Abstract
Adults and children with high-risk CRLF2-rearranged acute lymphoblastic leukemia (ALL) respond poorly to current cytotoxic chemotherapy and suffer unacceptably high rates of relapse, supporting the need to use alternative therapies. CRLF2 encodes the thymic stromal lymphopoietin (TSLP) receptor, which activates cell signaling in normal lymphocytes on binding its ligand, TSLP. We hypothesized that aberrant cell signaling occurs in CRLF2-rearranged ALL and can be targeted by signal transduction inhibitors of this pathway. In a large number of primary CRLF2-rearranged ALL samples, we observed increased basal levels of pJAK2, pSTAT5, and pS6. We thus characterized the biochemical sequelae of CRLF2 and JAK alterations in CRLF2-rearranged ALL primary patient samples via analysis of TSLP-mediated signal transduction. TSLP stimulation of these leukemias further induced robust JAK/STAT and PI3K/mTOR pathway signaling. JAK inhibition abrogated phosphorylation of JAK/STAT and, surprisingly, of PI3K/mTOR pathway members, suggesting an interconnection between these signaling networks and providing a rationale for testing JAK inhibitors in clinical trials. The PI3K/mTOR pathway inhibitors rapamycin, PI103, and PP242 also inhibited activated signal transduction and translational machinery proteins of the PI3K/mTOR pathway, suggesting that signal transduction inhibitors targeting this pathway also may have therapeutic relevance for patients with CRLF2-rearranged ALL and merit further preclinical testing.
Introduction
B-precursor acute lymphoblastic leukemia (ALL) is a heterogeneous group of diseases whose biologic behavior is largely driven by underlying genetic mutations. Initial risk stratification of ALL according to age and white blood cell count at diagnosis can be further refined by the identification of sentinel cytogenetic abnormalities.1 Therapy intensification for high-risk genetic subgroups has improved event-free survival dramatically during the past 50 years.2 However, ∼ 25%-30% of children have no known cytogenetic abnormality,3 and that percentage is even greater in adults with ALL.4 Currently, 80%-85% of children and 35%-50% of adults with ALL are cured with the use of modern multiagent therapeutic regimens,1,2,4,5 but most patients who do not respond to treatment will die.6 Thus, the identification of those patients who will ultimately relapse is a priority for basic and clinical investigators who seek discoveries that will lead to new therapies.7
Genomic profiling efforts via the use of high-density single-nucleotide polymorphism genotyping and gene expression profiling conducted by collaborative research groups in Europe and Israel and by the Children's Oncology Group (COG) via the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) Initiative of the National Cancer Institute (NCI) recently have identified novel gene alterations in high-risk (HR) adult and childhood ALL, including mutations in CRLF2 (ie, cytokine receptor-like factor 2),8–10 members of the JAK family of kinases (JAK1 and JAK2),11–14 and the IL-7 receptor α chain (IL-7Rα; IL7RA).15 JAK point mutations and CRLF2 alterations have been described in 60% of Down syndrome–associated ALL.9,11–14,16–18
CRLF2 alterations occur in up to 8% of unscreened childhood B-precursor ALL cases, in up to 15% of HR B-precursor ALL patients as defined by the Oxford Hazard Score (OHS) or the NCI-Rome risk criteria (age > 10 years and/or white blood cell count > 50 000/μL), and in ∼ 15% of adult B-precursor ALL patients.19–22 We and others have observed that NCI HR patients with CRLF2-rearranged (CRLF2r) ALL have high rates of minimal residual disease when measured by flow cytometry or PCR-based strategies at the end of induction chemotherapy.10,19,23 Retrospectively analyzed clinical studies note high rates of relapse and/or poor overall survival in NCI or OHS HR children and adults with CRLF2r ALL but an intermediate prognosis in standard-risk patients.19–23 Further biologic studies may facilitate the identification of factors contributing to these discrepant clinical outcomes, as well as provide additional rationale for the development of new therapies with potentially fewer toxicities, especially for patients (eg, adults) who are unable to tolerate traditional genotoxic chemotherapy.
Several alterations in CRLF2 resulting in its overexpression have been described in ALL: a focal ∼ 320-kilobase interstitial deletion of the pseudoautosomal region of the sex chromosomes (Xp22.23 or Yp11.32) that creates a fusion of CRLF2 to the G-protein–coupled purinergic receptor P2RY8 gene (P2RY8-CRLF2),8,9 translocation of CRLF2 to immunoglobulin heavy-chain transcriptional enhancers (14q32.3; IGH@-CRLF2),8,9,16,22 and a point mutation in CRLF2 resulting in a phenylalanine-to-cysteine substitution at amino acid 232 (F232C).18,22 Approximately 50% of CRLF2r ALLs also harbor JAK mutations, the most common of which gives rise to the ALL-specific JAK2 R683G alteration located in the JH2 pseudokinase domain, whereas virtually all cases with JAK mutations have concomitant CRLF2 alterations.8,11,14,16,22 Concurrent IKZF1 mutations also occur frequently,23 and IL-7RA mutations recently have been reported in a small number of CRLF2r B-precursor ALL cases.15
CRLF2 normally heterodimerizes with the IL-7Rα chain (also known as CD127) and binds its ligand, thymic stromal lymphopoietin (TSLP),24,25 an important physiologic signal for normal lymphopoiesis and for mediation of allergy and inflammation.25–27 Binding of TSLP to the TSLP receptor complex (TSLPR) activates STAT5 through currently unknown mechanisms but likely involves phosphorylation of JAK2 (pJAK2).26,28–30 Brown et al initially demonstrated murine TSLP-induced phosphorylation of STAT5, S6, and 4EBP1 in B-precursor ALL cell lines established from Eμ-RET transgenic mice.31,32 More recently, several groups in Europe and in the United States have observed constitutive and/or inducible phosphorylation of STAT5, S6, and/or ERK in Ba/F3 (murine pro-B cell lymphoma) cell lines transduced with human ALL-specific JAK and/or CRLF2 mutations, as well as inhibition of JAK/STAT signal transduction with chemical JAK inhibition or heat shock protein 90 inhibition.9,11,14–16,22,33,34
These data raise the possibility that multiple signaling cascades play a role in these leukemias. We hypothesized that CRLF2 rearrangements in primary human ALL result in perturbed signaling networks beyond the expected JAK/STAT signaling axis and thus sought to determine the phosphorylation status of intracellular signaling proteins in 2 human CRLF2r ALL cell lines, 46 CRLF2r ALL patient samples without and with JAK point mutations (JAKmut), and 33 CRLF2-wild type/JAK-wild type (CRLF2wt/JAKwt) patient samples after cytokine stimulation or exposure to signal transduction inhibitors (STIs). We demonstrated increased basal levels of pJAK2, pSTAT5, and pS6 and enhanced TSLP-mediated signal transduction via the JAK/STAT and PI3K/mammalian target of rapamycin (mTOR) pathways only in the CRLF2r ALL samples and could inhibit aberrant signaling in vitro with specific small molecule inhibitors of these pathways.
Methods
Cell culture
The human B-precursor ALL cell lines MUTZ5, MHH-CALL-4, RCH-ACV, and REH were purchased from the DSMZ. MUTZ5 overexpresses CRLF2 via an IGH@-CRLF2 translocation and has a JAK2 R683G mutation. MHH-CALL-4 has an IGH@-CRLF2 translocation and a JAK2 I682F mutation. RCH-ACV and REH are CRLF2wt B-precursor ALL cell lines with a t(1;19) and a t(12;21), respectively, that were used as negative controls. Cell lines were maintained in RPMI with FBS and penicillin/streptomycin/glutamine in a 37°C incubator with 5% CO2 and harvested in the logarithmic phase of growth with > 95% viability.
Patient samples and characteristics
We analyzed a total of 79 cryopreserved or fresh pretreatment primary ALL samples in a series of experiments, 46 of which had CRLF2 genomic alterations and 33 ALL samples that did not have CRLF2 rearrangements (Table 1 and supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Pretreatment leukemia samples were obtained from trials sponsored by the St Jude Children's Research Hospital (SJCRH) and the COG under institutional review board approval. Informed consent for tissue banking and research studies was obtained from patients and/or their guardians according to the Declaration of Helsinki. Patients were classified as NCI standard risk or HR on the basis of diagnostic clinical data.35 Thirty-two cryopreserved ALL samples were obtained from the SJCRH Tissue Bank and the COG Tissue Bank (Nationwide Children's Hospital).
Table 1.
CRLF2, JAK, and IL7RA mutation status of 79 patients with B-precursor ALL whose leukemias were analyzed by phosphoflow cytometry
| CRLF2 rearrangement | JAK mutation | IL7RA mutation | No. samples |
|---|---|---|---|
| No | No | No | 33 |
| Yes | No | No | 32 |
| Yes | Yes | No | 14 |
The SJCRH samples had been previously identified as CRLF2 overexpressors by gene expression analyses and subsequently characterized for specific CRLF2 and JAK alterations. Cells were thawed quickly, washed, and then assessed for postthaw cell counts and viability with a Vi-Cell analyzer (Beckman Coulter). Forty-seven (20 without and 27 with increased TSLPR) fresh ALL samples were obtained from the COG Core Flow Cytometry Laboratories at the Johns Hopkins University and the University of Washington, then submitted for phosphoflow cytometry analysis and for mutation testing. Mononuclear cell layers containing the ALL cells were isolated via standard density centrifugation. Six cryopreserved and 3 fresh samples were excluded from the phosphosignaling analyses because they did not meet the predetermined criterion of at least 25% shift of cells with the pervanadate positive control condition. Thus, 70 of the 79 cryopreserved or fresh leukemia samples were included in the final analyses.
PCR
DNA and RNA were extracted from isolated leukemia cells per standard methods. PCR was performed via the use of previously described cycling conditions and primers for JAK1 exons 13 and 1414; JAK2 exons 16, 20, and 2112,14; CRLF2 exon 622; and IL7RA exons 5 and 6.15 SuperScript III (Invitrogen) cDNA synthesis was performed per product instructions before RT-PCR for the P2RY8-CRLF2 fusion.9 Sanger sequencing was performed by Quintara Biosciences by the Hartwell Center at SJCRH. Mutations were analyzed by the use of Geospiza FinchTV Version 1.4 software.
FISH
Leukemia cell pellets were prepared in Carnoy fixative. FISH analyses to detect rearrangement and structural alterations of IGH@ and CRLF2 were performed as previously described.10
Western blotting
MUTZ5 cells were rested in serum-free media for 16 hours in IMDM with 1% BSA at 37°C, then incubated with 0.1% DMSO as a negative control or with STIs for 30 minutes at 37°C: the JAK1/JAK2 inhibitor ruxolitinib (1μM; Incyte), the mTOR inhibitor rapamycin (10nM; Calbiochem), the dual PI3K/mTOR inhibitor PI103 (1μM), the mTORC1/mTORC2 inhibitor PP242 (1μM), or the MEK inhibitor PD901 (1μM; Pfizer). Some cells were subsequently stimulated with recombinant human TSLP (25 ng/mL; R&D Systems) for an additional 30 minutes at 37°C. For noninhibitor conditions, cells were stimulated with TSLP for 30 minutes at 37°C. Pervanadate (125μM; prepared from sodium orthovanadate and 3% hydrogen peroxide; Sigma-Aldrich), an irreversible protein tyrosine phosphatase inhibitor, was also used as a positive control to elicit maximal phosphorylation of each signaling protein. Cells were immediately lysed in NP-40 cell lysis buffer (Invitrogen) supplemented with 1% protease and phosphatase inhibitors (Calbiochem). Lysates were separated via SDS-PAGE and immunoblotted on to nitrocellulose for probing with antibodies against total and phosphorylated (p) proteins STAT5, ERK 1/2, Akt, S6, 4EBP1, and eIF4E and against β-actin. Antibodies were from Cell Signaling Technologies (STAT5, pSTAT5Y694, ERK, pERKT42/44, Akt, pAktS473, S6, pS6S235/236, 4EBP1 and p4EBP1T37/46, eIF4E, and β-actin) and Invitrogen (peIF4ES209).
Phosphoflow cytometry
MUTZ5 cells and primary patient samples were prepared as described previously, rested without serum for 16 hours (MUTZ5) or 1 hour (patient samples) at 37°C and exposed to the identical series of agents at the same time points and concentrations as described previously. Some samples were also incubated with the JAK2 inhibitor XL019 (1μM; Exelixis) for 30 minutes at 37°C. The pervanadate exposure was used to delineate the potential dynamic signaling ranges and to identify subsets of cells that did not shift. After inhibition and/or stimulation, samples were immediately fixed with paraformaldehyde (Electron Microscopy Services) and permeabilized with ice-cold methanol (Electron Microscopy Services) per published methods.36 Samples were then stained with surface and/or phosphoprotein antibodies against human CD3, CD10, CD19, TSLPR, CD127, pSTAT5, pERK, pAkt, pS6, p4EBP1, and peIF4E before analysis on a Becton Dickinson LSRII flow cytometer. CRLF2r and CRLF2wt (control) ALL samples were gated on CD3−CD10+CD19+TSLPR+ cells and CD3−CD10+CD19+ cells, respectively, for phosphosignaling analyses. For cell line surface staining experiments, fluorescence-minus-one controls for each cell line were used.37 Antibodies were from BD Biosciences (CD3-Ax700 or CD3-Pacific Blue; CD10-PE-Cy7, CD19-Ax700 or CD19-APC-Cy7; CD127-Ax647, pSTAT5Y694-Ax647, pERKT42/44-Ax647, pAktS473-Ax488 or -V450; pS6S235/236-Ax488 or -V450; and isotype controls), Cell Signaling Technologies (unconjugated pS6S235/23 and p4EBP1T37/46), Invitrogen (unconjugated peIF4ES209 and Pacific Blue–conjugated secondary donkey anti–rabbit IgG), EBioscience (TSLPR-PE or -PerCP-eFluor-710 clone 1A6, CD127-FITC, CD132-PE), and Jackson ImmunoResearch (FITC-conjugated secondary donkey anti–rabbit IgG and goat anti–rat IgG). Data were analyzed via FlowJo Version 9.5 (TreeStar) and Cytobank38 software.
Proliferation and cytotoxicity assays
MUTZ5, MHH-CALL-4, RCH-ACV, and REH cells were incubated with media or increasing concentrations of TSLP (proliferation assays) or with DMSO or increasing concentrations of ruxolitinib, PI103, PP242, PD901, or ruxolitinib and rapamycin (cytotoxicity assays) in 96-well plates for 72 hours. MTT assays were performed as previously described.39 Experiments were performed 3 times with each experiment done in triplicate.
Statistical analyses
Digital phosphoflow cytometry data were recorded as raw data, as well as transformed with the inverse hyperbolic sine (arcsinh) equation via Cytobank to display and compare intensity values and to correct for large variances of some fluorophores.38 Data were then reported as the 75th percentile to minimize confounding effects of viable primary leukemia cells that signaled incompletely to pervanadate, then normalized to the basal phosphorylation levels for each patient sample for data display. Individual patient sample data were displayed as whisker plots via Prism Version 5.0 for Mac OS X (GraphPad Software). The distribution of patient sample data were tested and confirmed to be non-Gaussian. The 2-tailed Mann-Whitney U test with α = 0.05 was thus used to assess differences between the phosphorylation levels of basal states to those of the stimulation or inhibition conditions or between TSLP stimulation and STI with TSLP stimulation conditions. Data from MTT assays were displayed as bar graphs of mean data with SDs using Prism.
Results
Patients with CRLF2r ALL can be rapidly identified by flow cytometry
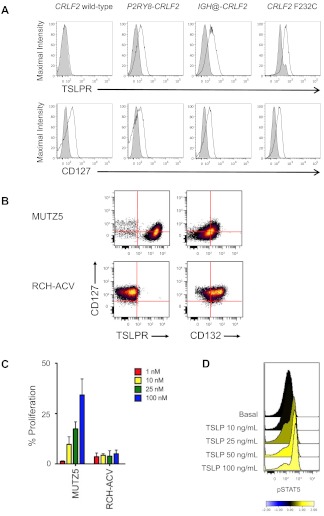
We first confirmed that primary CRLF2r ALL samples have increased surface levels of the TSLPR subunit,8 even in primary cells that had been fixed and permeabilized for phosphoflow cytometry analysis. The level of increased TSLPR staining was similar among leukemias with P2RY8-CRLF2, IGH@-CRLF2, and CRLF2 F232C (representative samples in Figure 1A). CD3−CD10+CD19+CRLF2r ALL samples were thus identified for these studies via their increased TSLPR staining in comparison with an isotype control (supplemental Figure 1). The presence or absence of a concomitant JAK or IL7RA mutation was not associated with changes in TSLPR surface staining (data not shown).
Figure 1.
Surface marker, proliferation, and intracellular signal transduction analyses of human CRLF2r and CRLF2wt leukemias. (A) Flow cytometry analysis of primary human CRLF2r and CRLF2wt ALL samples (N = 70; 4 representative samples shown) for the TSLPR and the IL-7Rα chain (CD127). CD10+CD19+ ALL cells with CRLF2 alterations demonstrated increased surface staining of the TSLPR regardless of the mechanism of CRLF2 rearrangement. No increased TSLPR staining was observed for leukemias without CRLF2 alterations. CD127 staining was similar in CRLF2r and CRLF2wt ALL cells. Shaded histograms indicate isotype control, and open histograms, stained cells. (B) Surface marker analysis of human B-precursor CRLF2r JAKmut ALL cell line MUTZ5 and CRLF2wt JAKwt cell line RCH-ACV. MUTZ5 cells stained uniformly for TSLPR, but were intermediate for CD132 (the common γ chain). RCH-ACV demonstrated bright CD127, absent TSLPR, and intermediate-to-bright CD132, suggesting the presence of the normal heterodimeric IL-7R. (C) Incubation of cells with human TSLP for 72 hours resulted in dose-dependent proliferation of MUTZ5, but not of RCH-ACV, measured by MTT assay. Red indicates 1 ng/mL; yellow, 10 ng/mL; green, 25 ng/mL; and blue, 100 ng/mL of TSLP. Data are normalized to the medium-only condition at 72 hours for each cell line. (D) Primary CRLF2r ALL cells were exposed to increasing concentrations of human TSLP for 30 minutes, then fixed and permeabilized for flow cytometric analysis of pSTAT5. Near-maximal STAT5 phosphorylation was elicited with 25 ng/mL of TSLP, the concentration used in the in vitro studies.
Surface staining of the IL-7Rα chain (CD127), the other subunit of the heterodimeric TSLPR, did not differ appreciably among CRLF2r (n = 17) and CRLF2wt (n = 24) ALL samples (representative data in Figure 1A). Analysis of MUTZ5 and MHH-CALL-4 demonstrated intermediate CD127, bright TSLPR, and dim-to-intermediate CD132 (the common γ chain, which associates with IL-7Rα to form the IL-7R) staining in contrast to RCH-ACV and REH, suggesting the possibility of preferential association of the IL-7Rα chain with the TSLPR subunit in CRLF2r ALL cells (Figure 1B and supplemental Figure 2). Isotype-stained normal mature human B lymphocytes and CD127- and CD132-stained normal T lymphocytes from peripheral blood were used as negative and positive controls, respectively (supplemental Figure 2).
TSLP, but not IL-7, induces growth and signaling in CRLF2r ALL
In proliferation assays, we observed TSLP dose-dependent increased proliferation in MUTZ5 and MHH-CALL-4 cells but not in RCH-ACV or REH (Figure 1C and supplemental Figure 3). In phosphoflow assays, in vitro stimulation of MUTZ5 cells and primary CRLF2r ALL cells with TSLP induced dose-dependent increases in STAT5 phosphorylation (n = 4; representative sample in Figure 1D). The 25 ng/mL TSLP concentration was selected for subsequent studies, which is concordant with concentrations used in other studies.27,40 TSLP stimulation of MUTZ5 robustly induced pJAK2 and pSTAT5 (Figure 2A and supplemental Figure 4). We did not observe increased TSLP-induced pSTAT1, pSTAT3, or pSTAT6 in MUTZ5, MHH-CALL-4, or 3 primary CRLF2r ALL samples (data not shown). TSLP stimulation also robustly induced pS6 and slightly increased pERK. PSTAT5 and pS6 were inhibited below basal levels by the JAK2-specific inhibitor XL019. Incubation of MUTZ5 or MHH-CALL-4 with IL-7 had no effect on pSTAT5, pS6, or pERK, which confirmed that IL-7 is not a ligand for the TSLPR (Figure 2A and supplemental Figure 5). In peripheral blood samples, we confirmed that IL-7 stimulation had no effect on CRLF2r primary ALL cells, but did induce pSTAT5 in CD3+ T cells of these patients, which provided a critical internal positive control (n = 12; representative sample in supplemental Figure 6).
Figure 2.
Constitutive and TSLP-inducible JAK/STAT and PI3K/mTOR signaling occurs in CRLF2r ALL. (A) Stimulation of MUTZ5 cells with IL-7 had no effect on JAK2, STAT5, S6, or ERK phosphorylation, as measured by phosphoflow cytometry. Stimulation with TSLP induced phosphorylation of pJAK2, pSTAT5, pS6, and, to a lesser extent, pERK. The JAK2-specific inhibitor XL019 inhibited pSTAT5 and pS6. Pervanadate was used as a positive control for measurement of maximal phosphorylation. Individual histograms are also displayed as a heat map for ease of comprehensive data visualization. Data were normalized to the basal phosphorylation level of each protein for colorimetric depiction of signaling changes. Blue indicates inhibition, and yellow is stimulation. (B) Basal levels of phosphorylation of JAK2, STAT5, S6, and ERK in primary CRLF2wt JAKwt (white bars, n = 27), CRLF2r JAKwt (light gray bars, n = 29), and CRLF2r JAKmut (dark gray bars, n = 14) were measured by phosphoflow cytometry, and median fluorescent intensity was calculated for each phosphoprotein. Data are displayed as whisker plots of 25th to 75th percentiles with means (central bars) and ranges (whiskers). PJAK2, pSTAT5, and pS6 were significantly elevated in the CRLF2r JAKwt and CRLF2r JAKmut samples in comparison with levels of the CRLF2wt JAKwt samples. ***P < .0005, **P < .005, *P < .05. Non-CRLF2–rearranged JAK wild-type = CRLF2wt JAKwt, CRLF2-rearranged JAK wild-type = CRLF2r JAKwt, CRLF2-rearranged JAK mutant = CRLF2r JAKmut. (C) Phosphoflow cytometry analysis of 41 primary human ALL samples without and with CRLF2 alterations and JAK mutations. Data are displayed as whisker plots of 25th to 75th percentiles with means (central bars) and ranges (whiskers). The dashed horizontal line represents normalized basal phosphorylation for each phosphoprotein. Stimulation with IL-7 (yellow bars) had no effect on phosphorylation in CRLF2wt JAKwt (n = 17), CRLF2r JAKwt (n = 12), or CRLF2r JAKmut (n = 12) ALL samples. TSLP stimulation (pink bars) induced pSTAT5 and pS6 in the CRLF2r samples regardless of concomitant JAK mutations (P < .05), but had no effect on CRLF2wt samples. XL019 (orange bars) inhibited phosphorylation below basal levels only for pS6 in the CRLF2r JAKmut subset (P = .013). Unlike in MUTZ5 cells, TSLP stimulation did not induce pERK significantly in these samples. *P < .05.
In our initial analyses of CRLF2wt and CRLF2r ALL patient samples, we observed greater basal levels of pJAK2, pSTAT5, and pS6 in samples with CRLF2 rearrangements than in those without rearrangements (P < .05 for all subgroups; Figure 2B). On the basis of the TSLP-inducible signaling we had observed in MUTZ5 and MHH-CALL-4, we hypothesized that in vitro TSLP stimulation would induce similar phosphorylation patterns in CRLF2r ALL patient samples. TSLP increased pSTAT5 and pS6 in the CRLF2r ALL cells regardless of JAK point mutation status (CRLF2r JAKwt, n = 14; CRLF2r JAKmut, n = 10; P < .05 for all subsets), but had no effect on CRLF2wt ALL (n = 17; Figure 2C). This phenomenon suggests that CRLF2 alterations are the main genetic event driving aberrant signal transduction. Interestingly, unlike in MUTZ5 and MHH-CALL-4, TSLP induced minimal pERK in the primary patient samples. As expected, IL-7 had no effect on pSTAT5, pS6, or pERK in the CRLF2r ALL patient samples. XL019 inhibited pS6 below basal levels in the CRLF2r JAKmut group (P = .013), but not in the CRLF2r JAKwt or CRLF2wt JAKwt groups, and it did not significantly inhibit pSTAT5 below basal levels in any of the 3 groups. Incubation of CRLF2wt JAKwt ALL cells with TSLP or IL-7 stimuli or inhibition with XL019 had no effect on any phosphoprotein examined (Figure 2C).
TSLP stimulation induces phosphorylation of multiple PI3K/mTOR pathway members, which can be abrogated by STIs
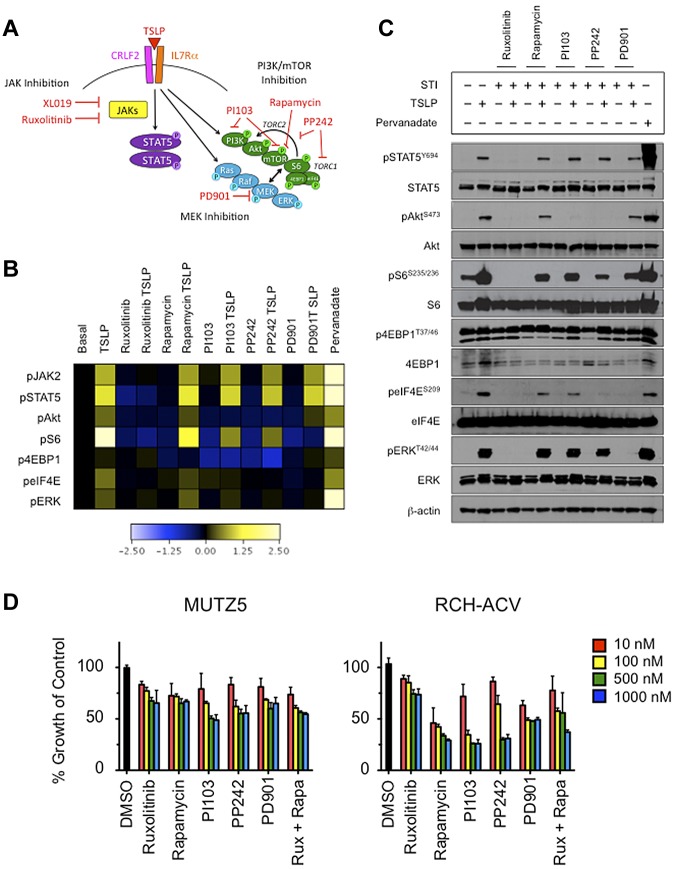
After we observed that TSLP stimulation of CRLF2r ALL cells also induces pS6, we then examined the phosphorylation status of other PI3K/mTOR pathway proteins, including Akt and the translational machinery proteins 4EBP1 and eIF4E, in MUTZ5 cells (Figure 3A). In addition to increases in pSTAT5, pS6, and pERK, TSLP stimulation of MUTZ5 also induced pAkt, p4EBP1, and peIF4E (Figure 3B and supplemental Figure 4). In addition to testing the JAK1/JAK2 inhibitor ruxolitinib and the MEK inhibitor PD901, we also tested the effects of several PI3K/mTOR pathway STIs (rapamycin, PI103, PP242) to delineate more precisely the role and potential interaction of the PI3K/mTOR, JAK/STAT, and Ras/MAPK pathways in TSLPR-mediated signaling (Figure 3A). Given our surprise that CRLF2r ALL cells were able to respond to in vitro TSLP stimulation, we then asked whether a “rescue” stimulation with TSLP could overcome the effects observed with inhibitors alone. Ruxolitinib inhibited pSTAT5, pAkt, pS6, and p4EBP1, which were not overcome by subsequent TSLP stimulation. As expected, there was no significant effect of rapamycin, PI103, or PP242 on pSTAT5, but pS6, p4EBP1, and (to a lesser degree) peIF4E were inhibited. Subsequent TSLP exposure partially rescued inhibition of pS6, whereas sustained inhibition was observed for p4EBP1. PD901 most appreciably inhibited pS6, and TSLP did not rescue cells from this pS6 inhibition (Figure 3B). These signaling phenomena were similarly recapitulated in MHH-CALL-4 (supplemental Figure 5).
Figure 3.
In vitro stimulation of CRLF2r ALL with TSLP-induced phosphorylation of STAT5 and PI3K pathway proteins and, to a lesser extent, ERK. Phosphorylation of signaling proteins was abrogated by specific STIs. (A) Hypothetical schema of aberrant signal transduction mediated by the TSLPR heterodimer in CRLF2r ALL and strategies for targeting the perturbed signaling nodes with STIs. (B) Phosphoflow cytometry analysis of fixed and permeabilized MUTZ5 cells demonstrated increased phosphorylation of JAK2, STAT5, ERK, Akt, S6, and eIF4E after stimulation with TSLP. Inhibition of basal and/or TSLP-induced phosphorylation of relevant proteins was observed with specific STIs. (C) Western blotting analysis of MUTZ5. Cells demonstrated constitutive phosphorylation of S6 and 4EBP1. TSLP-induced phosphorylation of signal transduction proteins and abrogation of phosphorylation by specific STIs was generally concordant with phosphoflow cytometry results in panel A, although more pronounced TSLP-induced pERK was observed by Western blotting. (D) Cytotoxicity assays of MUTZ5 and RCH-ACV cell lines treated with STIs. Cells were treated with the specified STIs for 72 hours before performing MTT assays. Ruxolitinib induced moderate dose-dependent cytotoxicity in MUTZ5. The PI3K/mTOR STIs induced dose-dependent cytotoxicity in both cell lines. Data are normalized to DMSO controls (black). Red indicates 10 nM; yellow, 100 nM; green, 500 nM; blue, 1000 nM of each STI. Rux indicates ruxolitinib, and Rapa is rapamycin.
We validated the phosphoflow cytometry results in MUTZ5 cells via Western blotting (Figure 3C). TSLP stimulation evoked phosphorylation of all examined phosphoproteins, including an increase in pERK. This induction of pERK was seen by phosphoflow cytometry to a lesser degree than by immunoblotting, which was most likely attributable to the different sensitivities of the antibodies used for each technique. In addition, the phosphoflow cytometry results are depicted as induced or inhibited signaling normalized to respective basal phosphorylation to visualize the dynamic signaling ranges above and below basal levels, respectively, whereas the Western blotting data are displayed independently as basal and induced or inhibited levels of phosphorylation. We also observed constitutively high basal p4EBP1 and total eIF4E levels via immunoblotting, which may account for the smaller increases in TSLP- or pervanadate-induced phosphorylation in comparison with other examined phosphoproteins. TSLP stimulation induced peIF4E and a slight increase in total 4EBP1 protein (but not p4EBP1), suggesting active protein translation and near-maximal phosphorylation of 4EBP1. Ruxolitinib shut down basal and TSLP-induced pSTAT5, pAkt, pS6, peIF4E, and pERK. Cells exposed to rapamycin, PI103, or PP242 demonstrated inhibition of pS6 and p4EBP1 with PP242 achieving the most potent and sustained inhibition. As observed in the phosphoflow cytometry assay (Figure 3B), rescue TSLP exposure partially overcame the rapamycin, PI103, and PP242-induced inhibition of pS6, but not the PI103- or PP242-induced inhibition of pAkt or the PD901-induced inhibition of pERK or peIF4E (Figure 3C). Rapamycin had no effect on pAkt.
Growth of CRLF2r ALL cell lines is inhibited by STIs
To assess the biologic effects of the STIs, we measured STI-induced cytotoxicity in short-term culture of ALL cell lines via MTT assay (Figure 3D and supplemental Figure 7). Dose-dependent cytotoxicity was observed with the STIs in all cell lines studied. Inhibition by ruxolitinib was modestly more pronounced in MUTZ5 and MHH-CALL-4 compared with RCH-ACV and REH, whereas rapamycin resulted in greater cytotoxicity in RCH-ACV and REH. The combination of ruxolitinib and rapamycin at 500 and 1000nM resulted in a mild increased cytotoxicity in MUTZ5 and MHH-CALL-4 (Figure 3D and supplemental Figure 7).
JAK/STAT and PI3K/mTOR STIs inhibit signal transduction preferentially in CRLF2r ALL patient samples
On the basis of our initial observations in cell lines and in primary ALL samples, we then examined our extended STI panel in an additional 7 CRLF2wt and 22 CRLF2r ALL samples. CRLF2r JAKwt and CRLF2r JAKmut samples were grouped for this analysis. No effects of the STIs were observed in the CRLF2wt samples with the exception of inhibition of pAkt, p4EBP1, and peIF4E by PP242 (P < .05; Figure 4A). As predicted, stimulation with TSLP induced pSTAT5, pAkt, pS6, p4EBP1, peIF4E, and pERK (P < .05 for all conditions) in the CRLF2r group (Figure 4B). Conversely, in the CRLF2r samples, ruxolitinib inhibited all 6 phosphoproteins examined (P < .05), and this inhibition was sustained despite subsequent TSLP stimulation. PI103 and PP242 inhibited TSLP-induced pAkt (P < .05). Similar to the MUTZ5 cells analyzed by Western blotting, the PI3K/mTOR pathway STIs and PD901 induced sustained inhibition of pS6 (P < .05), although TSLP restored p4EBP1 in rapamycin-, PI103-, PP242-, and PD901-exposed cells (Figure 4B).
Figure 4.
Expanded analysis of primary CRLF2r ALL samples demonstrates inhibition of JAK/STAT and PI3K pathway signaling. Analysis of JAK/STAT, PI3K/mTOR, and MAPK pathway STIs in 29 CRLF2wt (A) and CRLF2r (B) primary human leukemias. Data are displayed as whisker plots of 25th to 75th percentiles with means (central bars) and ranges (whiskers). The dashed horizontal line represents normalized basal phosphorylation for each phosphoprotein. Pink bars indicate TSLP; striped red, ruxolitinib, solid red, ruxolitinib + TSLP; striped green, rapamycin; solid green, rapamycin + TSLP; striped blue, PI103; solid blue, PI103 + TSLP; striped purple, PP242; solid purple, PP242 + TSLP; striped gray, PD901; solid gray, PD901 + TSLP. Minimal effects of TSLP stimulation or STIs were observed for the CRLF2wt samples. TSLP stimulation induced pSTAT5, pAkt, pS6, p4EBP1, peIF4E, and pERK in the CRLF2r samples (P < .05), and incubation with ruxolitinib resulted in continued inhibition of all aforementioned phosphoproteins (P < .05). PI103 inhibited TSLP-induced pAkt, pS6, and pERK (P < .05). PP242 abrogated TSLP-induced pAkt, pS6, and peIF4E (P < .05). PD901 inhibited TSLP-induced pS6, peIF4E, and pERK (P < .05).
Discussion
Our analyses are the first to provide biochemical characterization of the signaling consequences of CRLF2 rearrangements in a substantial cohort of primary human B-precursor ALL samples and highlight the potential utility of STI-based therapies for these patients. We observed basal activation of JAK/STAT and PI3K/mTOR signaling in CRLF2r ALL, as well as induced phosphorylation with TSLP stimulation of cell lines and primary patient samples. TSLP also increased proliferation of CRLF2r ALL cell lines, but not of CRLF2wt cell lines, in short-term culture. We further observed inhibition of aberrant JAK/STAT signaling in vitro by the JAK inhibitors XL019 and ruxolitinib and, intriguingly, suppression of S6 phosphorylation by JAK inhibition. Although the inhibition of pS6 by XL019 and ruxolitinib cannot exclude the possibility of off-target effects, we hypothesize that these data reflect interconnections between the JAK/STAT and PI3K/mTOR signaling networks given the increased Akt, S6, 4EBP1, and eIF4E phosphorylation in response to TSLP stimulation.41,42 We further explored the involvement of the PI3K/mTOR network via expanded phosphoflow cytometry analyses of cell lines and primary patient samples with rapamycin, PI103, and PP242. These STIs, particularly PP242, inhibited pAkt, pS6, p4EBP1, and/or peIF4E, suggesting that PI3K/mTOR pathway STIs may be an additional therapeutic strategy for patients with CRLF2r ALL. In MTT assays, ruxolitinib preferentially inhibited cell growth of the CRLF2r ALL cell lines, and incubation with PI3K/mTOR STIs also resulted in dose-dependent cytotoxicity.
Our results are generally concordant with those of other preclinical studies, although some differences exist. Although in one study investigators reported TSLP-induced cellular proliferation and induction of pSTAT5, pS6, and p4EBP1 in murine (presumably non-CRLF2 rearranged) B-precursor ALL cell lines and rapamycin-induced inhibition of pS6 and p4EBP1,31,32 we did not detect significant biochemical effects of IL-7, TSLP, or rapamycin in the 33 primary non-CRLF2r ALL samples in our studies. Murine and human TSLP share ∼ 43% sequence homology, however, and our discordant results may reflect inherent biologic differences of TSLP-mediated signaling in normal and malignant lymphopoiesis in mice versus humans.24,27,43,44 In addition, IL-7 is required for murine B-cell maintenance and growth, whereas the necessity of IL-7 in human B progenitor cell development remains less well-delineated.45,46
Recent studies in which the authors used Ba/F3 cells transduced with various ALL patient-derived CRLF2 and JAK mutations demonstrated constitutive and TSLP-inducible JAK/STAT signal transduction and cellular proliferation that could be abrogated by in vitro chemical JAK inhibition.8,9,14,16,22 Some groups also observed increased pS6 and/or pERK in some Ba/F3 cells transduced with ALL-specific CRLF2 and/or IL7RA mutations.15,22 Work by our group and by others thus demonstrates perturbed TSLP-induced JAK/STAT and PI3K/mTOR signal transduction in primary human CRLF2r ALL samples and the ability to inhibit these signaling networks via targeted inhibitors.8,9,14–16,22 Our STI data also suggest an interconnection between JAK/STAT and PI3K/mTOR signaling, although additional work is necessary to elucidate fully this connection. Although we were easily able to interrogate multiple surface markers and phosphoproteins in these studies, the use of more sophisticated flow cytometry technologies currently in development and conjugation of phosphoantibodies to additional distinguishable fluorochromes will greatly expand the scope of our ongoing studies.47,48
On the basis of our findings, we suggest that TSLPR surface staining could be included in routine clinical immunophenotyping for the identification of most newly diagnosed patients with CRLF2r ALL for swift treatment with STI-based therapies. Determination of the absence or presence of the TSLP-mediated phosphosignaling signature may be of particular diagnostic utility for TSLPR-dim samples with results obtainable within 24-48 hours of sample submission, but such approaches would require careful validation in clinical laboratory settings.
It is important to note the limitations of our in vitro studies. Primary patient samples are inherently heterogeneous, although most of the analyzed samples were from diagnostic BM specimens and were generally composed of > 90% leukemia cells with uniform surface staining. Because some samples had been cryopreserved and thawed, whereas the fresh samples were ∼ 48 hours old at time of processing because of unavoidable transit times, we gated on the live B-precursor lymphoblasts and used pervanadate as a positive control to ensure functional signaling of each sample because conventional propidium iodide staining to identify dead cells is not feasible in permeabilized cells. In addition, although optimized antibody cocktails and flow cytometer settings were standardized, we analyzed 2-6 samples per individual experiment on the basis of the availability of clinical samples and cannot exclude the possibility of minor experiment-to-experiment variability, although we did include MUTZ5 cells as a positive control cell line in each experiment. For this reason, we normalized the phosphorylation levels of each patient sample to its own basal levels for measurements of response to stimulation or inhibition.
In concordance with preclinical models, we observed constitutive activation of JAK/STAT and PI3K/mTOR signaling in primary ALL samples with CRLF2 rearrangements (Figure 2B). Although our study was not powered to detect potential signaling differences between the P2RY8-CRLF2 and IGH@-CRLF2 subgroups, we also noted a trend of greater basal levels of some phosphoproteins in some leukemia samples (eg, 0.5-1 log-fold greater basal pS6 for several IGH@-CRLF2 translocation samples than observed for P2RY8-CRLF2 fusion samples). The lack of obvious basal or TSLP-induced signaling differences between the CRLF2r JAKwt and the CRLF2r JAKmut groups, however, remains unsolved. Additional studies are ongoing to assess potential genetic differences among these subgroups. Furthermore, although we observed TSLP-inducible signaling in vitro, we do not know whether patients with CRLF2r ALL have elevated TSLP cytokine levels in vivo, which will require evaluation in future studies.
In summary, adults and children with NCI or OHS HR CRLF2r ALL treated with current regimens have unacceptably high rates of relapse, and new therapies are needed to improve clinical outcomes. These analyses are the first to our knowledge to begin to characterize the biochemical sequelae of CRLF2 rearrangements in primary human ALL patient samples. Our phosphoflow cytometry assays allowed simultaneous measurement of multiple phosphoproteins in small numbers and in heterogeneous subpopulations of primary leukemia cells. We demonstrated aberrant TSLP-mediated JAK/STAT and PI3K/mTOR pathway signal transduction in primary human CRLF2r ALLs, as well as the ability to inhibit these signaling networks with targeted STIs. Specifically, JAK inhibition was able to suppress both pathways, supporting the potential clinical utility of these agents. Our data provide rationale for further evaluation of these agents in preclinical and, eventually, clinical testing. Via identification of the specific perturbed signaling network(s) involved in individual leukemia samples, such assays may facilitate future selection of pertinent, tailored STI-based therapies. Precedent exists for the addition of STIs for treatment of pediatric leukemias, as evidenced by the dramatic improvement in overall survival for children with BCR-ABL1–positive ALL when imatinib was added to systemic chemotherapy.49
Additional studies are ongoing to assess the cytotoxic effects of JAK and PI3K/mTOR pathway STIs without and with conventional chemotherapy agents in short-term cultures of CRLF2r ALL primary samples. Results from these studies, as well as safety data from children with relapsed or refractory malignancies treated with ruxolitinib on the COG phase 1 trial ADVL1011, will likely facilitate the identification of potentially efficacious STIs for adults and children with CRLF2r ALL and will inform the development of subsequent clinical trials for these patients.
Supplementary Material
Acknowledgments
The authors thank Dr Douglas Clary (Exelixis) for providing XL019, Dr Jordan Fridman (Incyte) for providing ruxolitinib, and Dr Kevan Shokat (University of California, San Francisco) for providing PI103 and PP242. The authors acknowledge Ms Karen Bowles and Ms Stacey Middleditch (Johns Hopkins University) and Ms Eileen Stonerock (Nationwide Children's Hospital) for assistance with patient sample acquisition. The authors are grateful to the patients and their families for consenting to tissue banking and research studies. The authors also acknowledge Drs Jonathan Irish and Nikesh Kotecha at Stanford University for assistance with Cytobank and Dr Martin Carroll at the University of Pennsylvania for critical review of the manuscript.
Funding for this work was provided by National Institutes of Health training grants 5T32HD044331, 1T32CA128583, and K12CA076931 (to S.K.T.); NCI U10 CA098543 (COG, Chair's Award) and a supplement to that grant to support the TARGET Project, NCI GO 5RC2 CA14852902 (Targeted Therapies for Childhood Acute Lymphoblastic Leukemia: Translating Discovery into Practice); U10 CA98413, supporting the COG Statistical Center; U24 CA114766, supporting human specimen banking in NCI-supported cancer trials; a Conquer Cancer Foundation/American Society of Clinical Oncology Young Investigator Award (to S.K.T.); the TeamConnor Childhood Cancer Foundation (to S.K.T.); and the St Baldrick's Foundation (to M.L.L.). C.L.W. is the Maurice and Marguerite Liberman Chair in Cancer Research. S.P.H. is the Ergen Family Chair in Pediatric Cancer. C.G.M. is a Pew Scholar in the Biomedical Sciences and a St Baldrick's Scholar. M.L.L. is a Clinical Scholar of the Leukemia & Lymphoma Society.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.K.T. designed research, performed experiments, analyzed and interpreted data, and wrote the manuscript; M.Y.D. performed experiments and analyzed data; M.J.B., B.L.W., J.M.G.-F., S.P.H., and C.G.M. provided patient samples and interpreted data; I.-M.C. and R.C.H. performed experiments and analyzed data; C.L.W. interpreted data; M.L.L. designed research, interpreted data, and wrote the manuscript; and all authors critically reviewed the final manuscript before submission.
Conflict-of-interest disclosure: M.L.L. is a member of the Data Safety and Monitoring Committee for the Bristol-Myers Squibb clinical trial CA180226. The remaining authors declare no competing financial interests.
The current affiliation for S.K.T. is Department of Pediatrics, Division of Oncology, The Children's Hospital of Philadelphia, Philadelphia, PA.
Correspondence: Dr Mignon Loh, University of California, San Francisco School of Medicine, 513 Parnassus Ave, HSE 302, San Francisco, CA 94143; e-mail: lohm@peds.ucsf.edu.
References
- 1.Schultz KR, Pullen DJ, Sather HN, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children's Cancer Group (CCG). Blood. 2007;109(3):926–935. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowitz M, Devidas M, Hunger S, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children's Oncology Group study. Blood. 2008;111(12):5477–5485. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassan R, Hoelzer D. Modern therapy of acute lymphoblastic leukemia. J Clin Oncol. 2011;29(5):532–543. doi: 10.1200/JCO.2010.30.1382. [DOI] [PubMed] [Google Scholar]
- 5.Pui C, Robison L, Look A. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen K, Devidas M, Cheng SC, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children's Oncology Group study. Leukemia. 2008;22(12):2142–2150. doi: 10.1038/leu.2008.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunger SP, Raetz EA, Loh ML, Mullighan CG. Improving outcomes for high-risk ALL: translating new discoveries into clinical care. Pediatr Blood Cancer. 2011;56(6):984–993. doi: 10.1002/pbc.22996. [DOI] [PubMed] [Google Scholar]
- 8.Russell LJ, Capasso M, Vater I, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114(13):2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 9.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41(11):1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;116(23):4874–4884. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bercovich D, Ganmore I, Scott LM, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372(9648):1484–1492. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- 12.Gaikwad A, Rye CL, Devidas M, et al. Prevalence and clinical correlates of JAK2 mutations in Down syndrome acute lymphoblastic leukaemia. Br J Haematol. 2009;144(6):930–932. doi: 10.1111/j.1365-2141.2008.07552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearney L, Gonzalez De Castro D, Yeung J, et al. Specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood. 2009;113(3):646–648. doi: 10.1182/blood-2008-08-170928. [DOI] [PubMed] [Google Scholar]
- 14.Mullighan CG, Zhang J, Harvey RC, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106(23):9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shochat C, Tal N, Bandapalli OR, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med. 2011;208(5):901–908. doi: 10.1084/jem.20110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertzberg L, Vendramini E, Ganmore I, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115(5):1006–1017. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 17.Loudin MG, Wang J, Eastwood Leung HC, et al. Genomic profiling in Down syndrome acute lymphoblastic leukemia identifies histone gene deletions associated with altered methylation profiles. Leukemia. 2011;25(10):1555–1563. doi: 10.1038/leu.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapiro E, Russell L, Lainey E, et al. Activating mutation in the TSLPR gene in B-cell precursor lymphoblastic leukemia. Leukemia. 2010;24(3):642–645. doi: 10.1038/leu.2009.231. [DOI] [PubMed] [Google Scholar]
- 19.Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115(26):5393–5397. doi: 10.1182/blood-2009-11-256131. [DOI] [PubMed] [Google Scholar]
- 20.Chen IM, Harvey RC, Mullighan CG, et al. Outcome modeling with CRLF2, IKZF1, JAK and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group Study. Blood. 2012;119(15):3512–3522. doi: 10.1182/blood-2011-11-394221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ensor HM, Schwab C, Russell LJ, et al. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood. 2011;117(7):2129–2136. doi: 10.1182/blood-2010-07-297135. [DOI] [PubMed] [Google Scholar]
- 22.Yoda A, Yoda Y, Chiaretti S, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2010;107(1):252–257. doi: 10.1073/pnas.0911726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H, Chen IM, Wilson CS, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115(7):1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quentmeier H, Drexler HG, Fleckenstein D, et al. Cloning of human thymic stromal lymphopoietin (TSLP) and signaling mechanisms leading to proliferation. Leukemia. 2001;15(8):1286–1292. doi: 10.1038/sj.leu.2402175. [DOI] [PubMed] [Google Scholar]
- 25.Ziegler SF, Artis D. Sensing the outside world: TSLP regulates barrier immunity. Nat Immunol. 2010;11(4):289–293. doi: 10.1038/ni.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin SD, Koelling RM, Friend SL, et al. Thymic stromal lymphopoietin: a cytokine that promotes the development of IgM+ B cells in vitro and signals via a novel mechanism. J Immunol. 1999;162(2):677–683. [PubMed] [Google Scholar]
- 27.Scheeren FA, van Lent AU, Nagasawa M, et al. Thymic stromal lymphopoietin induces early human B-cell proliferation and differentiation. Eur J Immunol. 2010;40(4):955–965. doi: 10.1002/eji.200939419. [DOI] [PubMed] [Google Scholar]
- 28.Isaksen DE, Baumann H, Trobridge PA, Farr AG, Levin SD, Ziegler SF. Requirement for stat5 in thymic stromal lymphopoietin-mediated signal transduction. J Immunol. 1999;163(11):5971–5977. [PubMed] [Google Scholar]
- 29.Isaksen DE, Baumann H, Zhou B, et al. Uncoupling of proliferation and Stat5 activation in thymic stromal lymphopoietin-mediated signal transduction. J Immunol. 2002;168(7):3288–3294. doi: 10.4049/jimmunol.168.7.3288. [DOI] [PubMed] [Google Scholar]
- 30.Zhong J, Kim MS, Chaerkady R, et al. TSLP signaling network revealed by SILAC-based phosphoproteomics. Mol Cell Proteomics. 2012;11(6):M112.017764. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown VI, Hulitt J, Fish J, et al. Thymic stromal-derived lymphopoietin induces proliferation of pre-B leukemia and antagonizes mTOR inhibitors, suggesting a role for interleukin-7Ralpha signaling. Cancer Res. 2007;67(20):9963–9970. doi: 10.1158/0008-5472.CAN-06-4704. [DOI] [PubMed] [Google Scholar]
- 32.Brown VI, Seif AE, Reid GS, Teachey DT, Grupp SA. Novel molecular and cellular therapeutic targets in acute lymphoblastic leukemia and lymphoproliferative disease. Immunol Res. 2008;42(1-3):84–105. doi: 10.1007/s12026-008-8038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigert O, Lane AA, Bird L, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J Exp Med. 2012;209(2):259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tasian SK, Loh ML. Understanding the biology of CRLF2-overexpressing acute lymphoblastic leukemia. Crit Rev Oncog. 2011;16(1-2):13–24. doi: 10.1615/critrevoncog.v16.i1-2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith M, Arthur D, Camitta B, et al. Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. J Clin Oncol. 1996;14(1):18–24. doi: 10.1200/JCO.1996.14.1.18. [DOI] [PubMed] [Google Scholar]
- 36.Kotecha N, Flores NJ, Irish JM, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14(4):335–343. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243(1-2):77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 38.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. Chapter 10:Unit 10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teachey DT, Sheen C, Hall J, et al. mTOR inhibitors are synergistic with methotrexate: an effective combination to treat acute lymphoblastic leukemia. Blood. 2008;112(5):2020–2023. doi: 10.1182/blood-2008-02-137141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wohlmann A, Sebastian K, Borowski A, Krause S, Friedrich K. Signal transduction by the atopy-associated human thymic stromal lymphopoietin (TSLP) receptor depends on Janus kinase function. Biol Chem. 2010;391(2-3):181–186. doi: 10.1515/bc.2010.029. [DOI] [PubMed] [Google Scholar]
- 41.Bain J, Plater L, Elliott M, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertrand FE, Spengemen JD, Shelton JG, McCubrey JA. Inhibition of PI3K, mTOR and MEK signaling pathways promotes rapid apoptosis in B-lineage ALL in the presence of stromal cell support. Leukemia. 2005;19(1):98–102. doi: 10.1038/sj.leu.2403560. [DOI] [PubMed] [Google Scholar]
- 43.Carpino N, Thierfelder WE, Chang MS, et al. Absence of an essential role for thymic stromal lymphopoietin receptor in murine B-cell development. Mol Cell Biol. 2004;24(6):2584–2592. doi: 10.1128/MCB.24.6.2584-2592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 45.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 46.Jumaa H, Hendriks RW, Reth M. B cell signaling and tumorigenesis. Annu Rev Immunol. 2005;23:415–445. doi: 10.1146/annurev.immunol.23.021704.115606. [DOI] [PubMed] [Google Scholar]
- 47.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz KR, Bowman WP, Aledo A, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children's oncology group study. J Clin Oncol. 2009;27(31):5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.