher yielding and of better quality, directly impacting food security and human nutrition and health.
Massively parallel sequencing (MPS) technology is a high-throughput platform for genetic analysis based on ultra deep DNA sequencing17. Kharabian-Masouleh et al. (2011) discovered more than 501 SNPs and 113 In/dels in 17 starch synthesis genes in an Australian rice breeding population using a combination of a target-pooled long range PCR and MPS. By combining MPS with high throughput genotyping technologies such as multiplexed-MALDI-TOF (Sequenom), rapid polymorphism discovery followed by association analysis is now possible18.
In this study we investigated the role of 18 starch-related genes and their SNPs by assessing their contribution to variation in starch properties in a rice breeding population. We report a novel SNP in Glucose-6-Phosphate Translocator 1 (GPT1) gene which is associated with amylose content and retrogradation rate of resistant starch and establish an explicit-coherent gene by gene approach to unveil association of 18 starch-related genes and their SNP polymorphisms with rice starch physiochemical properties.
Results
Assays for 66 SNPs were designed and 233 individuals genotyped. The identification of genes (Figure 1) and code and coordinate of all SNPs studied appears in Table 1. SNP IDs starting with TBG or TBU were extracted from databases and the remainder, mainly starting with GA, were reported by Kharabian-Masouleh et al. (2011). No functional polymorphisms were found in this population in AGPS2b, SPHOL, SSIIb, SSIVb, ISA1 suggesting these genes have no effect on the phenotypes investigated in this population. A gene by gene approach was applied to find associations between individual genes and physiochemical and quality-related properties of rice grain.
Figure 1. Genes associated with variation in rice starch properties in a population of 233 Australian rice breeding lines.
The genes in red are those most correlated with starch properties, those in green do not explain variation in starch properties while genes in black have low to medium effects on rice starch quality.
Table 1. Name and characteristics of SNPs genotyped in 18 rice starch-related genes in a population of 233 Australian rice breeding lines.
| No | Gene | SNP ID* | Coordinates on gDNA | Expected SNP | SNP Assayed† | Association with Physiochemical traits | Status |
|---|---|---|---|---|---|---|---|
| 1 | AGPS2b | TBGU388647 | 233 | G/T | G/G | N/A | No polymorphism |
| 2 | AGPS2b | TBGI050742 | 1507 | T/C | T/T | N/A | No polymorphism |
| 3 | SPHOL | TBGU168031 | 2501 | G/T | G/G | N/A | No polymorphism |
| 4 | SPHOL | TBGU168032 | 2920 | C/T | C/C | N/A | No polymorphism |
| 5 | SPHOL | TBGU168027 | 1001 | C/A | C/C | N/A | No polymorphism |
| 6 | SPHOL | TBGU168024 | 176 | G/T | G/G | N/A | No polymorphism |
| 7 | SPHOL | TBGU168039 | 5514 | G/T | G/G | N/A | No polymorphism |
| 8 | GBSSI | WAXYEXIN1 | 246 | T/G | T/G | P1,BD,FV,SB,MT,AC,PN | Highly associated |
| 9 | GBSSI | WAXYEX6 | 2494 | A/C | A/C | SB,BD,MT,AC | Highly associated |
| 10 | GBSSI | WAXYEX10 | 3486 | C/T | C/T | T1,FV,SB,MT,AC,PN | Highly associated |
| 11 | GBSSII | GBSSII_GA_1638 | 1638 | G/A | G/A | PT, GT | Low-Medium association |
| 12 | SSI | TBGU272768 | 5153 | T/C | T/C | FV,SB,MT | Low-Medium association |
| 13 | SSIIa | SSIIa_GA_Ref631 | 631 | G/T | G/T | N/A | No association |
| 14 | SSIIa | ALKSSIIA4 | 4827–4828 | GC/TT | GC/TT | BDV,SB,PKT,PT,GT,CHK | Highly associated |
| 15 | SSIIb | TBGU116115 | 3416 | A/G | A/A | N/A | No polymorphism |
| 16 | SSIIb | TBGU116120 | 3948 | G/C | G/G | N/A | No polymorphism |
| 17 | SSIIb | TBGU116121 | 3979 | T/C | T/T | N/A | No polymorphism |
| 18 | SSIIb | TBGU116109 | 330 | G/A | A/A | N/A | No polymorphism |
| 19 | SSIIb | TBGU116119 | 3946 | C/T | C/C | N/A | No polymorphism |
| 20 | SSIIb | TBGU116116 | 3487 | T/G | T/T | N/A | No polymorphism |
| 21 | SSIIIa | GA_Ref1058 | 1058 | T/A | T/A | PT,MT, | Low-Medium association |
| 22 | SSIIIa | GA_Ref1680 | 1680 | G/A | G/A | SB,PT,MT,AC,PN,GT | Low associated |
| 23 | SSIIIa | GA_Ref3136 | 3136 | G/A | G/A | N/A | No association |
| 24 | SSIIIa | GA_Ref3391 | 3391 | T/A | T/A | N/A | No association |
| 25 | SSIIIa | GA_Ref3559 | 3559 | T/A | T/A | CHK | Low association |
| 26 | SSIIIa | GA_Ref4384 | 4384 | G/A | G/A | N/A | No association |
| 27 | SSIIIa | GA_Ref1379 | 1379 | A/C | A/C | FV,SB,PT,MT,AC,PN | Low-Medium association |
| 28 | SSIIIa | GA_Ref1708 | 1708 | G/A | G/A | MT,AC,PN,GT | Low-Medium association |
| 29 | SSIIIa | GA_Ref3274 | 3274 | G/A | G/A | N/A | No association |
| 30 | SSIIIa | GA_Ref6242 | 6242 | T/C | T/C | N/A | No association |
| 31 | SSIIIa | GA_Ref1457 | 1457 | A/C | A/C | N/A | No association |
| 32 | SSIIIa | GA_Ref1615 | 1615 | C/T | C/T | N/A | No association |
| 33 | SSIIIa | GA_Ref1834 | 1834 | C/T | C/T | N/A | No association |
| 34 | SSIIIa | GA_Ref2758 | 2758 | G/A | G/A | N/A | No association |
| 35 | SSIIIa | GA_Ref1722ER | 1722 | G/A | G/A | FV,SB,PT,MT,AC,PN,GT | Low-Medium association |
| 36 | SSIIIa | GA_Ref2488 | 2488 | C/T | C/T | N/A | No association |
| 37 | SSIIIa | GA_Ref3073 | 3073 | G/A | G/A | N/A | No association |
| 38 | SSIIIa | GA_Ref1357 | 1357 | G/A | G/A | MT | No association |
| 39 | SSIIIa | GA_Ref2080 | 2080 | C/T | C/T | N/A | No association |
| 40 | SSIIIa | GA_Ref3481 | 3481 | G/A | G/A | N/A | No association |
| 41 | SSIIIa | GA_Ref5466 | 5466 | G/A | G/A | FV,SB,PT,MT,AC,PN, | Low-Medium association |
| 42 | SSIIIa | GA_Ref10761 | 10761 | C/T | C/T | PT | Low association |
| 43 | SSIIIb | GA_Ref1315 | 1315 | T/C | T/C | PT | Medium association |
| 44 | SSIIIb | GA_Ref4543 | 4543 | C/A | C/A | PT | Medium association |
| 45 | SSIIIb | GA_Ref5451 | 5451 | T/C | T/C | PT | Medium association |
| 46 | SSIIIb | GA_Ref7232 | 3232 | T/G | T/G | PT | Medium-High association |
| 47 | SSIIIb | GA_Ref7255ER | 7255 | C/A | C/A | PKV | Medium association |
| 48 | SSIIIb | GA_Ref7437 | 7437 | A/C | A/C | PT | Low-Medium association |
| 49 | SSIVa | GA_Ref4048 | 4048 | C/T | C/T | PT,GT | Low-Medium association |
| 50 | SSIVa | GA_Ref7160 | 7160 | A/G | A/G | PKT,PT,AC,PN,GT | Low-Medium association |
| 51 | SSIVa | GA_Ref7506 | 7506 | A/T | A/T | PT,GT | Low-Medium association |
| 52 | SSIVa | GA_Ref7823 | 7823 | T/C | T/C | PT,GT | Low-Medium association |
| 53 | SSIVa | GA_Ref8383 | 8383 | C/A | C/A | PT,GT | Medium association |
| 54 | SSIVb | TBGU260749 | 5090 | G/C | G/G | N/A | No polymorphism |
| 55 | SSIVb | TBGU260765 | 9525 | G/A | G/G | N/A | No polymorphism |
| 56 | BEI | GA_Ref1558 | 1558 | C/T | C/T | PV,BDV,FV,SB,PT,MT,AC,PN | Low-Medium association |
| 57 | BEIIa | GA_Ref3266 | 3266 | T/G | T/G | N/A | No association |
| 58 | BEIIb | GA_Ref9035 | 9035 | C/T | C/T | N/A | No association |
| 59 | BEIIb | GA_Ref10068 | 10068 | C/A | C/A | N/A | No association |
| 60 | ISA1 | TBGU362347 | 1748 | G/A | G/G | N/A | No polymorphism |
| 61 | ISA1 | TBGU362346 | 1746 | C/G | C/C | N/A | No polymorphism |
| 62 | ISA2 | Iso2_GA_Ref960 | 960 | T/C | T/C | BDV, PT, CHK | Low association |
| 63 | ISA2 | Iso2_GA_Ref1712 | 1712 | C/A | C/A | BDV, PT, CHK | Low association |
| 64 | Pullulanase | TBGU185983 | 1938 | G/A | G/A | PT, GT | Low association |
| 65 | Pullulanase | TBGU185989 | 2380 | T/C | T/C | CHK | Low association |
| 66 | GPT1 | GPT1_GA_Ref_1188 | 1188 | T/C | T/C | AC, MT,BD,FV,SB | Highly associated |
*SNP identification can be found from Kharabian-Masouleh et al., 2011 (starting with GA code) or OryzaSNP MSU database (http://oryzasnp.plantbiology.msu.edu/) starting with TBG or TBU codes.
†Homozygosity of SNP calls mean no polymorphism in the corresponding allele.
MT = Martin test (retrogradation), PN = Predicted Nitrogen, CHK = Chalkiness (%).
GBSSI (Granule bound starch synthase I)
There was a strong correlation between the G/T SNP at the exon1/intron1 boundary and the RVA curve characteristics of PKV and BDV (Table 1 and Table 2). The highest F-value in this experiment was for this SNP and retrogradation rate (Martin test) (F-value = 223.29) and amylose content (F-value = 121.52). The R2 value for retrogradation and amylose content were 0.66 and 0.51, respectively. The second SNP in GBSSI associated with grain properties was the C/T SNP at co-ordinate 3486 (exon 10) which creates a P→S substitution and has a significant association with trough and final viscosity (FV), set back, retrogradation (Martin test) and amylose content. The R2 value for retrogradation and amylose content were 0.39 and 0.16, respectively.
Table 2. Association of 18 rice starch-related genes with rice starch physico-chemical traits in a population of 233 Australian rice breeding lines.
| Gene | Trait | Locus/SNP | F-test | p-adjusted value | R2_Marker | |
|---|---|---|---|---|---|---|
| AGPS2b | Section 1 | No functional polymorphism found in this gene | - | - | - | |
| SPHOL | Section 2 | No functional polymorphism found in this gene | - | - | - | |
| GBSSI | Section 3 | Peak1 | WAXYEXIN1 | 34.346 | 9.99E-04 | 0.23 |
| Trough1 | WAXYEX10 | 36.9498 | 9.99E-04 | 0.1384 | ||
| Breakdown | WAXYEXIN1 | 35.1893 | 9.99E-04 | 0.2343 | ||
| Breakdown | WAXYEX10 | 18.9223 | 9.99E-04 | 0.076 | ||
| Final Viscosity | WAXYEXIN1 | 15.0534 | 9.99E-04 | 0.1157 | ||
| Final Viscosity | WAXYEX10 | 106.068 | 9.99E-04 | 0.3156 | ||
| Setback | WAXYEXIN1 | 76.2739 | 9.99E-04 | 0.3988 | ||
| Setback | WAXYEX10 | 59.8068 | 9.99E-04 | 0.2064 | ||
| Martin_N | WAXYEXIN1 | 223.294 | 9.99E-04 | 0.6601 | ||
| Martin_N | WAXYEX10 | 147.783 | 9.99E-04 | 0.3912 | ||
| Martin_N | WAXYEX6 | 16.8014 | 9.99E-04 | 0.0681 | ||
| AC_percent | WAXYEXIN1 | 121.53 | 9.99E-04 | 0.5138 | ||
| AC_percent | WAXYEX10 | 44.0661 | 9.99E-04 | 0.1608 | ||
| AC_percent | WAXYEX6 | 16.2252 | 9.99E-04 | 0.0659 | ||
| predicted_N | WAXYEXIN1 | 121.543 | 9.99E-04 | 0.5138 | ||
| predicted_N | WAXYEX10 | 43.967 | 9.99E-04 | 0.1605 | ||
| predicted_N | WAXYEX6 | 16.3841 | 9.99E-04 | 0.0665 | ||
| GBSSII | Section 4 | Past_temp | GBSSII_GA_Ref1638 | 27.8519 | 9.99E-04 | 0.2028 |
| GT | GBSSII_GA_Ref1638 | 9.7254 | 9.99E-04 | 0.0938 | ||
| SSI | Section 5 | Trough1 | SSI_TBGU272768_5153 | 14.2713 | 9.99E-04 | 0.0592 |
| FinalVisc | SSI_TBGU272768_5153 | 43.6138 | 9.99E-04 | 0.1612 | ||
| Setback | SSI_TBGU272768_5153 | 28.8805 | 9.99E-04 | 0.1129 | ||
| Martin_N | SSI_TBGU272768_5153 | 45.7145 | 9.99E-04 | 0.1676 | ||
| AC_percent | SSI_TBGU272768_5153 | 20.5891 | 9.99E-04 | 0.0832 | ||
| predicted_N | SSI_TBGU272768_5153 | 20.4244 | 9.99E-04 | 0.0825 | ||
| SSIIa | Section 6 | Breakdown | ALKSSIIA4 | 22.4536 | 9.99E-04 | 0.1682 |
| PeakTime | ALKSSIIA4 | 53.0867 | 9.99E-04 | 0.3235 | ||
| Past_temp | ALKSSIIA4 | 199.652 | 9.99E-04 | 0.6427 | ||
| GT | ALKSSIIA4 | 32.806 | 9.99E-04 | 0.2547 | ||
| Chalk% | ALKSSIIA4 | 8.9273 | 9.99E-04 | 0.0744 | ||
| SSIIb | Section 7 | No functional polymorphism found in this gene | - | - | - | |
| SSIIIa | Section 8 | FinalVisc | SSIIIa_GA_Ref1379 | 9.0413 | 9.99E-04 | 0.0753 |
| FinalVisc | SSIIIa_GA_Ref1722ER | 8.8028 | 9.99E-04 | 0.0723 | ||
| FinalVisc | SSIIIa_GA_Ref5466 | 8.9423 | 9.99E-04 | 0.0736 | ||
| Setback | SSIIIa_GA_Ref1680 | 7.8821 | 9.99E-04 | 0.0655 | ||
| Setback | SSIIIa_GA_Ref1379 | 11.6269 | 9.99E-04 | 0.0948 | ||
| Setback | SSIIIa_GA_Ref1722ER | 10.1037 | 9.99E-04 | 0.0821 | ||
| Setback | SSIIIa_GA_Ref5466 | 9.0543 | 9.99E-04 | 0.0745 | ||
| Past_temp | SSIIIa_GA_Ref1058 | 8.7158 | 9.99E-04 | 0.0722 | ||
| Past_temp | SSIIIa_GA_Ref1680 | 7.4574 | 9.99E-04 | 0.0622 | ||
| Past_temp | SSIIIa_GA_Ref1379 | 7.9273 | 9.99E-04 | 0.0667 | ||
| Past_temp | SSIIIa_GA_Ref1722ER | 9.3315 | 9.99E-04 | 0.0763 | ||
| Past_temp | SSIIIa_GA_Ref10761 | 7.2026 | 9.99E-04 | 0.062 | ||
| Past_temp | SSIIIa_GA_Ref5466 | 8.756 | 9.99E-04 | 0.0722 | ||
| Martin_N | SSIIIa_GA_Ref1058 | 13.2478 | 9.99E-04 | 0.1058 | ||
| Martin_N | SSIIIa_GA_Ref1680 | 20.8545 | 9.99E-04 | 0.1564 | ||
| Martin_N | SSIIIa_GA_Ref1379 | 27.7893 | 9.99E-04 | 0.2002 | ||
| Martin_N | SSIIIa_GA_Ref1708 | 16.5211 | 9.99E-04 | 0.1301 | ||
| Martin_N | SSIIIa_GA_Ref1722ER | 20.6652 | 9.99E-04 | 0.1546 | ||
| Martin_N | SSIIIa_GA_Ref1357 | 7.6136 | 9.99E-04 | 0.0639 | ||
| Martin_N | SSIIIa_GA_Ref5466 | 20.4182 | 9.99E-04 | 0.1536 | ||
| AC_percent | SSIIIa_GA_Ref1680 | 10.3167 | 9.99E-04 | 0.084 | ||
| AC_percent | SSIIIa_GA_Ref1379 | 14.2201 | 9.99E-04 | 0.1136 | ||
| AC_percent | SSIIIa_GA_Ref1708 | 9.3351 | 9.99E-04 | 0.0779 | ||
| AC_percent | SSIIIa_GA_Ref1722ER | 10.6866 | 9.99E-04 | 0.0864 | ||
| AC_percent | SSIIIa_GA_Ref5466 | 11.1556 | 9.99E-04 | 0.0902 | ||
| predicted_N | SSIIIa_GA_Ref1680 | 10.2716 | 9.99E-04 | 0.0837 | ||
| predicted_N | SSIIIa_GA_Ref1379 | 14.2099 | 9.99E-04 | 0.1135 | ||
| predicted_N | SSIIIa_GA_Ref1708 | 9.3091 | 9.99E-04 | 0.0777 | ||
| predicted_N | SSIIIa_GA_Ref1722ER | 10.6615 | 9.99E-04 | 0.0862 | ||
| predicted_N | SSIIIa_GA_Ref5466 | 11.1281 | 9.99E-04 | 0.09 | ||
| GT | SSIIIa_GA_Ref1680 | 10.0271 | 9.99E-04 | 0.0946 | ||
| GT | SSIIIa_GA_Ref1708 | 30.2791 | 9.99E-04 | 0.2436 | ||
| GT | SSIIIa_GA_Ref1722ER | 15.2535 | 9.99E-04 | 0.1365 | ||
| Chalk% | SSIIIa_GA_Ref3559 | 8.9878 | 9.99E-04 | 0.0821 | ||
| SSIIIb | Section 9 | Peak Viscosity | SSIIIb_GA_Ref7255ER | 7.7442 | 9.99E-04 | 0.0666 |
| Past_temp | SSIIIb_GA_Ref4543 | 21.3553 | 9.99E-04 | 0.2251 | ||
| Past_temp | SSIIIb_GA_Ref5451 | 23.0673 | 9.99E-04 | 0.176 | ||
| Past_temp | SSIIIb_GA_Ref1315 | 25.0653 | 9.99E-04 | 0.1849 | ||
| Past_temp | SSIIIb_GA_Ref7232 | 41.4018 | 9.99E-04 | 0.3151 | ||
| Past_temp | SSIIIb_GA_Ref7255ER | 29.1937 | 9.99E-04 | 0.212 | ||
| Past_temp | SSIIIb_GA_Ref7437 | 21.0809 | 9.99E-04 | 0.1572 | ||
| SSIVa | Section 10 | PeakTime | SSIva_GA_Ref7160 | 10.7899 | 9.99E-04 | 0.0875 |
| Past_temp | SSIva_GA_Ref4048 | 27.6864 | 9.99E-04 | 0.1989 | ||
| Past_temp | SSIva_GA_Ref7160 | 39.5053 | 9.99E-04 | 0.2599 | ||
| Past_temp | SSIva_GA_Ref7823 | 30.2856 | 9.99E-04 | 0.2159 | ||
| Past_temp | SSIva_GA_Ref8383 | 30.8007 | 9.99E-04 | 0.2227 | ||
| Past_temp | SSIva_GA_Ref7506 | 29.3874 | 9.99E-04 | 0.205 | ||
| AC_percent | SSIva_GA_Ref7160 | 9.1222 | 9.99E-04 | 0.075 | ||
| predicted_N | SSIva_GA_Ref7160 | 9.077 | 9.99E-04 | 0.0747 | ||
| GT | SSIva_GA_Ref4048 | 8.5371 | 9.99E-04 | 0.0825 | ||
| GT | SSIva_GA_Ref7160 | 19.7873 | 9.99E-04 | 0.1709 | ||
| GT | SSIva_GA_Ref7823 | 10.209 | 9.99E-04 | 0.098 | ||
| GT | SSIva_GA_Ref8383 | 10.4426 | 9.99E-04 | 0.1014 | ||
| GT | SSIva_GA_Ref7506 | 10.6137 | 9.99E-04 | 0.0982 | ||
| SSIVb | Section 11 | No polymorphism detected in this gene | ||||
| BEI | Section 12 | Peak Viscosity | BEI_GA_Ref1558 | 9.5546 | 9.99E-04 | 0.0796 |
| Breakdown | BEI_GA_Ref1558 | 11.2003 | 9.99E-04 | 0.092 | ||
| FinalViscosity | BEI_GA_Ref1558 | 13.0129 | 9.99E-04 | 0.1054 | ||
| Setback Viscosity | BEI_GA_Ref1558 | 32.1812 | 9.99E-04 | 0.2255 | ||
| Past_temp | BEI_GA_Ref1558 | 8.4131 | 9.99E-04 | 0.0708 | ||
| Martin_N | BEI_GA_Ref1558 | 34.5608 | 9.99E-04 | 0.2383 | ||
| AC_percent | BEI_GA_Ref1558 | 38.8652 | 9.99E-04 | 0.2602 | ||
| predicted_N | BEI_GA_Ref1558 | 39.1031 | 9.99E-04 | 0.2614 | ||
| BEIIa | Section 13 | No significant association was observed with starch traits | - | - | - | |
| BEIIb | Section 14 | No significant association was observed with starch traits | - | - | - | |
| Iso1 | Section 15 | No polymorphism detected in this gene | - | - | - | |
| Iso2 | Section 16 | Breakdown | Iso2_GA_Ref1712 | 8.2378 | 9.99E-04 | 0.0768 |
| Breakdown | Iso2_GA_Ref960 | 9.0028 | 9.99E-04 | 0.076 | ||
| Past_temp | Iso2_GA_Ref1712 | 7.8355 | 9.99E-04 | 0.0733 | ||
| Past_temp | Iso2_GA_Ref960 | 7.8341 | 9.99E-04 | 0.0668 | ||
| Chalk% | Iso2_GA_Ref1712 | 7.2855 | 9.99E-04 | 0.0685 | ||
| Chalk% | Is02_GA_Ref960 | 8.2391 | 9.99E-04 | 0.07 | ||
| Pullulanase | Section 17 | Past_temp | Pullu_TBGU185983_1938 | 23.5989 | 9.99E-04 | 0.1747 |
| GT | Pullu_TBGU185983_1938 | 19.1496 | 9.99E-04 | 0.167 | ||
| Chalk% | Pullu_TBGU185989_2380 | 7.5266 | 9.99E-04 | 0.0666 | ||
| GPT1 | Section 18 | Peak Viscosity | GPT1_GA_Ref_1188 | 21.1979 | 9.99E-04 | 0.092 |
| Break down | GPT1_GA_Ref_1188 | 37.1798 | 9.99E-04 | 0.148 | ||
| Final viscosity | GPT1_GA_Ref_1188 | 31.1074 | 9.99E-04 | 0.126 | ||
| Set back viscosity | GPT1_GA_Ref_1188 | 83.2826 | 9.99E-04 | 0.282 | ||
| Martin_N | GPT1_GA_Ref_1188 | 292.143 | 9.99E-04 | 0.577 | ||
| AC_percent | GPT1_GA_Ref_1188 | 123.047 | 9.99E-04 | 0.365 | ||
| Predicted N | GPT1_GA_Ref_1188 | 122.543 | 9.99E-04 | 0.364 | ||
The exon 6 SNP also revealed some significant association according to p-values≤ 0.01 but did not show any remarkable F and R2 values (which suggest it has little control on critical pasting properties). In combination, the results suggest this gene is responsible for determining a significant proportion of the variation in retrograded-resistant rice starch.
GBSSII (Granule bound starch synthase II)
GBSSII synthesises amylose and is found exclusively bound to starch granules in green tissues. During pre-heading, about 1–3 days after flowering, this gene/enzyme is expressed in leaf, leaf sheaths, culm, and pericarp tissue at a low level19. The synthesised amylose is subsequently consumed by the plant or mobilised to the endosperm20. One non-synonymous SNP (nsSNP) found at position 1638 of this gene was tested for association with starch physiochemical traits (Table 1). Only one association with PT with R2 value of 0.20 was observed for this SNP, although some minor association also calculated with GT and Peak time (Table 2).
SSI
Only one 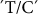 nsSNP at position 5153 of this gene showed minor associations with FV, SB and Martin test (MT), with R2 values of 0.16, 0.11, 0.16, respectively (Table 1).
nsSNP at position 5153 of this gene showed minor associations with FV, SB and Martin test (MT), with R2 values of 0.16, 0.11, 0.16, respectively (Table 1).
SSIIa
Highly significant associations were found between SNP of SSIIa and PT, peak time (PKT), GT and breakdown viscosity. The highest F-test value of 199.65 was observed for the [GC/TT] SNP at position 4827–4828 of SSIIa and PT. This SNP is associated with PT, PKT and BDV with R2 values of 0.642, 0.323 and 0.168, respectively. This SNP has one of the strongest associations among the physiochemical properties studied in this rice population (R2 = 0.642). The G/T SNP at position 631 showed no singnificant association with any traits.
SSIIIa
The highest polymorphism was observed in this gene with 22 SNPs in the coding region causing amino acid changes. Polymorphism in this gene showed association with a FV, SB, PT, MT, AC, predicted N, GT and chalkiness. However, most revealed very low R2values of less than 0.1, indicating that although they are associated, they do not have a highly significant effect on physiochemical properties (Table 2). The highest R2 values for GT, MT and AC were 0.243, 0.200, and 0.113, respectively (Table 2).
SSIIIb
The main effect of SSIIb was observed on PT. Associations were found between 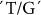 and
and 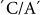 SNPs at positions 7232 and 4543 with R2 values of 0.315 and 0.225, respectively. These relatively high R2 values suggest SNPs in the coding regions of this gene influence PT, although a minor association was found with peak viscosity (PKV). These SNPs at positions 207 and 756 alter the corresponding amino acids Lys→Asn and Ser→Ile, respectively. This gene is a major gene contributing to PT as some other SNPs also exhibited significant associations with PT (Table 2).
SNPs at positions 7232 and 4543 with R2 values of 0.315 and 0.225, respectively. These relatively high R2 values suggest SNPs in the coding regions of this gene influence PT, although a minor association was found with peak viscosity (PKV). These SNPs at positions 207 and 756 alter the corresponding amino acids Lys→Asn and Ser→Ile, respectively. This gene is a major gene contributing to PT as some other SNPs also exhibited significant associations with PT (Table 2).
SSIVa
Five SNPs were examined in this gene (Table 3), of which four showed significant association with PT (Table 2). There was a relatively high R2 of 0.259 for the functional 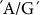 SNP at position 7160, which influences PT. In addition, four other SNPs, with R2 values ranging from 0.198–0.222, also have an influence on PT. A large portion of phenotypic variation of PT in this rice population seems to be explained by SNP in SSIVa. Some minor associations were observed with GT, PKT, AC and predicted nitrogen (PN). SSIIIb and SSIVa in combination contribute to PT in this rice population.
SNP at position 7160, which influences PT. In addition, four other SNPs, with R2 values ranging from 0.198–0.222, also have an influence on PT. A large portion of phenotypic variation of PT in this rice population seems to be explained by SNP in SSIVa. Some minor associations were observed with GT, PKT, AC and predicted nitrogen (PN). SSIIIb and SSIVa in combination contribute to PT in this rice population.
Table 3. Range of phenotypic values (variation) for measured physiochemical properties in 233 Australian rice genotypes.
| Traits | Range |
|---|---|
| Peak 1 | 2168–3669 |
| Trough 1 | 1312–2372 |
| Breakdown Viscosity | 667–1913 |
| Final Viscosity | 2560–4386 |
| Set Back | −658–+1203 |
| Peak Time | 5.7–6.3 |
| Pasting Temperature | 65.65–78.40 |
| Martin Test (N) | 0.405–3.612 |
| Amylose content (%) | 14.10–28.85 |
| Predicted N (N) | 0.31–1.82 |
| Gelatinization Temperature (°C) | 62.00–82.98 |
| Chalkiness (%) | 0.709–44.55 |
SSIVb
No polymorphism was detected in this gene. Therefore, it could be concluded that there is no association between this gene and the studied traits in this population.
BEI
Only one C/T SNP at position 1558 of this gene was discovered21. Nine out of 13 studied physiochemical traits were associated with this SNP at a medium level with the highest R2 values observed for AC, MT, SB and FV. The relatively high R2 values of 0.260 and 0.238 for AC and MT respectively suggests this gene has a prominent effect on amylose content and retrogradation. Minor associations were also found between this SNP and PV, BDV and FV (Table 2).
BEIIa
BEIIa is a leaf expressed gene involved in amylopectin synthesis. The 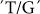 SNP at position 3266 displayed no significant association, confirming BEIIa as a green tissue-specific gene with no impact on grain starch properties (Table 2).
SNP at position 3266 displayed no significant association, confirming BEIIa as a green tissue-specific gene with no impact on grain starch properties (Table 2).
BEIIb
BEIIb is known as amylose extender (ae) in maize and other cereals (Yun and Matheson, 1993). Two SNPs in this gene were examined (Table 2) but no significant association was found with grain starch properties in this population (Table 1).
ISA1 (Isoamylase 1)
No polymorphism was detected in ISA1 in this population.
ISA2 (Isoamylase 2)
Two SNPs were assessed in this gene and all R2 values were less than 0.1, indicating a very low association with breakdown viscosity and chalkiness traits (Table 2).
Pullulanase
A recent association study between pullulanase and RVA profile parameters in glutinous rice has shown strong relations of this gene with PKV, HPV, BDV, PKT14. In this study only weak associations with the two assayed SNPs in pullulanase, PT, GT and CHK with R2 values of 0.174, 0.167 and 0.066, respectively were found (Table 2).
GPT1 (Glucose-6-Phosphate Translocator)
For the first time we report that the GPT1 gene, early in the biochemical pathway of starch synthesis, encoding the glucose-6-phosphate translocator enzyme, has a major association with resistant starch production in rice. A 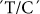 SNP at position 1188 of the GPT1 gene, alters Leu24 to Phe, and is highly associated with resistant-retrograded starch and amylose content (Table 2). The
SNP at position 1188 of the GPT1 gene, alters Leu24 to Phe, and is highly associated with resistant-retrograded starch and amylose content (Table 2). The 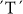 and
and 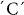 alleles produce high and low levels of retrograded starch, respectively. An association study of 233 genotypes demonstrated a highly significant correlation (R2) of 0.577 and 0.365 (P = 0.00099) between this SNP and retrogradation degree and apparent amylose content, respectively (Table 2).
alleles produce high and low levels of retrograded starch, respectively. An association study of 233 genotypes demonstrated a highly significant correlation (R2) of 0.577 and 0.365 (P = 0.00099) between this SNP and retrogradation degree and apparent amylose content, respectively (Table 2).
Discussion
GBSSI and SSIIa are major genes involved in many grain quality properties such as amylose content and gelatinization temperature (Figure 1 and Figure 2). Highly significant associations were found between GBSSI and retrogradation and amylose content although this gene showed more significant relations with properties such as BDV, SB and FV. A number of authors have already reported the importance of this enzyme in determining the starch physiochemical properties in rice and other cereals. SNPs at the intron/exon 1 junction site, exon 6 and 10 in rice GBSSI (waxy gene) have the most significant impact on amylose content and by extension, starch quality8,9,10. This study confirms the 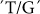 SNP at the intron1/exon1 junction site has a major influence on a number of physiochemical properties.
SNP at the intron1/exon1 junction site has a major influence on a number of physiochemical properties.
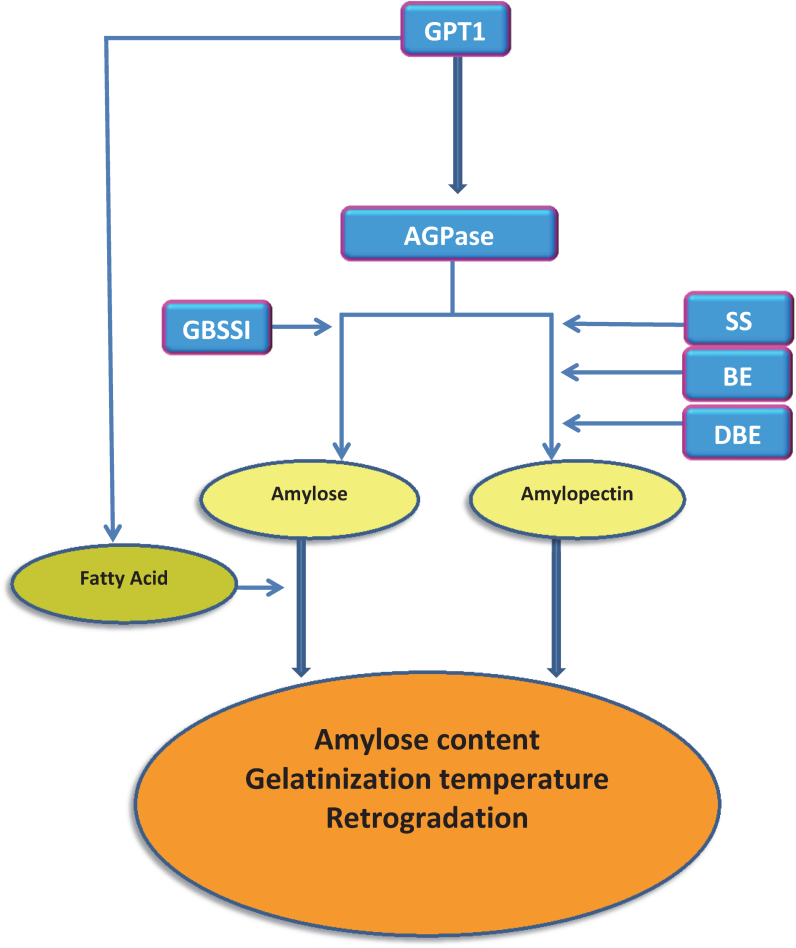
Figure 2. Simplified pathway of starch synthesis in rice and interaction with starch properties.
SSIIa had a high association with pasting temperature, gelatinization temperature and peak time. The effect of this gene on cooking quality and starch texture has been extensively studied by many authors11,12. Umemoto and Aoki, (2005) found alkali disintegration and eating quality of rice starch were explained by two SNP, [A/G] and [GC/TT], within the exon 8 of alk locus. These SNPs also have significantly associated with starch GT13. Two SNPs at positions 631 and 4827–4828 (ALKSSIIA4) respectively were tested for association (Table 1 and 2). The effect of the [GC/TT] SNP on alkali disintegration and rice starch eating quality has already been explained by many authors22,23. Highly significant associations were found between SSIIa SNPs and important physiochemical properties such as PT, PKT, GT and BDV. Melting of starch crystalline regions is measured by pasting and gelatinisation temperature and peak time signifies the end of the melting process. The highest F-test value of 199.65 was observed for ALKSSIIA4 [GC/TT] SNP and PT. This SNP clearly controls PT, PKT and BDV with R2 values of 0.642, 0.323 and 0.168, respectively. This SNP has one of the strongest associations among the physiochemical properties of rice studied in this population (R2 = 0.642). The G/T SNP at position 631 showed no significant association with any traits.
SSIVa is one of the least well characterized starch genes in rice. This study showed a significant influence of this gene on PT and GT. In total, five SNPs were examined in this gene (Table 1), of which four SNPs showed significant association with PT (Table 2).
Six genes, GBSSII, SSI, SSIIIa, SSIIIb, SSIVa and BE, had low to medium effects on variation in starch traits. SNPs in these genes had association with a number of characters with low to medium R2 values. The effect of these genes on starch traits have been studied at the gene level20,15,24,11. Here for the first time, SSIIIb and SSIVa have been identified as PT-associated (pasting temperature-associated) at a relatively medium to high level.
SSI transcript level has been measured at different seed developmental stages. A high expression level was reported at 1–3 days after flowering (DAF), peaking at 5 DAF, and then remaining almost constant during starch synthesis in the endosperm. This suggests that SSI is a major SS form in cereals25. Only one nsSNP in SSI, 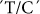 at position 5153, in this gene showed minor associations with FV, SB and MT, with R2 values of 0.16, 0.11, 0.16, respectively (Table 2).
at position 5153, in this gene showed minor associations with FV, SB and MT, with R2 values of 0.16, 0.11, 0.16, respectively (Table 2).
Pullulanase had low associations with PT, GT and CHK in the population studied here. In contrast, a recent association study in glutinous rice has shown strong relationships between pullulanase and RVA profile parameters, PKV, HPT, BDV and PKT11. The differing observations are most likely due to the structure of each population. Minor genes are very population-specific and the analysis of Yan et al. (2010) was undertaken within a glutinous population composed of rice varieties which have very low amylose content and this would have revealed the role of pullulanse in this genetic background.
Seven genes of 18 did not contribute to starch physiochemical properties in this population. No polymorphisms were detected in five genes, AGPS2b, SPHOL, SSIIb, SSIVb and ISA1 while BEIIa and BEIIb displayed polymorphism but these were not associated with any physiochemical properties measured in this study. In contrast, other studies have suggested some of these genes are important in determining rice starch physiochemical properties and quality. For example, Kawagoe et al. 2005 found the AGPS2b subunit plays an important role in starch granule synthesis and is associated with rice shrunken mutants26. SPHOL is reported to be involved in starch degradation and biosynthesis by phosphorylation of some starch-related enzymes and proteins such as starch branching enzymes (SBEs) and starch synthase (SSIIa)27. Almost all of these studies have been based on mutants totally deficient in enzyme activity28 which abolish the gene function and therefore have a significant effect on the content of soluble sugars, structure and appearance of starch granules and endosperm quality in rice and other species.
SSIIb and BEIIa are mostly expressed in green tissues and theoretically do not have major impact on grain quality traits29. In this study we confirm that SNPs in green-tissue related genes have no or very small effects on grain starch properties. No significant association was found between the two BEIIb SNPs and quality traits in this population. The differing results can be attributed to the different structure of each population, in each population each gene has a particular impact which is determined by the presence of the range of alleles present at other starch biosynthesis loci.
This study found BEIIb (amylose extender) and ISA1 had no association with any of the physiochemical properties of rice starch measured despite previous reports that these genes in several cereal species impact starch properties30,31. We examined two SNP in this gene (Table 1) but no significant association was found with starch properties. Previous biochemical analysis of rice (Oryza sativa) amylose-extender (ae) mutants revealed the influence of this gene on gelatinization properties through the structural alteration of amylopectin by reducing short chains and degree of polymerization32. However, these studies focused on mutant populations where a large segment of the gene has been deleted. Therefore, the results of those experiments are not comparable with our variation study at SNP level. Antisense inhibition of rice ISA1 has altered the structure of endosperm amylopectin and the starch physiochemical properties33. The ISA genes also contribute to the degree of setback on glutinous rice cultivars14.
Philpot et al. (2006)5 reported removal of lipid increased the rate of retrogradation and the firmness of gels significantly in rice. Analysis of O. sativa cultivar Koshihikari grown in Japan and Australia found individuals grown in Japan had a lower retrogradation rate, despite the fact that flour from both origins contained 18% amylose. Removal of the lipids from these samples resulted in retrogradation rates which were not significantly different. The amount of amylose complexed with lipids affects starch retrogradation34 and so it was suggested this phenomenon can be attributed to the amount of lipid complexed with long amylose chains, the higher concentration of lipid linked to long amylose chains explained the lower retrogradation in the Japanese grown rice. GPT1 is required for transportation of reduced carbon into plastids which is ultimately utilised for both lipid and amylose synthesis (Figure 2). It has been suggested amylose content is correlated with lipid content35 and it is thought lipids play a structural role as a core scaffold in holding together the helical architecture of amylose. GTP1 is involved in determining plastid fatty acid concentration36 and this may influence the formation of lipid-amylose complexes. GPT1 is associated with amylose content and retrogradation rate in this set of germplasm. In addition to its impact on amylose content, GPT1 may also affect retrogradation rate by influencing lipid content in rice grain.
This study has found the genes which have an impact upon starch traits within the Australian rice breeding program display relatively low levels of diversity. This set of genes is bounded by two sets of genes, one which has no diversity and another which has high levels of diversity. Australian rice breeders are managing a small number of genes and alleles which have an impact on starch quality in order to achieve desirable starch quality within the breeding program. Construction of new quality classes may require access to a wider range of alleles, genes and germplasm.
Methods
Plant materials
Plant material was supplied by Department of Primary Industries NSW, Yanco Agricultural Institute, Australia. A population of 233 temperate (japonica-type) F6 rice breeding lines was selected from pedigree rows . Selection had taken place on capacity of lines to flower and set seed and morphological traits of plant height, grain size and shape. No selection had taken place for grain starch quality traits.
Physiochemical properties
Physiochemical traits measured were apparent amylose content (AC), gelatinization temperature (GT) quantified according to standard differential scanning calorimetry methods (DSC)37. Percent grain chalk was estimated by a FOSS Cervitec according to the manufacturer's instruction. Retrogradation rate (Martin test5) was estimated by measuring the force in Newtons (N) required to push a probe into a gel derived from flour samples post viscosity measurements and stored overnight at 20°C (Lloyd texture analyser TAPlus, Hemisphere United Kingdom). Peak viscosity (PKV), trough viscosity (TV), final Viscosity (FV), breakdown viscosity (BDV), setback (SB), peak time (PKT) and pasting temperature (PT) were measured by a Rapid Visco Analyser to evaluate rheological properties of starch structure (Perten RVA 4500, Segeltorp, Sweden) according to the manufacturer's instructions. The range of values observed for these traits is shown in Table 3 and a list of all data in supplementary materials (Supplementary data 1).
Designation of starch-synthesis genes
The available literature was used to identify the most likely candidate genes associated with rice starch quality19,23,24,38. The general entries of nucleotide sequences (gDNA) and full-length cDNAs of important gene classes which were presumed to be involved in starch biosynthesis were retrieved from the NCBI (http://www.ncbi.nlm.nih.gov/) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/cgi-bin/putative_function_search.pl) databases and then re-sequenced using integrated long range PCR in combination with massively parallel sequencing (Illumina) to find novel SNPs/Indels in the studied population21. A consensus sequence alignment was generated for each candidate gene to design the amplification primers.
Candidate genes/enzymes for SNP genotyping
Eighteen genes representing seven groups of enzymes, namely ADP-glucose pyrophosphorylase (AGPase), granule bound starch synthases (GBSSI and GBSSII), starch synthases (SSI, SSIIa, SSIIb, SSIIIa, SSIIIb, SSIVa, SSIVb), branching enzymes (BEI, BEIIa, BEIIb), debranching enzyme (ISA1, ISA2, Pullullanase), starch phosphorylase (SPHOL) and glucose-6-phosphate- translocator (GPT1) were selected for SNP genotyping (Figure 1).
SNP dataset
SNP data were primarily retrieved through SNP discovery within the population of 233 rice breeding lines21. The functional polymorphisms discovered within the studied population were then compared to SNPs available within the OryzaSNP MSU database (http://oryzasnp.plantbiology.msu.edu/) and extra SNPs harvested to minimise the possibility non-synonymous SNP (nsSNP) were missed. In total, 65 nsSNPs were chosen for genotyping, of which 48 were polymorphic or existing in the population (Table 1). The remaining 17 SNP which were not polymorphic in this population were mainly retrieved from data bases.
Primer design and SNP genotyping
Multiplexed assays were designed by Sequenom MassARRAY Assay design 3.1 software to cover all available SNPs. The optimal amplicon size containing the polymorphic site was set to 80–120 bp. A 10-mer tag (5-ACGTTGGATG-3) was added to the 5′end of each amplification primer to avoid confusion in the mass spectrum and to improve PCR performance18 (Supplementary data 2).
Capture PCR protocol, primer extension and mass spectrometry
The steps of PCR capture, primer extension, resin cleanup and mass spectrometry were undertaken according to the manufacturer's instructions (Sequenom MassARRAY).
Association analysis
Assays were constructed for 110 polymorphisms defining each of the alleles of 18 genes controlling starch quality traits and retrogradation. SNP data of genotyped polymorphic alleles (Supplementary data 3) along with phenotypic data were analysed by TASSEL v2.139 software to find SNP associated with physiochemical properties. A gene by gene approach was employed to understand association of individual gene/SNP with each trait.
Statistical analysis
Genotypic and phenotypic files were prepared according to Bradbury et al. (2007) and then imported to TASSEL v2.1. The general linear model (GLM) was used for alignment of data with 1000 permutations. Critical statistics such as F-test, p-value, adjusted p-value and R2 were calculated to measure associations. P-values ≤ 0.01 were considered to have a significant effect on each trait. After identifying significantly contributing SNP, F-test values were used for comparison, larger F values were interpreted as exhibiting a higher association between SNP and its corresponding trait. Finally, R2 is the portion of total variation explained by the full model39.
Author Contributions
RH, DW and RR designed the project. A K-M and RW performed the experiments. A K-M, DW and RH wrote the paper. All authors commented on the manuscript.
Supplementary Material
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 3
Acknowledgments
We are grateful to Stirling Bowen of Southern Cross Plant Genomics for providing technical support in Sequenom MassARRAY analysis and Timothy Sexton for his valuable assistance in genotyping. This project was funded by Australian Research Council (ARC). Supply of germplasm from Department of Primary Industries NSW is gratefully acknowledged.
References
- Asp N. G. & Björck I. Resistant starch. Trends Food Sci Tech 3, 111–114 (1992). [Google Scholar]
- Fan J. & Marks B. Retrogradation kinetics of rice flours as influenced by cultivar. Cereal Chem 75, 153–155 (1998). [Google Scholar]
- Faisant N. et al. Structural features of resistant starch at the end of the human small intestine. Eur J Clin Nutr 47, 285 (1993). [PubMed] [Google Scholar]
- Ring S. G., Gee J. M., Whittam M., Orford P. & Johnson I. T. Resistant starch: its chemical form in foodstuffs and effect on digestibility in vitro. Food Chem 28, 97–109 (1988). [Google Scholar]
- Philpot K., Martin M., Butardo Jr V., Willoughby D. & Fitzgerald M. Environmental factors that affect the ability of amylose to contribute to retrogradation in gels made from rice flour. J Agric Food Chem 54, 5182–5190 (2006). [DOI] [PubMed] [Google Scholar]
- Hu P., Zhao H., Duan Z., Linlin Z. & Wu D. Starch digestibility and the estimated glycemic score of different types of rice differing in amylose contents. J Cereal Sci 40, 231–237 (2004). [Google Scholar]
- Chung H. J., Lim H. S. & Lim S. T. Effect of partial gelatinization and retrogradation on the enzymatic digestion of waxy rice starch. J Cereal Sci 43, 353–359 (2006). [Google Scholar]
- Chen M. H., Bergman C., Pinson S. & Fjellstrom R. Waxy gene haplotypes: Associations with apparent amylose content and the effect by the environment in an international rice germplasm collection. J Cereal Sci 47, 536–545 (2008). [Google Scholar]
- Cai X. L., Wang Z. Y., Xing Y. Y., Zhang J. L. & Hong M. M. Aberrant splicing of intron 1 leads to the heterogeneous 5 UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J 14, 459–465 (1998). [DOI] [PubMed] [Google Scholar]
- Larkin P. D. & Park W. D. Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol Breed 12, 335–339 (2003). [Google Scholar]
- Umemoto T. et al. Effects of Variations in Starch Synthase on Starch Properties and Eating Quality of Rice. Plant Prod Sci 11, 472–480 (2008). [Google Scholar]
- Umemoto T. et al. Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct Plant Biolo 31, 671–684 (2004). [DOI] [PubMed] [Google Scholar]
- Waters D. L. E., Henry R. J., Reinke R. F. & Fitzgerald M. A. Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol J 4, 115–122 (2006). [DOI] [PubMed] [Google Scholar]
- Yan C. J. et al. Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.). Theor Appl Genet 1–14 (2010). [DOI] [PubMed] [Google Scholar]
- Fujita N. et al. Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140, 1070 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry B. S. SNP alleles in human disease and evolution. J Hum Genet 47, 561–566 (2002). [DOI] [PubMed] [Google Scholar]
- Imelfort M., Duran C., Batley J. & Edwards D. Discovering genetic polymorphisms in next generation sequencing data. Plant Biotechnol J 7, 312–317 (2009). [DOI] [PubMed] [Google Scholar]
- Masouleh A. K., Waters D. L. E., Reinke R. F. & Henry R. J. A high throughput assay for rapid and simultaneous analysis of perfect markers for important quality and agronomic traits in rice using multiplexed MALDI TOF mass spectrometry. Plant Biotechnol J 7, 355–363 (2009). [DOI] [PubMed] [Google Scholar]
- Ohdan T. et al. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56, 3229 (2005). [DOI] [PubMed] [Google Scholar]
- Dian W., Jiang H., Chen Q., Liu F. & Wu P. Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm. Planta 218, 261–268 (2003). [DOI] [PubMed] [Google Scholar]
- Kharabian Masouleh A., Waters D. L. E., Reinke R. F. & Henry R. J. Discovery of polymorphisms in starch related genes in rice germplasm by amplification of pooled DNA and deeply parallel sequencing†. Plant Biotechnol J 9(9),1074–1085 (2011). [DOI] [PubMed] [Google Scholar]
- Umemoto T. & Aoki N. Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct Plant Biol 32, 763–768 (2005). [DOI] [PubMed] [Google Scholar]
- Waters D. L. E. & Henry R. J. Genetic manipulation of starch properties in plants: Patents 2001–2006. Recent Pat Biotechnol 1, 252–259 (2007). [DOI] [PubMed] [Google Scholar]
- Hirose T., Ohdan T., Nakamura Y. & Terao T. Expression profiling of genes related to starch synthesis in rice leaf sheaths during the heading period. Physiol Plantarum 128, 425–435 (2006). [Google Scholar]
- Cao H. et al. Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120, 205–216 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe Y., Kubo A., Satoh H., Takaiwa F. & Nakamura Y. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J 42, 164–174 (2005). [DOI] [PubMed] [Google Scholar]
- Tetlow I. J. et al. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. Plant Cell Online 16, 694–708 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H. et al. Antisense inhibition of the plastidial glucose 6 phosphate/phosphate translocator in Vicia seeds shifts cellular differentiation and promotes protein storage. Plant J 51, 468–484 (2007). [DOI] [PubMed] [Google Scholar]
- Hirose T. & Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220, 9–16 (2004). [DOI] [PubMed] [Google Scholar]
- Sun C., Sathish P., Ahlandsberg S., Deiber A. & Jansson C. Identification of four starch-branching enzymes in barley endosperm: partial purification of forms I, IIa and IIb. New Phytol 137, 215–222 (1997). [DOI] [PubMed] [Google Scholar]
- Yamakawa H., Ebitani T. & Terao T. Comparison between locations of QTLs for grain chalkiness and genes responsive to high temperature during grain filling on the rice chromosome map. Breed Sci 58, 337–343 (2008). [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N. & Satoh H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127, 459 (2001). [PMC free article] [PubMed] [Google Scholar]
- Fujita N. et al. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol 44, 607 (2003). [DOI] [PubMed] [Google Scholar]
- Yao Y. & Ding X. L. Pulsed nuclear magnetic resonance (PNMR) study of rice starch retrogradation. Cereal Chem. 2002, (6),751–756. [Google Scholar]
- Morrison W. R., Tester R. F., Gidley M. J. & Karkalas J. Resistance to acid hydrolysis of lipid-complexed amylose and lipid-free amylose in lintnerised waxy and non-waxy barley starches. Carbohyd Res 245, 289–302 (1993). [Google Scholar]
- Niewiadomski P., Knappe S., Geimer S., Fisher K., Schulz B., Unte U. S., Rosso M. G., Ache P., Flugge U.-I. & Schneider A. The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GTP1 is essential for pollen maturation and enbryo sac development. Plant Cell 17, 760–774 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand F. & Marshall W. Differential scanning calorimetry of whole grain milled rice and milled rice flour. Cereal Chem 66, 317–320 (1989). [Google Scholar]
- Rahman S. et al. Genetic alteration of starch functionality in wheat. J Cereal Sci 31, 91–110 (2000). [Google Scholar]
- Bradbury P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 3