Abstract
Summary
Two approaches commonly used to deal with missing data are multiple imputation (MI) and inverse-probability weighting (IPW). IPW is also used to adjust for unequal sampling fractions. MI is generally more efficient than IPW but more complex. Whereas IPW requires only a model for the probability that an individual has complete data (a univariate outcome), MI needs a model for the joint distribution of the missing data (a multivariate outcome) given the observed data. Inadequacies in either model may lead to important bias if large amounts of data are missing. A third approach combines MI and IPW to give a doubly robust estimator. A fourth approach (IPW/MI) combines MI and IPW but, unlike doubly robust methods, imputes only isolated missing values and uses weights to account for remaining larger blocks of unimputed missing data, such as would arise, e.g., in a cohort study subject to sample attrition, and/or unequal sampling fractions. In this article, we examine the performance, in terms of bias and efficiency, of IPW/MI relative to MI and IPW alone and investigate whether the Rubin’s rules variance estimator is valid for IPW/MI. We prove that the Rubin’s rules variance estimator is valid for IPW/MI for linear regression with an imputed outcome, we present simulations supporting the use of this variance estimator in more general settings, and we demonstrate that IPW/MI can have advantages over alternatives. IPW/MI is applied to data from the National Child Development Study.
Keywords: Marginal model, Missing at random, Survey weighting, 1958 British Birth Cohort
1. Introduction
Datasets collected for medical or social research contain missing values. One approach for dealing with this problem is simply to exclude individuals with missing data. This “complete-case” analysis is valid when data are missing completely at random but not necessarily when missing at random (MAR) (Little and Rubin, 2002). It can also be inefficient. Two alternatives are inverse-probability weighting (IPW) (Höfler et al., 2005) and multiple imputation (MI) (Little and Rubin, 2002). In IPW, again only complete cases are included in the analysis (excepting analysis of repeated measures, which we do not treat here), but weights are used to rebalance the set of complete cases so that it is representative of the whole sample. Inverse-probability weights can also be used to adjust for different sampling fractions in a survey. They are then known as sampling weights and rebalance the sample to make it representative of the population.
In MI, missing data are replaced by data drawn from an imputation model. This is done
M times, generating M complete datasets. Each
is analyzed and an estimate of the model parameters,  , calculated. Let
, calculated. Let
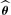 denote the
complete-data estimator of
denote the
complete-data estimator of  , and
, and
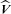 its estimated
variance. Let
its estimated
variance. Let 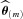 and
and
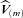 be their values for
the mth imputed dataset (m= 1, … ,
M). Rubin (1987)
proposed
be their values for
the mth imputed dataset (m= 1, … ,
M). Rubin (1987)
proposed  be estimated by
be estimated by
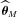 and
and
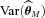 by
by
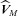 , where
, where
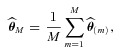 |
(1) |
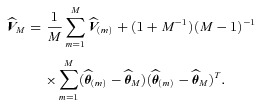 |
(2) |
IPW and MI yield consistent estimators of  when the data are MAR
and the imputation and weighting models, respectively, are correctly specified. The
variance of the IPW estimator is consistently estimated provided the weighting is
taken into account, e.g., using a sandwich estimator (Robins, Rotnitzky, and Zhao, 1994). For MI, when
when the data are MAR
and the imputation and weighting models, respectively, are correctly specified. The
variance of the IPW estimator is consistently estimated provided the weighting is
taken into account, e.g., using a sandwich estimator (Robins, Rotnitzky, and Zhao, 1994). For MI, when
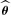 is the maximum
likelihood estimator (MLE),
is the maximum
likelihood estimator (MLE), 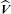 is the inverse Fisher
information, and missing data are sampled from their Bayesian posterior predictive
distribution,
is the inverse Fisher
information, and missing data are sampled from their Bayesian posterior predictive
distribution, 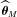 is asymptotically
normally distributed with variance
is asymptotically
normally distributed with variance 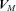 , and
, and
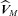 is an asymptotically
unbiased estimator of
is an asymptotically
unbiased estimator of 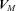 and is consistent
when M=∞ (Rubin,
1987; Wang and Robins, 1998; Nielsen, 2003).
and is consistent
when M=∞ (Rubin,
1987; Wang and Robins, 1998; Nielsen, 2003).
MI is often preferred to IPW, as it is usually more efficient. If the imputation model is correctly specified, MI should work well. However, if many data are being imputed, any inadequacies in the imputation model may lead to considerable bias. If few variables are missing on an individual, it may be considered desirable to impute them, rather than exclude the individual. On the other hand, if many variables are missing on the same individual, the imputation model must describe the joint distribution of all these variables, and if many individuals have many missing variables, the analyst may be nervous about relying on this complex and possibly misspecified imputation model. This situation could arise, for example, in a longitudinal study when whole blocks of data are missing on some of the individuals due to missed visits, or in a survey when some individuals have declined to answer whole sets of related questions. In such situations, the analyst may feel more confident using IPW.
Another possibility is to combine MI and IPW. A rule is specified for when to include
an individual in the analysis: e.g., if they attended a follow-up visit, or if more
than a certain percentage of their data is observed. Missing values in included
individuals are multiply imputed and each resulting dataset (which we call a
“quasi-complete dataset” because the data are complete for the
included, but not excluded, individuals) is analyzed using IPW to account for the
exclusion of individuals not satisfying the inclusion rule and for different
sampling fractions (if any). The “quasi-complete-data” estimator
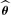 is then the IPW
estimator using the data on included individuals in a single quasi-complete dataset
and
is then the IPW
estimator using the data on included individuals in a single quasi-complete dataset
and 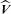 is the corresponding
sandwich variance estimator. We call this method “IPW/MI.” By imputing
in individuals with few missing values but excluding individuals with more missing
data, IPW/MI could inherit some of the efficiency advantage of MI while avoiding
bias resulting from incorrectly imputing larger blocks of data. IPW/MI is also
needed when sampling weights are used together with MI, even if all individuals are
included in the analysis.
is the corresponding
sandwich variance estimator. We call this method “IPW/MI.” By imputing
in individuals with few missing values but excluding individuals with more missing
data, IPW/MI could inherit some of the efficiency advantage of MI while avoiding
bias resulting from incorrectly imputing larger blocks of data. IPW/MI is also
needed when sampling weights are used together with MI, even if all individuals are
included in the analysis.
Several authors have used IPW/MI. Caldwell et al. (2008) and Stansfeld et al. (2008a,2008b) analyzed data from the National Childhood Development Study (NCDS). They regressed outcomes measured at age 45 on predictors measured at the same or earlier visits. Attrition of the cohort over time meant that 41% missed the age 45 visit. Weights were used to adjust for attrition, while missing values in those who attended the visit were multiply imputed. Priebe et al. (2004) multiply imputed missing data in a logistic regression with sampling weights.
It is not obvious that Rubin’s rules will give valid variance estimators for
IPW/MI. IPW estimators are inefficient. Robins and
Wang (2000) and Nielsen (2003)
show for MI that when 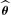 is inefficient,
is inefficient,
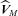 can be asymptotically
biased, even if
can be asymptotically
biased, even if 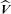 is a consistent
estimator of the complete-data variance and imputation is from the correct posterior
predictive distribution. The purpose of the present article is twofold: to examine
asymptotic bias in
is a consistent
estimator of the complete-data variance and imputation is from the correct posterior
predictive distribution. The purpose of the present article is twofold: to examine
asymptotic bias in 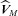 when
when
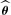 is an IPW estimator
and to show when IPW/MI is useful.
is an IPW estimator
and to show when IPW/MI is useful.
In Section 2, we define IPW/MI and show it gives consistent estimation of
 . In Section 3, we
show
. In Section 3, we
show 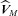 is asymptotically
unbiased for IPW/MI with linear regression and imputed outcomes. Section 4 describes
a simulation study verifying this and demonstrating IPW/MI can have advantages over
MI or IPW alone. Section 5 is a simulation with imputed covariate, suggesting
is asymptotically
unbiased for IPW/MI with linear regression and imputed outcomes. Section 4 describes
a simulation study verifying this and demonstrating IPW/MI can have advantages over
MI or IPW alone. Section 5 is a simulation with imputed covariate, suggesting
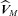 is approximately
unbiased in this case. Section 6 is an application to NCDS.
is approximately
unbiased in this case. Section 6 is an application to NCDS.
2. IPW/MI and Consistency of 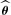
In this section, we describe IPW/MI for the situation where there are no sampling weights. The inclusion of sampling weights is covered in the Web Appendix available online.
An independent random sample of size N is drawn from the population.
Let 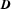 denote, for an
individual, the vector of the set of variables included in the analysis model as
well as possibly other variables that will be used to impute missing values in that
set of variables. Let R denote the missingness pattern in
denote, for an
individual, the vector of the set of variables included in the analysis model as
well as possibly other variables that will be used to impute missing values in that
set of variables. Let R denote the missingness pattern in
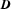 (i.e., which elements
of
(i.e., which elements
of 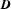 are missing), and
write
are missing), and
write 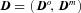 , where
, where
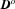 and
and
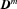 denote the observed
and missing parts of
denote the observed
and missing parts of 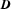 , respectively.
Subscript i denotes individual i in the sample;
e.g.,
, respectively.
Subscript i denotes individual i in the sample;
e.g., 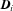 denotes
denotes
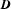 for individual
i.
for individual
i.
The IPW/MI method is as follows. Let 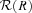 be a binary function
of R chosen by the analyst.
be a binary function
of R chosen by the analyst. 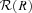 is the rule
determining whether an individual is included in the analysis. An example of
is the rule
determining whether an individual is included in the analysis. An example of
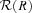 is
is
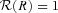 if fewer than a
certain percentage of variables in the analysis model are missing and
if fewer than a
certain percentage of variables in the analysis model are missing and
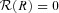 otherwise. Let
otherwise. Let
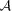 denote the set of
indices of individuals with
denote the set of
indices of individuals with 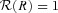 . As formalized below,
we estimate
. As formalized below,
we estimate  by fitting the
analysis model only to individuals
by fitting the
analysis model only to individuals 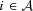 , using
inverse-probability weights to account for the selection by
, using
inverse-probability weights to account for the selection by
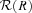 . Missing values in
individuals
. Missing values in
individuals 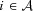 are multiply
imputed.
are multiply
imputed.
To impute 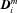 in individuals
in individuals
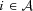 , we assume a model
, we assume a model
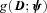 for the conditional
distribution of
for the conditional
distribution of 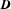 given
given
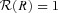 with parameters
with parameters
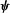 . We say this model is
correctly specified if
. We say this model is
correctly specified if 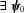 such that
such that
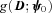 is the true
distribution of
is the true
distribution of 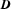 given
given
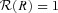 .
.
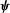 is estimated by
is estimated by
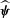 , its MLE using only
the data on individuals
, its MLE using only
the data on individuals 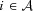 . Imputation may be
proper or improper. Let
. Imputation may be
proper or improper. Let 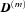 denote the
mth imputed value of
denote the
mth imputed value of 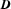 (m= 1, … , M). Note that if some
elements of
(m= 1, … , M). Note that if some
elements of 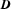 are observed in all
individuals with
are observed in all
individuals with 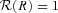 , the imputation model
can be a model for the distribution of the remaining elements of
, the imputation model
can be a model for the distribution of the remaining elements of
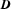 given these elements
and
given these elements
and 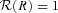 .
.
Let 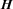 be a vector of fully
observed variables that predict whether
be a vector of fully
observed variables that predict whether 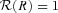 . Assume a model
. Assume a model
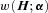 for
for
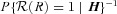 , where
, where
 are parameters. We
say this model is correctly specified if
are parameters. We
say this model is correctly specified if 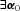 such that
such that
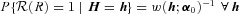 . Let
. Let
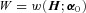 . Assume
∃δ > 0 such that
P(W−1 > δ)
= 1. Typically,
. Assume
∃δ > 0 such that
P(W−1 > δ)
= 1. Typically, 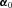 , the true value of
, the true value of
 , will be unknown. Let
, will be unknown. Let
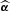 equal
equal
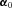 if
if
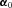 is known and denote
the MLE of
is known and denote
the MLE of  otherwise.
otherwise.
Let 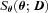 denote an
individual’s contribution to the (unweighted) complete-data estimating
equations of the analysis model. Let
denote an
individual’s contribution to the (unweighted) complete-data estimating
equations of the analysis model. Let 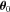 denote the solution
of
denote the solution
of 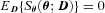 . Therefore,
. Therefore,
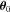 is the
“true” value of
is the
“true” value of  : it is the value to
which the solution to estimating equations
: it is the value to
which the solution to estimating equations 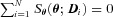 would converge as
N→∞. Based just on data from individuals
would converge as
N→∞. Based just on data from individuals
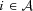 , let
, let
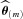 be the solution to
(weighted) estimating equations
be the solution to
(weighted) estimating equations 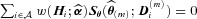 and let
and let
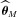 be given by equation (1). Theorem 1 and its
corollary state that under specified conditions
be given by equation (1). Theorem 1 and its
corollary state that under specified conditions 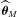 is a consistent
estimator of
is a consistent
estimator of  . Proofs are given in
the Web Appendix.
. Proofs are given in
the Web Appendix.
theorem 1
Assume (i) model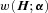 is correctly
specified, (ii)
is correctly
specified, (ii)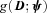 is correctly
specified, (iii)
is correctly
specified, (iii)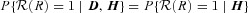 ,
(iv)
,
(iv)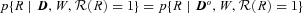 , and
(v)
, and
(v)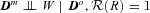 . Then,
when
. Then,
when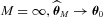 as
N→∞.
as
N→∞.
Condition (iii) states that the probability an individual is used in the fitting of
the imputation and analysis models does not depend on his values of the variables
(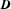 ) used in those
models given the covariates (
) used in those
models given the covariates (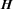 ) in the weighting
model. Condition (iv) states that among individuals to whom the imputation model is
fitted,
) in the weighting
model. Condition (iv) states that among individuals to whom the imputation model is
fitted, 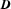 is MAR given the
true weight W. Condition (v) adds to this that among these
individuals the missing variables in the imputation model must be conditionally
independent of W given the observed variables. Note condition (v)
can be satisfied by including W or
is MAR given the
true weight W. Condition (v) adds to this that among these
individuals the missing variables in the imputation model must be conditionally
independent of W given the observed variables. Note condition (v)
can be satisfied by including W or 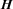 in
in
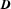 . The necessity for
condition (v) can be understood by considering how imputation will work if it is not
satisfied. Set
. The necessity for
condition (v) can be understood by considering how imputation will work if it is not
satisfied. Set 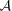 is enriched for
individuals with small values of W (and contains fewer with large
values) compared to the entire sample. If (v) is false, the distribution of
is enriched for
individuals with small values of W (and contains fewer with large
values) compared to the entire sample. If (v) is false, the distribution of
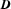 given
W depends on W, and when the imputation model
is fitted to set
given
W depends on W, and when the imputation model
is fitted to set 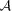 , the resulting
estimate of the marginal distribution of
, the resulting
estimate of the marginal distribution of 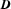 will be biased
toward the conditional distribution of
will be biased
toward the conditional distribution of 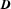 given small values
of W. Missing data in all individuals in
given small values
of W. Missing data in all individuals in
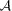 are then imputed
using the same model, a model that has been estimated giving too much weight to
individuals with small W. Including W (or
are then imputed
using the same model, a model that has been estimated giving too much weight to
individuals with small W. Including W (or
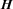 ) in the imputation
model avoids this problem: individuals with different W are imputed
differently.
) in the imputation
model avoids this problem: individuals with different W are imputed
differently.
The following corollary shows that an alternative to including the true weights
(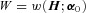 ) or the covariates
that predict the weights (
) or the covariates
that predict the weights (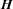 ) in the imputation
model is to include the estimated weights (
) in the imputation
model is to include the estimated weights (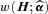 ). The latter may be
appealing because true weights are typically unknown and the dimension of
). The latter may be
appealing because true weights are typically unknown and the dimension of
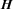 may be large.
may be large.
corollary 1
Suppose the imputation model includes, in addition
to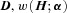 . Assume
conditions (i), (iii), and (iv) of Theorem 1 are satisfied, the imputation
model
. Assume
conditions (i), (iii), and (iv) of Theorem 1 are satisfied, the imputation
model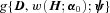 is correctly
specified,
is correctly
specified,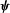 is estimated
by its MLE
is estimated
by its MLE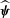 at
at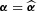 using only
individuals
using only
individuals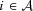 ,
and
,
and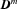 is imputed
using
is imputed
using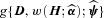 . Then,
when
. Then,
when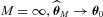 as
N→∞.
as
N→∞.
3. Linear Regression with Imputed Outcome
Consider the special case of linear regression with an imputed outcome. As in Section
2, we assume that there are no sampling weights; the generalization to sampling
weights is given in the Web Appendix. Write 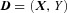 and let
and let
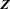 be
be
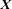 or a subvector of
or a subvector of
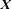 . Below,
Y and
. Below,
Y and 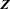 will be the response
and covariates, respectively, in the analysis model. Let
will be the response
and covariates, respectively, in the analysis model. Let
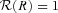 if
if
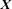 is complete;
is complete;
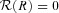 otherwise. Let
RY= 1 if
otherwise. Let
RY= 1 if 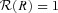 and
Y is observed; RY= 0
otherwise. We assume weights W are known and ∃δ
> 0 such that P(W−1
> δ) = 1.
and
Y is observed; RY= 0
otherwise. We assume weights W are known and ∃δ
> 0 such that P(W−1
> δ) = 1.
We estimate  in the analysis
model
in the analysis
model
| (3) |
by
linear regression of Y on 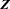 . Therefore,
. Therefore,
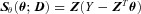 . The true value of
. The true value of
 is the solution of
is the solution of
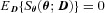 , which is
, which is
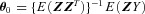 . We say the analysis
model is correctly specified if equation
(3) holds
. We say the analysis
model is correctly specified if equation
(3) holds 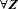 when
when
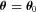 ; otherwise it is
misspecified.
; otherwise it is
misspecified.
The quasi-complete-data estimator, 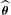 , is the solution to
weighted estimating equations
, is the solution to
weighted estimating equations 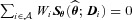 , which is the
weighted least squares estimator
, which is the
weighted least squares estimator 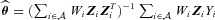 . The
quasi-complete-data variance estimator
. The
quasi-complete-data variance estimator 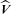 is the sandwich
estimator
is the sandwich
estimator 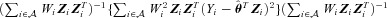 . Missing
Y values in individuals
. Missing
Y values in individuals 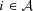 are multiply imputed
using
are multiply imputed
using 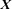 as predictors,
as predictors,
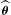 and
and
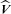 are calculated for
each imputed dataset, and
are calculated for
each imputed dataset, and 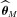 and
and
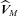 are calculated from
equations (1) and (2).
are calculated from
equations (1) and (2).
theorem 2 Let missing Y be imputed from their posterior predictive distributions using the regression imputation procedure of Schenker and Welsh (1988) (p. 1560) with imputation model
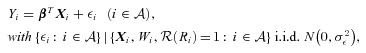 |
(4) |
and improper prior density for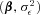 proportional
toσ−2ε. Assume this
model is correctly specified, i.e., there exists
a
proportional
toσ−2ε. Assume this
model is correctly specified, i.e., there exists
a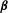 for
which
equation (4)
holds, and that
for
which
equation (4)
holds, and that
| (5) |
Then (i) 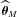 is a consistent
estimator of
is a consistent
estimator of  ; (ii) if
; (ii) if
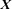 includes
includes
 (i.e.,
(i.e.,
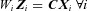 for some matrix of
constants
for some matrix of
constants  ),
),
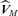 is an asymptotically
(N→∞) unbiased estimator of
is an asymptotically
(N→∞) unbiased estimator of
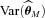 ; and iii) if
; and iii) if
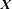 includes
includes
 and
and
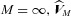 is a consistent
estimator of
is a consistent
estimator of 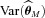 .
.
Including  in
in
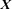 means including the
pairwise interactions between the weight and all the variables in
means including the
pairwise interactions between the weight and all the variables in
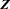 , as well as (if the
analysis model includes an intercept term) the weights themselves. Proofs of parts
(i) and (ii) come from extending the proof of Kim et
al. (2006), which shows (ii) is true in the special case where
, as well as (if the
analysis model includes an intercept term) the weights themselves. Proofs of parts
(i) and (ii) come from extending the proof of Kim et
al. (2006), which shows (ii) is true in the special case where
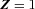 ; that of part (iii)
comes from applying Theorem 2 of Robins and Wang
(2000). Details are in the Web Appendix.
; that of part (iii)
comes from applying Theorem 2 of Robins and Wang
(2000). Details are in the Web Appendix.
The reason  needs to be in
needs to be in
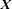 is to avoid the
imputer assuming more than the analyst. Consider the simple case where
is to avoid the
imputer assuming more than the analyst. Consider the simple case where
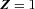 (so θ is the
population mean) and there are two values of W: a
and b. The complete-data estimator of θ corresponds to
stratifying the sample by W, calculating the mean in each of the
two strata and then calculating a weighted average of these two means. Thus, the
analysis model does not assume the population mean is the same in the two strata. If
the imputation model does not include W, it assumes the population
mean is the same in the two strata, with the result that the imputer is assuming
more than the analyst, which is known to lead to overestimation of the variance of
(so θ is the
population mean) and there are two values of W: a
and b. The complete-data estimator of θ corresponds to
stratifying the sample by W, calculating the mean in each of the
two strata and then calculating a weighted average of these two means. Thus, the
analysis model does not assume the population mean is the same in the two strata. If
the imputation model does not include W, it assumes the population
mean is the same in the two strata, with the result that the imputer is assuming
more than the analyst, which is known to lead to overestimation of the variance of
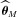 when the extra
assumption made by the imputer is correct (Meng,
1994). If the true value of the coefficients of
when the extra
assumption made by the imputer is correct (Meng,
1994). If the true value of the coefficients of
 is zero, because the
imputation model is correctly specified without the
is zero, because the
imputation model is correctly specified without the  terms, it is
probably better not to include these terms and instead accept some overestimation of
terms, it is
probably better not to include these terms and instead accept some overestimation of
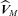 : imputation will be
more efficient if they are set to zero rather than estimated.
: imputation will be
more efficient if they are set to zero rather than estimated.
Note that, because 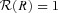 only if
only if
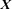 is complete,
individuals with incomplete
is complete,
individuals with incomplete 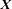 are excluded, even
if their Y and
are excluded, even
if their Y and 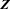 are complete. For
this reason, it would not be appealing to use this method if the sample contained
more than a few such individuals.
are complete. For
this reason, it would not be appealing to use this method if the sample contained
more than a few such individuals.
An alternative to IPW/MI is what we call “IPW/CC.” Here
Y is regressed on 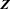 only in complete
cases (those with RY= 1), again using weights
W. This estimator is unbiased if
only in complete
cases (those with RY= 1), again using weights
W. This estimator is unbiased if
| (6) |
and the analysis model is correctly
specified. If weights W are all equal, and
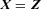 and the imputation
and analysis models are the same, there is no benefit to IPW/MI over IPW/CC: it is
more efficient to exclude individuals with missing Y (unless
M=∞, in which case exclusion and imputation are
equivalent) (White and Carlin, 2010).
However, there are two reasons for preferring IPW/MI to IPW/CC. These apply whether
or not weights are equal. First, if (6) does not hold or if the analysis model is misspecified, the complete-case
estimator may be inconsistent, whereas, as Theorem 2 states, IPW/MI gives consistent
estimators if equation (5) holds (and
assuming the imputation model is correctly specified). Equation (5) may be satisfied even if (6) is not, as (5) allows the probability that
Y is observed to depend on a larger set of variables
and the imputation
and analysis models are the same, there is no benefit to IPW/MI over IPW/CC: it is
more efficient to exclude individuals with missing Y (unless
M=∞, in which case exclusion and imputation are
equivalent) (White and Carlin, 2010).
However, there are two reasons for preferring IPW/MI to IPW/CC. These apply whether
or not weights are equal. First, if (6) does not hold or if the analysis model is misspecified, the complete-case
estimator may be inconsistent, whereas, as Theorem 2 states, IPW/MI gives consistent
estimators if equation (5) holds (and
assuming the imputation model is correctly specified). Equation (5) may be satisfied even if (6) is not, as (5) allows the probability that
Y is observed to depend on a larger set of variables
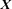 . Second, even if
(6) holds and the analysis model
is correctly specified, it may be more efficient to use all the available
information (i.e.,
. Second, even if
(6) holds and the analysis model
is correctly specified, it may be more efficient to use all the available
information (i.e., 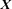 ) to impute
Y.
) to impute
Y.
4. Simulation Study: Imputed Outcome
In this section, we explore IPW/MI for linear regression with imputed outcome. As in
Section 3, the analysis model is fitted only to individuals with complete
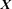 and missing
Y in these individuals are imputed.
and missing
Y in these individuals are imputed.
Analysis of the sample must deal with two stages of missingness: stage 1 is the
missingness in 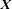 ; stage 2,
missingness in Y. At stage 1, one could either exclude individuals
with incomplete
; stage 2,
missingness in Y. At stage 1, one could either exclude individuals
with incomplete 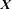 (
(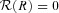 ) or impute missing
) or impute missing
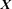 . Similarly, each
individual with missing Y not already excluded at stage 1
(
. Similarly, each
individual with missing Y not already excluded at stage 1
(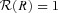 ) could either be
excluded at stage 2 or have Y imputed. At each stage, if exclusion
is used, one can either adjust for the exclusion using IPW or not adjust. Thus,
there are three possibilities at each stage, giving 3 × 3 = 9 possible
strategies in total. Denote a strategy by ST1/ST2, where ST1 and ST2 are each CC
(exclude and do not weight), IPW (exclude and weight) or MI (impute). In IPW/MI, the
focus of this article, individuals with missing
) could either be
excluded at stage 2 or have Y imputed. At each stage, if exclusion
is used, one can either adjust for the exclusion using IPW or not adjust. Thus,
there are three possibilities at each stage, giving 3 × 3 = 9 possible
strategies in total. Denote a strategy by ST1/ST2, where ST1 and ST2 are each CC
(exclude and do not weight), IPW (exclude and weight) or MI (impute). In IPW/MI, the
focus of this article, individuals with missing 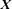 (
(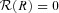 ) are excluded and
weights used to adjust for this; individuals with complete
) are excluded and
weights used to adjust for this; individuals with complete
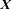 but missing
Y (
but missing
Y (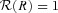 ) have
Y imputed. CC/CC uses only individuals with complete
) have
Y imputed. CC/CC uses only individuals with complete
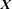 and
Y and there is no weighting. IPW/IPW uses the same individuals,
but weights them by the inverse of their probability of being a complete case. In
MI/MI all missing values are imputed. We also consider CC/IPW, CC/MI, and IPW/CC,
but not MI/CC or MI/IPW, which combine the disadvantage of having to specify an
imputation model for
and
Y and there is no weighting. IPW/IPW uses the same individuals,
but weights them by the inverse of their probability of being a complete case. In
MI/MI all missing values are imputed. We also consider CC/IPW, CC/MI, and IPW/CC,
but not MI/CC or MI/IPW, which combine the disadvantage of having to specify an
imputation model for 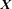 with that of losing
out on the potential efficiency gains of imputing Y.
with that of losing
out on the potential efficiency gains of imputing Y.
The purpose of the following simulation is three-fold: to verify
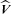 is approximately
unbiased for IPW/MI; to show IPW/MI can be more efficient than IPW/IPW; and to show
MI/MI can yield biased parameter estimators when the stage 1 (for
is approximately
unbiased for IPW/MI; to show IPW/MI can be more efficient than IPW/IPW; and to show
MI/MI can yield biased parameter estimators when the stage 1 (for
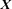 ) or stage 2 (for
Y given
) or stage 2 (for
Y given 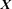 ) imputation model is
misspecified, and that IPW/MI remains approximately unbiased or at least less biased
than MI/MI in these situations. The data-generating mechanism has been chosen to
illustrate these points. It will now be described and then its features
elucidated.
) imputation model is
misspecified, and that IPW/MI remains approximately unbiased or at least less biased
than MI/MI in these situations. The data-generating mechanism has been chosen to
illustrate these points. It will now be described and then its features
elucidated.
Data 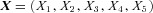 and
Y were generated for N= 1000
individuals. For each individual, X1 was one with
probability 0.5 and zero otherwise, X2,
X3, and X4 were
independent and identically distributed N(0, 1) and, finally,
X5 was sampled from
N(X2×X3,
1). Response Y was generated from
and
Y were generated for N= 1000
individuals. For each individual, X1 was one with
probability 0.5 and zero otherwise, X2,
X3, and X4 were
independent and identically distributed N(0, 1) and, finally,
X5 was sampled from
N(X2×X3,
1). Response Y was generated from
| (7) |
where ε∼N(0, 1). X1 was observed for all N individuals. With probability 0.8 − 0.6X1, (X2, X3, X4, X5) was observed; otherwise it was missing. If (X2, X3, X4, X5) was observed, Y was observed with probability {1 + exp (−1.5 + 0.6X2X4)}−1; otherwise Y was missing.
The analysis model was Y = θ0 +
θ2X2 +
θ3X3 +
θ23X2X3
+ e, where E(e
∣X2, X3) =
0. Therefore, 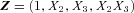 . By integrating
(7) with respect to
X1, X4, and
X5, it can be shown that this analysis model is
correctly specified and the true
. By integrating
(7) with respect to
X1, X4, and
X5, it can be shown that this analysis model is
correctly specified and the true  is
(θ0, θ2, θ3,
θ23) = (− 3, 0.5, 0.5, 1).
is
(θ0, θ2, θ3,
θ23) = (− 3, 0.5, 0.5, 1).
This data-generating mechanism was chosen for three reasons. First, the
X1X2 and
X1X3 interactions in
(7) mean the relation between
Y and (X2,
X3) is different in the two strata defined by
X1. Also, the probability that
(X2, X3) is observed
differs: in one stratum it is 0.2; in the other, 0.8. Thus, the relation between
Y and (X2,
X3) is different in individuals with complete
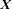 and incomplete
and incomplete
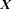 . Failure to adjust
for the missingness at stage 1, by weighting or imputation, will therefore lead to
bias in θ2 and θ3. Therefore, CC/IPW, CC/MI, and
CC/CC will be biased. Second, for individuals with observed
(X2, X3,
X4, X5) the probability
Y is observed depends on X4, which
is not in the analysis model but is associated with Y. This causes
the relation between Y and
. Failure to adjust
for the missingness at stage 1, by weighting or imputation, will therefore lead to
bias in θ2 and θ3. Therefore, CC/IPW, CC/MI, and
CC/CC will be biased. Second, for individuals with observed
(X2, X3,
X4, X5) the probability
Y is observed depends on X4, which
is not in the analysis model but is associated with Y. This causes
the relation between Y and 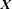 described by the
analysis model to be different in the set of complete cases from in the set with
complete
described by the
analysis model to be different in the set of complete cases from in the set with
complete 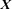 but missing
Y. In particular, because the probability of Y
being missing depends on
X2X4, the relation
between Y and X2 will be different in
the two sets. Failure to adjust for the missingness at stage 2 will therefore lead
to bias (specifically in θ2). Therefore, IPW/CC, MI/CC, and CC/CC
will be biased. Third, X5 is included in the
data-generating mechanism for Y to show that using MI at stage 1
can cause bias if the imputation model for
but missing
Y. In particular, because the probability of Y
being missing depends on
X2X4, the relation
between Y and X2 will be different in
the two sets. Failure to adjust for the missingness at stage 2 will therefore lead
to bias (specifically in θ2). Therefore, IPW/CC, MI/CC, and CC/CC
will be biased. Third, X5 is included in the
data-generating mechanism for Y to show that using MI at stage 1
can cause bias if the imputation model for 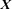 is misspecified (see
results for MI*/MI below).
is misspecified (see
results for MI*/MI below).
A total of 1000 datasets were generated and the seven methods applied to each. For each of θ0, θ2, θ3, and θ23 and each method, the mean of the 1000 parameter estimates and of the 1000 estimated variances was calculated. The empirical SE was calculated as the standard deviation of the parameter estimates. Where a method involved imputation, 10 imputations were performed.
For MI/MI, the (correctly specified) imputation model at stage 1 was (X2, X3, X4) ∼N{(γ2, γ3, γ4), Σ1} and X5∣X2, X3∼N(γ5+γ6X2+γ7X3+γ8X2X3 , Σ2). Noninformative normal and inverse-Wishart priors were used, yielding normal and inverse-Wishart posteriors (Gelman et al., 2004, p. 88). For CC/MI, IPW/MI, and MI/MI, the (correctly specified) imputation model used at stage 2 was Y=β0+β1X1+β2X2+β3X3+β4X4+β5X5+β12X1X2+β13X1X3+β23X2X3+β123X1X2X3+ε.
For IPW/CC, IPW/IPW, and IPW/MI, weights were estimated by fitting the (correctly
specified) missingness model for stage 1:
P(X2,
X3, X4 and
X5 observed)
=δ0+δ1X1.
Note that, because X1 is binary,
W=
(δ0+δ1X1)−1=δ−10−X1δ1{δ0(δ0+δ1)}−1
is a linear function of X1. Hence, as the stage 2
imputation model includes 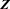 and
and
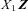 , it implicitly
includes
, it implicitly
includes  . For CC/IPW and
IPW/IPW, weights were estimated using the (correctly specified) model for stage 2:
. For CC/IPW and
IPW/IPW, weights were estimated using the (correctly specified) model for stage 2:
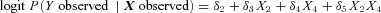 . For IPW/IPW, the
probability of being a complete case is the product of these two probabilities.
. For IPW/IPW, the
probability of being a complete case is the product of these two probabilities.
Table 1 shows mean parameter estimates, empirical SEs, and square roots of the mean estimated variances. It can be seen that IPW/MI yields approximately unbiased estimators of parameters and SEs, as expected from Theorem 2. As explained above, CC/IPW, CC/MI, CC/CC, and IPW/CC are biased for one or more parameters. IPW/IPW and MI/MI are both approximately unbiased. The former is less efficient than IPW/MI because the imputation model at stage 2 uses auxiliary information, i.e. covariates (notably X4 and X5) not included in the analysis model. The most efficient unbiased method is MI/MI, confirming that imputation is the best method when the imputation models are correct.
Table 1.
Mean parameter estimate (“mean”), square root of mean
estimated variance (“aSE”), and empirical SE
(“eSE”) for four parameters and 10 analysis methods. The
true value of is (θ0,
θ2, θ3, θ23)
= (−3, 0.5, 0.5, 1).
is (θ0,
θ2, θ3, θ23)
= (−3, 0.5, 0.5, 1).
| θ0 | θ2 | θ3 | θ23 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Mean | aSE | eSE | Mean | aSE | eSE | Mean | aSE | eSE | Mean | aSE | eSE |
| True | −3.000 | .500 | .500 | 1.000 | ||||||||
| CC/CC | −2.995 | .080 | .079 | .090 | .081 | .087 | .200 | .080 | .086 | 1.005 | .082 | .091 |
| CC/IPW | −2.993 | .082 | .079 | .199 | .092 | .091 | .200 | .086 | .089 | 1.004 | .094 | .100 |
| CC/MI | −2.994 | .075 | .076 | .202 | .081 | .081 | .201 | .079 | .083 | 1.004 | .084 | .086 |
| IPW/CC | −2.993 | .102 | .101 | .382 | .110 | .112 | .495 | .109 | .114 | 1.008 | .114 | .119 |
| IPW/IPW | −2.990 | .106 | .104 | .489 | .120 | .124 | .494 | .112 | .117 | 1.006 | .121 | .132 |
| IPW/MI | −2.992 | .097 | .096 | .498 | .105 | .105 | .497 | .104 | .107 | 1.006 | .110 | .113 |
| MI/MI | −3.000 | .089 | .081 | .503 | .092 | .087 | .497 | .090 | .088 | 1.006 | .092 | .082 |
| MI*/MI | −2.998 | .092 | .085 | .498 | .095 | .093 | .496 | .094 | .094 | .749 | .100 | .083 |
| MI/MI* | −2.999 | .108 | .101 | .100 | .088 | .054 | .099 | .088 | .051 | .391 | .091 | .055 |
| IPW/MI* | −2.998 | .107 | .100 | .492 | .119 | .122 | .495 | .117 | .115 | .776 | .131 | .127 |
However, when the imputation model at stage 1 or stage 2 is misspecified, MI/MI may
be biased. First, suppose that the imputation model at stage 1 is misspecified as
(X2, X3,
X4,
X5)T∼N{(γ2,
γ3, γ4,
γ5)T , Σ}. As
X2, X3,
X4 and X5 are
uncorrelated (though not independent), Σ will be estimated as an
approximately diagonal matrix. Therefore, for individuals with incomplete
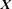 the imputed values
of X5 will be approximately independent of
X2 and X3; the relation
between X5 and the interaction of
X2 and X3
(E(X5)
=X2X3) is not
present in the imputed data. The missing Y values of these
individuals will then be imputed in such a way that the interaction between
X2 and X3 is only 0.5,
half what it should be. As half the individuals have incomplete
the imputed values
of X5 will be approximately independent of
X2 and X3; the relation
between X5 and the interaction of
X2 and X3
(E(X5)
=X2X3) is not
present in the imputed data. The missing Y values of these
individuals will then be imputed in such a way that the interaction between
X2 and X3 is only 0.5,
half what it should be. As half the individuals have incomplete
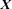 , fitting the
analysis model to the whole sample results in an estimate of θ23
of about 0.75. This is seen in Table 1 in
the row MI*/MI.
, fitting the
analysis model to the whole sample results in an estimate of θ23
of about 0.75. This is seen in Table 1 in
the row MI*/MI.
Second, suppose the imputation model at stage 1 is correct but that at stage 2 is misspecified by leaving out the β23X2X3, β123X1X2X3, and β5X5 terms. Missing Y values will now be imputed in such a way that there is no interaction between X2 and X3. As approximately 60% of Y values are missing, θ23 will be underestimated by about 60%. This result is shown in Table 1 in the row MI/MI*. The row IPW/MI* shows the result of IPW/MI with the same misspecified imputation model at stage 2. This method is considerably less biased than MI/MI*, because fewer Y values are being imputed. Therefore, the IPW element of IPW/MI provides some protection against misspecification of the imputation model.
5. Simulation Study: Imputed Covariate
In this section, we investigate the bias of 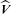 for IPW/MI in the
case of linear regression with an imputed covariate. In the simulation study below,
we find that the bias is small. This study also demonstrates again that IPW/MI can
be more efficient than IPW/IPW, and that MI/MI can yield biased estimators when the
imputation model for stage 1 is misspecified. Only brief details are presented here;
full details can be found in the Web Appendix.
for IPW/MI in the
case of linear regression with an imputed covariate. In the simulation study below,
we find that the bias is small. This study also demonstrates again that IPW/MI can
be more efficient than IPW/IPW, and that MI/MI can yield biased estimators when the
imputation model for stage 1 is misspecified. Only brief details are presented here;
full details can be found in the Web Appendix.
The (correctly specified) analysis model was Y=θ0+θ2X2+θ3X3+θ4X4+θ23X2X3+e, where E(e ∣X2, X3, X4) = 0. Variables X1 and Y were always observed; X2 and X3 were both observed or both missing. The probability they were observed depended on Y and X1. If (X2, X3) was missing, so was X4; otherwise the probability X4 was observed depended on Y. The two stages of missingness are that stage 1 is missingness in (X2, X3) and stage 2 is missingness in X4.
For MI/MI, the imputation model used at stage 1 (to impute X2 and X3) falsely assumed that (Y, X2, X3) was trivariate normal. Although misspecified, this imputation model might easily be used in practice. As the stage 1 imputation model is misspecified, we call this method MI*/MI. For IPW/MI and MI*/MI, the imputation model used at stage 2 (to impute X4) was correctly specified in terms of X1, X2, X3, Y, and certain interactions. The covariates (X1 and Y) that determine the weights are included in this model. IPW/MI* and MI*/MI* used a stage 2 imputation model that was misspecified because interaction terms were omitted.
Table 2 shows the results. IPW/IPW and IPW/MI are approximately unbiased, and SE estimators for IPW/MI are approximately unbiased. SEs for IPW/MI are smaller than for IPW/IPW: it is more efficient to impute missing X4 for individuals with otherwise complete data than to exclude them.
Table 2.
Mean parameter estimate (mean), square root of mean estimated
variance (aSE), and empirical SE (eSE) for five parameters and 10
analysis methods. Results forθ2are
omitted because, apart from Monte Carlo error, they are the same as
forθ3. The true value
of is (θ0,
θ2, θ3, θ4,
θ23) = (0, 0.5, 0.5, 0.5, 1).
is (θ0,
θ2, θ3, θ4,
θ23) = (0, 0.5, 0.5, 0.5, 1).
| θ0 | θ3 | θ4 | θ23 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Mean | aSE | eSE | Mean | aSE | eSE | Mean | aSE | eSE | Mean | aSE | eSE |
| True | .000 | .500 | .500 | 1.000 | ||||||||
| CC/CC | .238 | .060 | .056 | .196 | .061 | .065 | .183 | .060 | .064 | .992 | .064 | .077 |
| IPW/IPW | .020 | .095 | .102 | .485 | .103 | .113 | .479 | .108 | .124 | .990 | .108 | .119 |
| IPW/MI | .002 | .075 | .075 | .495 | .084 | .084 | .490 | .092 | .089 | 1.001 | .089 | .088 |
| MI*/MI | −.086 | .051 | .061 | .663 | .100 | .129 | .372 | .071 | .072 | .976 | .079 | .117 |
| MI*/MI* | −.087 | .051 | .060 | .674 | .100 | .126 | .337 | .077 | .081 | .970 | .080 | .112 |
| IPW/MI* | −.003 | .078 | .076 | .504 | .086 | .091 | .427 | .096 | .089 | .978 | .092 | .095 |
| IPWe/MI | .003 | .061 | .060 | .497 | .081 | .083 | .491 | .089 | .087 | 1.001 | .088 | .089 |
MI*/MI gives biased estimation, because the imputation model at stage 1 is misspecified. Misspecification also of the imputation model at stage 2 (MI*/MI*) adds to the bias, especially in θ4. Bias also occurs when IPW is used at stage 1 instead of MI (IPW/MI*), but is smaller than that of MI*/MI*, and indeed of MI*/MI.
Theorems 1 and 2 of Robins and Wang (2000)
enable the asymptotic (N→∞) percentage bias in
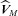 to be calculated
when M=∞ (see Web Appendix). The asymptotic
percentage bias in
to be calculated
when M=∞ (see Web Appendix). The asymptotic
percentage bias in 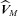 was 3.7% for
θ4 and less than 1% for θ0,
θ2, θ3, and θ23, which is
in line with the finding above that
was 3.7% for
θ4 and less than 1% for θ0,
θ2, θ3, and θ23, which is
in line with the finding above that 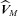 was approximately
unbiased for finite N and M.
was approximately
unbiased for finite N and M.
The results above were obtained using the true weights. In practice, weights would
usually be estimated. Row IPWe/MI in Table
2 shows the results when weights are estimated. The variance estimators
are approximately unbiased. Note that for IPWe/MI, 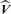 was replaced by a
sandwich estimator that accounts for uncertainty in the weights (Robins et al., 1994). When
was replaced by a
sandwich estimator that accounts for uncertainty in the weights (Robins et al., 1994). When
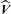 was instead used,
the variance for θ0 was overestimated.
was instead used,
the variance for θ0 was overestimated.
6. Application
The NCDS consists of 17,638 individuals born in Britain during one week in 1958 (Power and Elliott, 2006). 920 immigrants added later are not considered here. Data were collected at birth and at ages 7, 11, 16, 23, 33, and 45. A total of 16,334 nonimmigrants were still alive and free from type 1 diabetes at age 45 and of these, 8953 (55%) participated in a biomedical survey.
Thomas, Hypponen, and Power (2007) investigated the effect of characteristics measured at birth and adult adiposity (body mass index [BMI] and waist size at 45) on glucose metabolism at age 45. Subjects were classified as having high blood glucose if their glycosylated hemoglobin (A1C) was greater than 6% or they had type 2 diabetes. Immigrants and individuals with type 1 diabetes were excluded. Data on blood glucose, BMI and waist size at 45 were available for 7518 of the 8953 participants. Of these, 1845 (25%) had incomplete data on the factors measured at birth. Thomas et al., using the ice command in STATA (Royston, 2005), performed MI by chained equations (Van Buuren, 2007) on the 7518 subjects, producing 10 complete datasets. These 7518 were then analyzed as though representative of all 16,334 nonimmigrants alive and free from type 1 diabetes at age 45. Thomas et al. concluded that the factors measured at birth were related to blood glucose at 45 and that, moreover, some of these effects were largely mediated through adult adiposity.
We repeated this analysis but used IPW to allow the relation between glucose and the predictors to differ in the 7518 subjects with complete age 45 data from the other 8816 cohort members. Here stage 1 missingness refers to the age 45 data and stage 2 refers to the data measured at or before birth. Thomas et al. used a CC/MI analysis (i.e., used complete cases at stage 1 and MI at stage 2), whereas we use IPW/MI.
In the missingness model for stage 1, i.e., for the probability that at least one of glucose, BMI and waist size is missing, we used the potential predictors of missingness recorded at birth or age 7 identified by Atherton et al. (2008) and listed in their Table 3. We also used gestational age (< 38 versus ≥ 38 weeks) and a set of variables recorded at age 11: math and reading scores (normal/low), internalizing and externalizing problems (normal/intermediate/problem), and verbal and nonverbal scores (normal/low). All predictors were categorical, and most binary.
Table 3.
LOR and SEs for predictors of high blood glucose. Binary predictors are gestational age < 38 weeks, preeclampsia, smoking during pregnancy, prepregnancy BMI≥ 25Kg/m2, and manual socioeconomic position (SEP) at birth. Ordinal and continuous predictors are birth weight for gestational age (tertile), BMI at age 45 (Kg/m2), and waist circumference at age 45 (cm). Adjustment was also made for sex and family history of diabetes.
| CC/MI | IPW/MI | MI/MI | ||||
|---|---|---|---|---|---|---|
| LOR | SE | LOR | SE | LOR | SE | |
| Short gestation | 0.46 | 0.22 | 0.48 | 0.23 | 0.44 | 0.20 |
| Preeclampsia | 0.46 | 0.27 | 0.55 | 0.27 | 0.47 | 0.25 |
| Mother overweight | 0.29 | 0.15 | 0.36 | 0.16 | 0.18 | 0.12 |
| Smoke in pregnancy | 0.02 | 0.14 | 0.04 | 0.14 | 0.04 | 0.14 |
| Manual SEP | 0.37 | 0.17 | 0.44 | 0.18 | 0.39 | 0.17 |
| Birth weight | −0.31 | 0.09 | −0.31 | 0.09 | −0.32 | 0.09 |
| BMI age 45 | 0.04 | 0.02 | 0.02 | 0.02 | 0.03 | 0.02 |
| Waist size age 45 | 0.07 | 0.01 | 0.07 | 0.01 | 0.07 | 0.01 |
Not everyone attended the age 7 and age 11 visits, and even those who did had some missing values. Therefore, some predictors of missingness at stage 1 were themselves missing. To deal with visit missingness, we partitioned the sample into four strata according to which of the age 7 and age 11 visits were attended. A different logistic regression was fitted to each stratum, using only predictors from the visits attended by individuals in that stratum. Missing values in these predictors were dealt with by introducing missing indicator variables. The missing indicator method can cause bias when used for variables in an analysis model (Jones, 1996). Although we are using it to calculate weights, not in the analysis model, this method is imperfect and we do not recommend it for general use. Therefore, we also calculated a second set of weights by multiply imputing missing predictors of missingness. The results obtained using this second set of weights were very similar to those (reported below) obtained using missing indicators.
The mean weight was 2.5; 5th and 95th percentiles were 1.6 and 5.2; the maximum was 23.1. As found by Atherton et al. (2008), disadvantaged individuals were more likely to be missing at stage 1. In the stratum who attend both age 7 and 11 visits, the following variables were significant at the 5% level: breastfed <1 month; mother leaving school at or before statutory age; short stature, overweight, internalizing, and externalizing problems at age 7; internalizing and externalizing problems, low math, low reading, and low nonverbal scores at age 11.
For stage 2 we used the same imputation model as Thomas et al., except that we included the weights. Following guidelines of White, Royston, and Wood (2010), 25 imputations were used. This MI model used only the variables in the analysis model and the weights. We also tried adding variables used as predictors in the missingness model to the imputation model, but this made very little difference to the results below.
Table 3 shows the estimated log odds ratios (LOR) and SEs. Due to the stochastic nature of MI and the inclusion of weights in the imputation model, the results for CC/MI are slightly different (maximum difference 0.03) from those reported by Thomas et al. (2007). As can be seen, using IPW at stage 1 (IPW/MI) does not substantially change the results. The biggest differences are that ORs for preeclampsia, mother overweight, and manual class have risen slightly, and the first two have changed from being almost significant to just significant. SEs are also slightly larger.
We investigated why these ORs increased slightly when weighting was used. The missingness model indicated that disadvantaged individuals were more likely to be missing at stage 1. Therefore, using IPW gives more weight to disadvantaged individuals. We partitioned the stratum who attended both age 7 and age 11 visits into two groups, advantaged and disadvantaged, using the following rule: individuals with at least three of the following indicators of disadvantage were classified as disadvantaged: breastfed < 1 month; mother leaving school early; short stature, overweight, internalizing, and externalizing problems at age 7; and internalizing and externalizing problems, and poor math, reading, and nonverbal scores at age 11. Using this rule, the disadvantaged group contained 29% of individuals. The other 71% were classified as advantaged. The analysis model was fitted to the two groups separately. The LORs for preeclampsia, mother overweight, and social class were 0.59, 0.70, and 0.33, respectively, in the disadvantaged group, and −0.04, −0.04, and 0.41 in the advantaged group. Therefore, the observed relation between glucose and preeclampsia/overweight is stronger in the disadvantaged individuals. It seems likely therefore that the reason why ORs for preeclampsia and overweight in the whole cohort are greater when IPW is used (IPW/MI versus CC/MI) is that IPW gives more weight to the disadvantaged group. The relation between manual class and glucose, however, is slightly weaker in the disadvantaged group, leaving its increased OR unexplained.
Assuming then that the probability that glucose, BMI and waist size at 45 years are complete does not depend on variables in the analysis model given available predictors of missingness, the associations found by Thomas et al. in the sample of 7518 individuals do generalize to the population of nonimmigrants still alive and free from type 1 diabetes at age 45.
Finally, we used MI/MI, i.e., imputed all missing values for all 16,334 individuals. Included in the imputation were the variables in the analysis model and the predictors in the missingness model of IPW/MI. A total of 100 imputed datasets were created. Table 3 shows the results. They do not differ substantially from those of IPW/MI. Some SEs are slightly smaller. The small increases in the ORs of preeclampsia and mother overweight seen in IPW/MI relative to CC/MI are not replicated. In fact, the OR of overweight is lower in MI/MI than in CC/MI.
To investigate why, we partitioned the 12,501 individuals who attended both age 7 and age 11 visits into four groups, using the same rule for disadvantage as before: disadvantaged with observed glucose; disadvantaged with imputed glucose; advantaged with observed glucose; and advantaged with imputed glucose. The analysis model was fitted to each group separately. It was found that, whereas the relation between blood glucose and its predictors differed considerably between the advantaged and disadvantaged groups in the set of individuals whose glucose was observed, this difference was not seen in those with imputed glucose. In particular, the LORs for overweight were 0.57 and −0.17 in the disadvantaged and advantaged groups with observed glucose, respectively, but were 0.15 and 0.18 for those with imputed glucose. Interaction terms are needed in the imputation model, e.g., imputation could be done separately in the two groups. Careful assessment of the imputation model might have revealed this, but such assessment might not always be made.
7. Discussion
Robins and Wang (2000) derive a general formula for the asymptotic variance of an MI estimator based on a complete-data estimator solving a set of estimating equations. This formula applies when improper imputation and a parametric imputation model are used. IPW/MI could be carried out in this way and the Robins and Wang (2000) variance formula used. The formula is, however, complicated and has not been implemented in standard software. Using proper imputation with Rubin’s rules is appealing because it is simpler and can be used with nonparametric imputation procedures. Robins and Wang (2000) also give a formula for the asymptotic bias of the Rubin’s rules variance estimator when M=∞. We used this to show that, in the case of linear regression with MI of a missing outcome, the Rubin’s rules variance estimator for IPW/MI is consistent when M=∞. We also used it in the setting described in Section 5, where a missing covariate is imputed. The expression derived for the asymptotic bias in the Rubin’s rules variance estimator for IPW/MI was complicated and did not reduce to zero. However, both the asymptotic and finite-sample biases were found to be small in this study. In the Web Appendix, we describe two simulation studies of logistic regression, one with an imputed outcome and one with an imputed covariate. In both, the Rubin’s rules variance estimator was approximately unbiased. Schafer (2003) comments that “although we may find it difficult to prove good performance for [MI using a nonmaximum likelihood estimator], that does not imply that good performance will not be seen in practice. Experience suggests that Bayesian MI does interact well with a variety of semi- and nonparametric estimation procedures.”
If the weights are just sampling weights, they will be known, but if they are used to account for missing data, they will need to be estimated. A limitation of our proof in Section 4 is that the complete-data variance estimator assumes that weights are known and ignores any estimation uncertainty about them. This uncertainty is commonly ignored, thus overestimating the variance (Robins et al., 1994), as we saw in Section 5. If software allows, we recommend using a sandwich estimator that accounts for the uncertainty in the weights (Robins et al., 1994).
Some researchers may prefer to use straightforward MI (what we called MI/MI). Provided that the imputation models are correctly specified, this will be more efficient than IPW/MI. However, our (admittedly contrived) simulations and (not contrived) real data example have shown that those who prefer IPW/MI have some justification for their caution. A possible use for IPW/MI is as a check, or diagnostic, for MI/MI. If the results of IPW/MI and MI/MI are very different, further exploration would be warranted, possibly leading to refinement of the imputation model. We have not considered the effect of misspecified missingness models. Such misspecification would typically cause bias, just like misspecification of the imputation model in MI/MI. However, the fit of the missingness model, which is a model for a univariate response, is easier to assess, and more able to be assessed (Vansteelandt, Carpenter, and Kenward, 2010), than that of a complex multivariate imputation model. Furthermore, IPW/MI is needed when sampling weights are used, even if all missing values are imputed.
IPW/MI will be most appealing when the model for the weights is relatively simple compared with the imputation model. This will not always be so. Also, a limitation of all IPW methods is their difficulty in handling nonmonotone missingness in the predictors in the missingness model. Robins and Gill (1997) propose a procedure for handling such missingness, but this is complicated to use and limited in practice to a small number of missing predictors.
Another alternative to IPW/MI is IPW/IPW. This is simpler, but has the disadvantage that an individual is excluded from an analysis even if he/she is missing just one variable. Furthermore, if multiple analyses are being performed with different variables, either a different set of weights is needed for each analysis (because an individual who is complete for one analysis may be incomplete for another) or a single set of weights is calculated but only for individuals who are complete cases for all the analyses (Goldstein, 2009). IPW/MI, on the other hand, would allow a single set of weights to be used, as imputation could ensure that the set of complete cases were the same for each analysis.
8. Supplementary Materials
The Web Appendix referenced in Sections 2, 3, 5, and 7 is available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
acknowledgements
We thank Chris Power for valuable discussion and assistance in obtaining the NCDS data, Claudia Thomas for preparing the variables we used, and two anonymous reviewers. The Centre for Longitudinal Studies provided the official NCDS data. SS and IW were funded by MRC grants MC_US_A030_0014 and MC_US_A030_0015 and LL by an MRC Career Development Award.
References
- Atherton K, Fuller E, Shepherd P, Strachan DP, Power C. Loss and representativeness in a biomedical survey at age 45 years: 1958 British Birth Cohort. Journal of Epidemiology and Community Health. 2008;62:216–223. doi: 10.1136/jech.2006.058966. [DOI] [PubMed] [Google Scholar]
- Caldwell TM, Rodgers B, Clark C, Jefferis BJMH, Stansfeld SA, Power C. Lifecourse socioeconomic predictors of midlife drinking patterns, problems and abstention: Findings from the 1958 British Birth Cohort study. Drug and Alcohol Dependence. 2008;95:269–278. doi: 10.1016/j.drugalcdep.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Rubin DB, editors. Bayesian Data Analysis. London: Chapman and Hall/CRC; 2004. [Google Scholar]
- Goldstein H. Handling attrition and non-response in longitudinal data. Longitudinal and Life Course Studies. 2009;1:63–72. [Google Scholar]
- Höfler M, Pfister H, Lieb R, Wittchen H. The use of weights to account for non-response and drop-out. Social Psychiatry and Psychiatric Epidemiology. 2005;40:291–299. doi: 10.1007/s00127-005-0882-5. [DOI] [PubMed] [Google Scholar]
- Jones MP. Indicator and stratification methods for missing explanatory variables in multiple linear regression. Journal of the American Statistical Association. 1996;91:222–230. [Google Scholar]
- Kim JK, Brick JM, Fuller WA, Kalton G. On the bias of the multiple-imputation variance estimator in survey sampling. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2006;68:509–521. [Google Scholar]
- Little RJA, Rubin DB, editors. Statistical Analysis with Missing Data. New Jersey, NJ:: Wiley; 2002. [Google Scholar]
- Meng X-L. Multiple-imputation inferences with uncongenial sources of input. Statistical Science. 1994;9:538–573. [Google Scholar]
- Nielsen SF. Proper and improper multiple imputation. International Statistical Review. 2003;71:593–627. [Google Scholar]
- Power C, Elliott J. Cohort profile: 1958 British Birth Cohort (National Child Development Study) International Journal of Epidemiology. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- Priebe S, Fakhoury W, White I, Watts J, Bebbington P, Billings J, Burns T, Johnson S, Muijen M, Ryrie I, Wright C P.L.A.O.S. Group. Characteristics of teams, staff and patients: Associations with outcomes of patients in assertive outreach. British Journal of Psychiatry. 2004;185:306–311. doi: 10.1192/bjp.185.4.306. [DOI] [PubMed] [Google Scholar]
- Robins JM, Gill RD. Non-response models for the analysis of non-monotone ignorable missing data. Statistics in Medicine. 1997;16:39–56. doi: 10.1002/(sici)1097-0258(19970115)16:1<39::aid-sim535>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Robins J, Wang N. Inference for imputation estimators. Biometrika. 2000;87:113–24. [Google Scholar]
- Robins J, Rotnitzky A, Zhao L. Estimation of regression coefficients when some regressors are not always observed. Journal of the American Statistical Association. 1994;89:846–866. [Google Scholar]
- Royston J. Multiple imputation of missing values: Update of ice. Stata Journal. 2005;5:527–536. [Google Scholar]
- Rubin DB, editor. Multiple Imputation for Nonresponse in Surveys. New York, NJ:: Wiley; 1987. [Google Scholar]
- Schafer JL. Multiple imputation in multivariate problems when the imputation and analysis models differ. Statistica Neerlandica. 2003;57:19–35. [Google Scholar]
- Schenker N, Welsh AH. Asymptotic results for multiple imputation. Annals of Statistics. 1988;16:1550–1566. [Google Scholar]
- Stansfeld SA, Clark C, Caldwell TM, Rodgers B, Power C. Psychosocial work characteristics and anxiety and depressive disorders in midlife: The effects of prior psychological distress. Occupational and Environmental Medicine. 2008a;65:634–642. doi: 10.1136/oem.2007.036640. [DOI] [PubMed] [Google Scholar]
- Stansfeld SA, Clark C, Rodgers B, Caldwell TM, Power C. Childhood and adulthood socio-economic position and midlife depressive and anxiety disorders. Drug and Alcohol Dependence. 2008b;95:269–278. doi: 10.1016/j.drugalcdep.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Thomas C, Hypponen E, Power C. Prenatal exposures and glucose metabolism in adulthood. Diabetes Care. 2007;30:918–924. doi: 10.2337/dc06-1881. [DOI] [PubMed] [Google Scholar]
- Van Buuren S. Multiple imputation of discrete and continuous data by fully conditional specification. Statistical Methods in Medical Research. 2007;16:219–242. doi: 10.1177/0962280206074463. [DOI] [PubMed] [Google Scholar]
- Vansteelandt S, Carpenter J, Kenward MG. Analysis of incomplete data using inverse probability weighting and doubly robust estimators. Methodology. 2010;6:37–48. [Google Scholar]
- Wang N, Robins JM. Large-sample theory for parametric multiple imputation procedures. Biometrika. 1998;85:935–948. [Google Scholar]
- White IR, Carlin JB. Bias and efficiency of multiple imputation compared with complete-case analysis for missing covariate values. Statistics in Medicine. 2010;29:2920–2931. doi: 10.1002/sim.3944. [DOI] [PubMed] [Google Scholar]
- White IR, Royston P, Wood AM. Multiple imputation using chained equations: Issues and guidance for practice. Statistics in Medicine. 2010;30:377–399. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]


