Abstract
Bortezomib (Velcade) is used widely for the treatment of various human cancers; however, its mechanisms of action are not fully understood, particularly in myeloid malignancies. Bortezomib is a selective and reversible inhibitor of the proteasome. Paradoxically, we find that bortezomib induces proteasome-independent degradation of the TRAF6 protein, but not mRNA, in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) cell lines and primary cells. The reduction in TRAF6 protein coincides with bortezomib-induced autophagy, and subsequently with apoptosis in MDS/AML cells. RNAi-mediated knockdown of TRAF6 sensitized bortezomib-sensitive and -resistant cell lines, underscoring the importance of TRAF6 in bortezomib-induced cytotoxicity. Bortezomib-resistant cells expressing an shRNA targeting TRAF6 were resensitized to the cytotoxic effects of bortezomib due to down-regulation of the proteasomal subunit α-1 (PSMA1). To determine the molecular consequences of loss of TRAF6 in MDS/AML cells, in the present study, we applied gene-expression profiling and identified an apoptosis gene signature. Knockdown of TRAF6 in MDS/AML cell lines or patient samples resulted in rapid apoptosis and impaired malignant hematopoietic stem/progenitor function. In summary, we describe herein novel mechanisms by which TRAF6 is regulated through bortezomib/autophagy–mediated degradation and by which it alters MDS/AML sensitivity to bortezomib by controlling PSMA1 expression.
Introduction
Myelodysplastic syndrome (MDS) is a hematologic malignancy defined by blood cytopenias due to ineffective hematopoiesis, a predisposition to acute myeloid leukemia (AML), and genomic instability.1,2 Molecular-targeted therapies do not exist for MDS and the mechanisms of current therapies are largely unknown. More recently, bortezomib (Velcade), which is widely used for the treatment of multiple myeloma (MM) and lymphomas, is being evaluated as a single agent or in combination with chemotherapy in certain MDS and AML patients.3–5 Bortezomib is a selective and reversible inhibitor of the 26S proteasome, and mechanistic studies have revealed that inhibition of the proteasome complex leads to accumulation of lysine (K)–48 ubiquitin-linked proteins and consequently to cytotoxic effects in malignant cells.6 Proapoptotic and cell-cycle inhibitor proteins are stabilized after proteasome inhibition and thought to contribute to the anticancer effect by inducing apoptosis and inhibiting the cell cycle, respectively.6 Nevertheless, the molecular and cellular mechanisms of bortezomib-induced cytotoxicity remain unknown, particularly in MDS/AML. Whereas the role of bortezomib in regulating cell-cycle entry and survival have been characterized partially in MDS/AML,7–9 recent evidence has pointed to a more general cellular effect: bortezomib treatment results in the accumulation of nondegraded proteins, leading to endoplasmic reticulum stress and autophagy in cancer.10–12 Under normal cellular stresses, autophagy, a catabolic pathway, degrades long-lived proteins and defective and superfluous organelles.13 However, under conditions of extreme cellular stress, autophagy is used by the cell to undergo death.14
Human miR-146a, a candidate gene in del(5q) MDS/AML, is reduced significantly in del(5q) and normal karyotype MDS/AML patients.15–17 TRAF6 is a key target of miR-146a15,18,19 and, as expected, miR-146a–knockout mice have a dramatic increase in TRAF6 protein within the hematopoietic compartment.20,21 Retroviral overexpression of TRAF6 in mouse hematopoietic stem/progenitor cells results in MDS-like hematopoietic defects and progression to AML.15 Bortezomib has been shown previously to be effective for an MDS patient with del(5q) and was also reported to reduce directly TRAF6 mRNA and protein in osteoclast precursors from MM patients.22,23 Because TRAF6 is implicated in MDS/AML and bortezomib has been shown to be effective in del(5q) MDS and to inhibit TRAF6 in MM, we hypothesized that one mechanism of bortezomib action is through inhibition of TRAF6.
In the present study, we identified TRAF6 as a relevant target of bortezomib-induced cytotoxicity in MDS/AML (independent of chromosome 5q status). Paradoxically, we found that bortezomib induced the degradation of the TRAF6 protein, but not mRNA, in MDS/AML cells. The reduction in TRAF6 protein coincided with bortezomib-induced autophagy, and subsequently with apoptosis in MDS/AML cells. The addition of an autophagy inhibitor, 3-methyladenine (3-MA), to bortezomib-treated AML cells restored TRAF6 protein expression and enhanced cell viability. These findings suggest that a mechanism of bortezomib-induced cell death in myeloid malignancies involves elimination of the TRAF6 protein by autophagic lysosomes. RNAi-mediated depletion of TRAF6 in MDS and AML samples resulted in reduced malignant leukemic progenitor function and rapid apoptosis. To determine the molecular consequences of the loss of TRAF6, in the present study, we applied gene-expression profiling and identified genes relevant to the survival of MDS and AML cells. One significantly down-regulated gene encodes the α-subunit of the proteasome (PSMA1). Reduced expression of PSAM1 resensitizes bortezomib-resistant leukemia cells. In summary, the results of the present study show that bortezomib initiates apoptosis in MDS/AML cells by autophagic/lysosome–mediated degradation of the TRAF6 protein. These findings implicate TRAF6 in bortezomib-induced cell death and in the survival of MDS/AML progenitors.
Methods
Cell lines and CD34+ cells
The AML cell lines TF-1, THP1, and HL60 were from ATCC. The MDS cell line MDS-L was provided by Dr Kaoru Tohyama (Kawasaki Medical School, Okayama, Japan).24 Detailed cytogenetics for the cell lines has been described previously.17 CD34+ cells were positively selected from cryopreserved BM or cord blood cells by immunomagnetic separation (at the Cincinnati Children's Hospital Medical Center core facility). Bortezomib-resistant (Bort-R) and bortezomib-sensitive (Bort-S) THP1 cells were described previously.25 Culture conditions for each cell line are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Patient MDS and AML samples
The primary MDS samples (MDS-01 and MDS-02) are mononuclear cells from BM aspirates obtained at diagnosis as part of a multicenter phase 2 trial based in Italy evaluating the safety and efficacy of lenalidomide. The patients were diagnosed as low-risk MDS based on International Prognostic Scoring System criteria. Cytogenetic assays confirmed del(5q) in both patients. The primary AML-03 cells were from a patient with relapsed/refractory M4 AML with t(8;18)(q22:q23) and t(11;13)(q21:q12).26 BM cells were cultured in StemSpan SFEM (StemCell Technologies) supplemented with human SCF, Flt3 ligand, erythropoietin, IL-3, and IL-6 at final concentrations of 10 ng/mL. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The study was approved by the Cincinnati Children's Hospital Medical Center.
Reagents
Bortezomib was from Millennium Pharmaceuticals and LC Laboratories (B-1408). MG132 (C2211), 3-MA, bafilomycin-A (BafA; B1793), rapamycin (R8781), and thapsigargin (T9033) were from Sigma-Aldrich. Flag-tagged TRAF6 cDNA was provided by Dr Aly Karsan (Vancouver, BC) and cloned into pLEGO-iG2 for stable overexpression. The pLKO.1 (OpenBiosystems) constructs were obtained from the lentiviral core at Cincinnati Children's Hospital Medical Center.
Survival analysis
Annexin V analysis has been described previously.15 For the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, cell growth was measured with the MTT reduction assay (ATCC) according to the manufacturer's protocol in the presence of bortezomib or DMSO for 24 hours. For the shRNA knockdown experiment, the cell lines were transduced with virus encoding shTRAF6 or shPSMA1 and control vector at a multiplicity of infection of approximately 0.5-1. After 48 hours, transduced cells were treated with bortezomib or DMSO for 24 hours before analysis.
Abs for immunoblotting
PVDF membranes were blocked in 1 × TBS with 0.1% Tween 20 and 5% fat-free milk, and blotted with GAPDH (5174; Cell Signaling Technology), TRAF6 (sc-7221; Santa Cruz Biotechnology), p62 (GP62-C; PROGEN), LC3 (4108; Cell Signaling Technology), or ubiquitin (sc-8017; Santa Cruz Biotechnology). Quantification of bands was determined using Adobe Photoshop Version CS5 software.
Microarray analysis
TF-1 cells were transduced with lentivirus targeting human TRAF6 or a nontargeting control. At 2.5 days after transduction, green fluorescent protein–positive populations were isolated by flow cytometry. Total RNA was extracted and purified with the Quick-RNA MiniPrep Kit. RNA quality was tested using the Agilent Bioanalyzer 2100 (Hewlett Packard). Total RNA was reverse transcribed, labeled, and hybridized onto the GeneChip Human Gene 1.0 ST Array (Affymetrix). Scanning was performed with the Affymetrix GeneChip Version 3.2 Scanner 3000 7G and evaluated with GeneChip operating software (Affymetrix). Data mining was performed with GeneSpring GX 11.5 software (Agilent Technologies). Gene set enrichment analysis was performed on a JAVA-based dataset supported by the Broad Institute.27 For evaluation of TRAF6 and PSMA1 coexpression in MDS and AML, previously published datasets were used.28,29 All microarray data are available on the Gene Expression Omnibus public database under accession no. GSE38519.
Statistical analysis
Results are depicted as the means ± SEM. Statistical analyses were performed using Student t test. Prism Version 4 software (GraphPad) was used for statistical analysis.
For additional experimental details, please see supplemental Methods.
Results
Bortezomib suppresses MDS/AML cell survival and progenitor function by inhibiting the proteasome
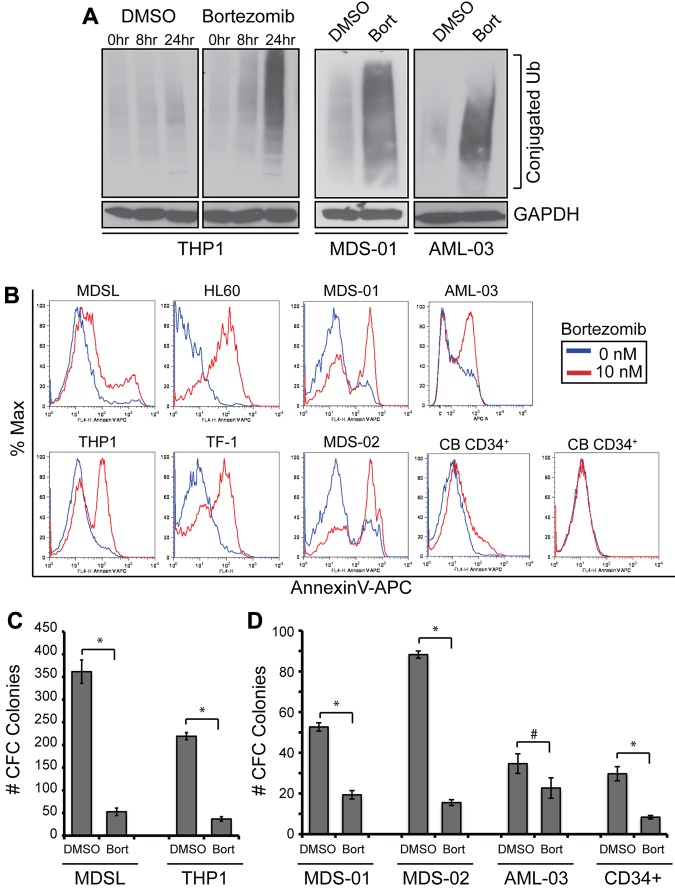
Bortezomib has been shown to be effective for suppressing the proliferation and survival of malignant cells by inhibiting proteasomal degradation of K48-linked ubiquitinated proteins directly.30 In the present study, an accumulation of ubiquitinated proteins, which is an indication of suppressed proteasome function, was evident after 10nM bortezomib treatment in THP1 cells or primary MDS and AML BM cells (Figure 1A). MDS/AML cell lines and primary cells cultured with bortezomib exhibited increased apoptosis (Figure 1B) and impaired cell proliferation (supplemental Figure 1) within 24 hours of treatment. In contrast, 2 independent cord blood–derived CD34+ cells did not undergo apoptosis in the presence of bortezomib (Figure 1B). To determine whether bortezomib also suppresses leukemic progenitor activity, MDS/AML cells were evaluated for colony formation in methylcellulose (Figure 1C-D). MDS/AML cell lines and primary MDS and AML BM cells treated with bortezomib exhibited a 30%-90% reduction in colony formation. We observed similar effects of bortezomib when cells were cultured without supplemented cytokines, excluding the possibility that exogenous growth factors affect bortezomib sensitivity in vitro (supplemental Figure 2A). These findings indicate that bortezomib can target effectively leukemic cells and progenitors, a relevant population of disease-initiating cells.
Figure 1.
Inhibitory effect of bortezomib on MDS and AML cell viability and progenitor function. (A) THP1 cells were treated with 10nM bortezomib or DMSO for the indicated times. Protein lysates were immunoblotted for ubiquitin-conjugated proteins and GAPDH (left panel). BM cells from MDS (MDS-01) and AML (AML-03) patients were treated with 10nM bortezomib or DMSO for 24 hours and evaluated by immunoblotting for ubiquitin and GAPDH (right panels). (B) The indicated cell lines and primary patient samples were treated with bor-tezomib for 24 hours and analyzed for cell survival by staining for annexin V+ cells. (C) Hematopoietic stem/progenitor cell colony-forming potential of THP1 and MDSL cell lines treated with bortezomib (10nM) was determined in methylcellulose. (D) Hematopoietic stem/progenitor cell colony-forming potential of control CD34+ cells and BM cells from MDS (MDS-01 and MDS-02) and AML (AML-03) patients treated with bortezomib (10nM) was determined in methylcellulose. Colonies were evaluated after 10 days. *P < .05; #P < .1.
Bortezomib targets TRAF6 for lysosomal-mediated protein degradation
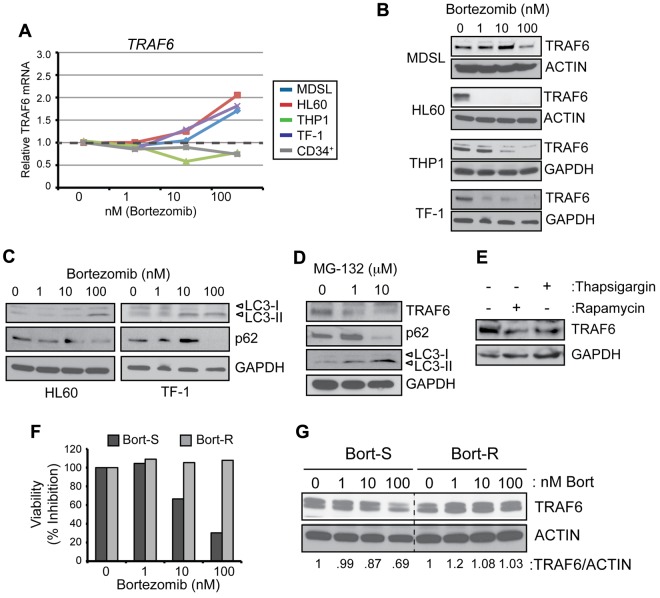
A recent study indicated that bortezomib inhibits TRAF6 mRNA and protein expression in osteoclast precursors from myeloma patients.22 To further investigate this finding, in the present study, a panel of 4 MDS/AML cell lines were cultured with increasing concentrations of bortezomib for 24 hours. In contrast to the effects seen in osteoclast precursors, bortezomib did not suppress TRAF6 transcription in MDS/AML cells (Figure 2A). However, bortezomib reduced protein levels of TRAF6 dramatically and in a dose-dependent manner in MDS/AML cells (Figure 2B). The paradoxical observation that the TRAF6 protein was diminished despite blocked proteasome activity indicates that other cellular mechanisms contribute to TRAF6 protein degradation.
Figure 2.
Reduced TRAF6 protein expression in bortezomib-treated cells coincides with autophagy activation. (A) TRAF6 mRNA was measured by quantitative PCR in the indicated cell lines and CD34+ cells treated with increasing concentrations of bortezomib for 24 hours. (B) TRAF6 protein was measured by immunoblotting of the indicated cell lines treated with increasing concentrations of bortezomib for 24 hours. (C) HL60 and TF-1 cell lines cultured with bortezomib for 24 hours were evaluated by immunoblotting for LC3-I/II, p62, and GAPDH. (D) TF-1 cells were treated with the proteasome inhibitor, MG-132 (0, 1, and 10μM) for 24 hours and evaluated by immunoblotting for TRAF6, LC3-I/II, p62, and GAPDH. (E) TF-1 cells were treated with an autophagy inducer (100nM rapamycin) and an ER stress inducer (100nM thapsigargin). (F) Cell viability of THP-1 cells that have acquired resistance to bortezomib (Bort-R) and that are sensitive (Bort-S) was determined after treatment with increasing concentrations of bortezomib for 24 hours. (G) TRAF6 protein was measured by immunoblotting Bort-S and Bort-R cells treated with increasing concentrations of bortezomib for 24 hours. Values below the gel represent the intensity of TRAF6 relative to ACTIN.
Previous studies have suggested that bortezomib can induce autophagy,31 a process resulting in lysosomal-mediated degradation of select cellular components under stressed conditions. To determine whether bortezomib-mediated degradation of TRAF6 occurs as a consequence of autophagy, common measures of autophagy were assessed in MDS/AML cells. Increasing concentrations of bortezomib (1, 10, and 100nM for 24 hours) resulted in activation of autophagy in MDS/AML cells, as indicated by the conversion of LC3-I to LC3-II and the degradation of p62 (Figure 2C), irrespective of cytokine conditions (supplemental Figure 2B). In most MDS/AML cell lines examined, elimination of the TRAF6 protein coincided with conversion of LC3-I and p62 degradation (Figure 2B-C and supplemental Figure 3A-C). To determine whether degradation of TRAF6 and induction of autophagy are specific to bortezomib, another proteasome inhibitor (MG-132) was evaluated. As shown in Figure 2D, treatment with MG-132 (10μM for 24 hours) similarly resulted in degradation of the TRAF6 protein and p62 and accumulation of LC3-II. In addition, activation of autophagy by rapamycin (100nM for 24 hours) mediated the degradation of TRAF6, albeit not as significantly as with the proteasome inhibitors (Figure 2E). A potential explanation for bortezomib-induced autophagy is the accumulation of nondegraded proteins contributing to endoplasmic reticulum (ER) stress. Treatment of MDS/AML cells with thapsigargin (100nM for 24 hours), an inducer of ER stress, coincided with decreased TRAF6 protein (Figure 2E).
To further explore the effects of bortezomib-induced degradation of TRAF6, we used THP1 cells that have acquired resistance to bortezomib as a result of a mutation in a highly conserved bortezomib-binding pocket within the proteasome β5-subunit (PSMB5) protein.25,32 As expected, increasing concentrations of bortezomib reduced the viability of Bort-S cells but did not impair the viability of Bort-R cells even when treated with 200nM bortezomib (Figure 2F and supplemental Figure 4). In support of our observations that inhibition of the proteasome results in degradation of TRAF6, Bort-S cells exhibited reduced levels of the TRAF6 protein by > 30%, whereas Bort-R cells maintained normal TRAF6 levels in the presence of bortezomib (Figure 2G). These observations suggest that inhibition of the proteasome induces ER stress, leading to autophagy and selective degradation of the TRAF6 protein.
TRAF6 protein expression and cell viability in bortezomib-treated MDS/AML cells is restored when autophagy is inhibited
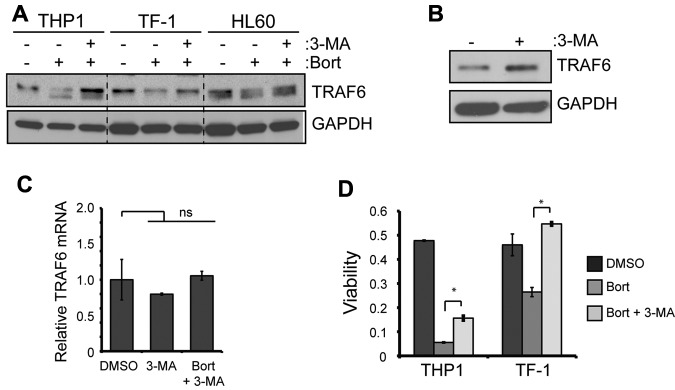
3-MA, an inhibitor of autophagy, was used to evaluate whether TRAF6 protein degradation is reversible in the presence of bortezomib. This early-stage autophagic lysosome and type I/III PI3K inhibitor33 blocked bortezomib-induced autophagy (LC3-II and p62 accumulation) and restored TRAF6 protein expression significantly in all MDS/AML cell lines examined (Figure 3A and supplemental Figure 3C). 3-MA treatment restored TRAF6 protein expression effectively even at the highest doses of bortezomib (supplemental Figure 3C). Surprisingly, BafA, a late-stage autophagy inhibitor that blocks vacuolar-type H+-ATPase, did not prevent the degradation of TRAF6 (supplemental Figure 3C). In the absence of bortezomib, MDS/AML cells treated with 3-MA also exhibited increased TRAF6 protein, indicating that steady-state TRAF6 protein is regulated by autophagic lysosomes (Figure 3B). Given the dramatic effect of 3-MA on TRAF6 protein, we also investigated whether 3-MA may regulate TRAF6 mRNA concomitantly. TRAF6 mRNA expression in MDS/AML cells did not change significantly in the presence of 3-MA or with the combination of 3-MA and bortezomib (Figure 3C), further supporting our hypothesis that a posttranscriptional mechanism regulates TRAF6 protein levels.
Figure 3.
TRAF6 is degraded by the autophagy lysosome pathway. (A) The indicated cell lines were cultured with 10nM bortezomib and cotreated with 5mM 3-MA for 24 hours. TRAF6 and GAPDH protein was determined by immunoblot analysis. (B) In the absence of proteasome inhibition, TF-1 cells were treated with 5mM 3-MA for 24 hours, and TRAF6 protein expression was measured by immunoblot analysis. (C) cDNA from TF-1 cells treated with 3-MA (5mM) or 3-MA and bortezomib (10nM) was evaluated for TRAF6 mRNA expression. Data are relative to GAPDH and normalized to cells treated with DMSO. (D) Cell viability of THP1 and TF-1 cells was determined after treatment with DMSO, 10nM bortezomib, or bortezomib and 5mM 3-MA for 24 hours. *P < .05.
We assessed cell viability to investigate the cellular consequences of 3-MA on bortezomib-treated MDS/AML cells. Compared with cells treated with bortezomib alone, the addition of 3-MA to bortezomib-treated cells restored cell viability in 2 independent cell lines (Figure 3D). BafA did not restore cell viability of bortezomib-treated cells, but rather synergized to induce cell death (supplemental Figure 3D), consistent with our observation that BafA does not revert TRAF6 degradation. We conclude that inhibiting early-stage autophagy with 3-MA suppresses the cytotoxic effects of bortezomib on MDS/AML cells by preventing the lysosomal degradation of TRAF6.
Quantitative changes in TRAF6 protein affect bortezomib sensitivity by regulating proteasome subunit expression
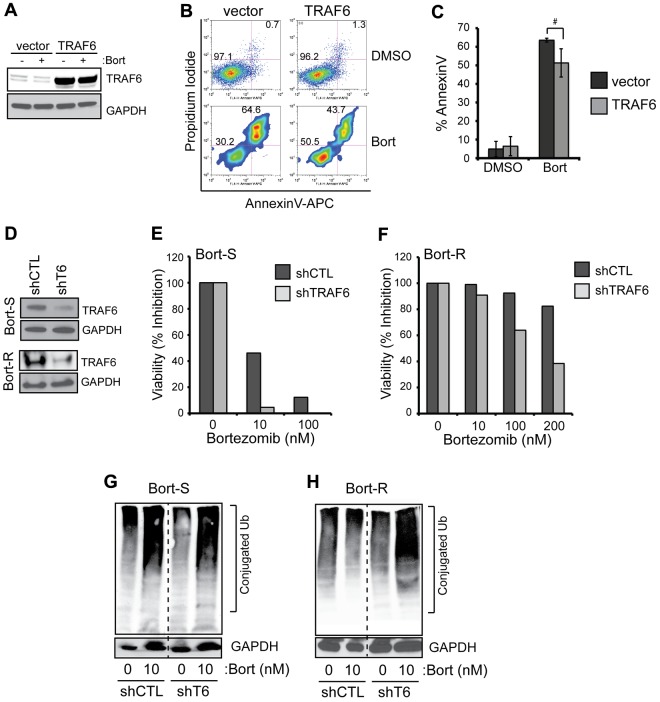
To investigate whether restoring TRAF6 can rescue bortezomib-induced cell death, TRAF6 was overexpressed using a lentiviral vector in THP1 cells (Figure 4A). THP1 cells expressing either empty vector or TRAF6 were then treated with 10nM bortezomib and evaluated for cell survival. As shown in Figure 4B, overexpression of TRAF6 ameliorated the cytotoxic effects of bortezomib: THP1 cells transduced with TRAF6 exhibited a trend to lower apoptosis compared with vector-transduced cells when treated with bortezomib (51% ± 7.6% vs 64% ± 1.1%; P = .1; Figure 4C). These data indicate that TRAF6 overexpression provides moderate protection from bortezomib-induced apoptosis, but does not suffice to reverse the proapoptotic effect of bortezomib in MDS/AML. Conversely, we knocked down TRAF6 to determine whether depletion of TRAF6 can sensitize cells to bortezomib (Figure 4D-F). For these experiments, a control shRNA (shCTL) or an shRNA targeting TRAF6 (shTRAF6 #48) were transduced into Bort-S cells (Figure 4D). Knockdown of TRAF6 reduced the viability of bortezomib-treated cells by approximately 90% compared with cells expressing the control shRNA (10nM, P = .0044, and 100nM, P = .019; Figure 4E). These findings suggest that TRAF6 loss of function sensitizes MDS/AML cells to bortezomib treatment.
Figure 4.
Levels of TRAF6 expression affect the cytotoxic effects of bortezomib. (A) THP1 cells were transduced with a bicistronic lentiviral vector encoding TRAF6 and green fluorescent protein (GFP) or an empty vector expressing only GFP. Transduced cells were sorted for GFP expression, cultured with bortezomib (10nM) for 24 hours, and protein lysates were evaluated for TRAF6 expression. (B) Cell viability of bortezomib-treated THP1 cells transduced with vector or TRAF6 was measured by annexin V/propidium iodide staining. (C) Summary of replicate experiments from panel B. (D) Bort-S and Bort-R THP1 cells were transduced with shRNA lentiviral vectors targeting TRAF6 or shCTL. Knockdown of the TRAF6 protein was confirmed by immunoblot analysis. (E-F) Cell viability of Bort-S (E) and Bort-R (F) cells transduced with shTRAF6 or control vector was determined by MTT assay after treatment with increasing concentrations of bortezomib for 24 hours (P < .05). (G-H) Proteasome inhibition in bortezomib-treated Bort-S (G) and Bort-R (H) cells transduced with shTRAF6 or control vector was determined by immunoblotting for ubiquitinated proteins (Ub) on total cell lysates. #P < .1; *P < .05.
We next determined the effects of TRAF6 depletion in Bort-R cells (Figure 4D,F). After knockdown of TRAF6, Bort-R cells were resensitized to as low as 100nM bortezomib (P = .023) and exhibited a > 50% reduction in cell viability when cultured with 200nM bortezomib (Figure 4F and supplemental Figure 4). In contrast, at 200nM bortezomib, Bort-R cells expressing the control shRNA (shCTL) maintained cell viability comparable to untreated cells (Figure 4F). To determine the consequences of TRAF6 deletion on proteasome function in Bort-R cells, we examined the accumulation of ubiquitinated proteins (Figure 1A). As expected, knockdown of TRAF6 in Bort-S cells did not result in significant changes in the accumulation of ubiquitinated proteins when cultured with bortezomib (Figure 4G). Similarly, Bort-R cells transduced with shCTL did not accumulate ubiquitinated proteins when treated with bortezomib (Figure 4H). In stark contrast, knockdown of TRAF6 in Bort-R cells treated with bortezomib resulted in an accumulation of ubiquitinated proteins (Figure 4H). This finding indicates that knockdown of TRAF6 restores bortezomib-mediated inhibition of proteasome function in Bort-R cells.
TRAF6 regulates the expression of PSMA1
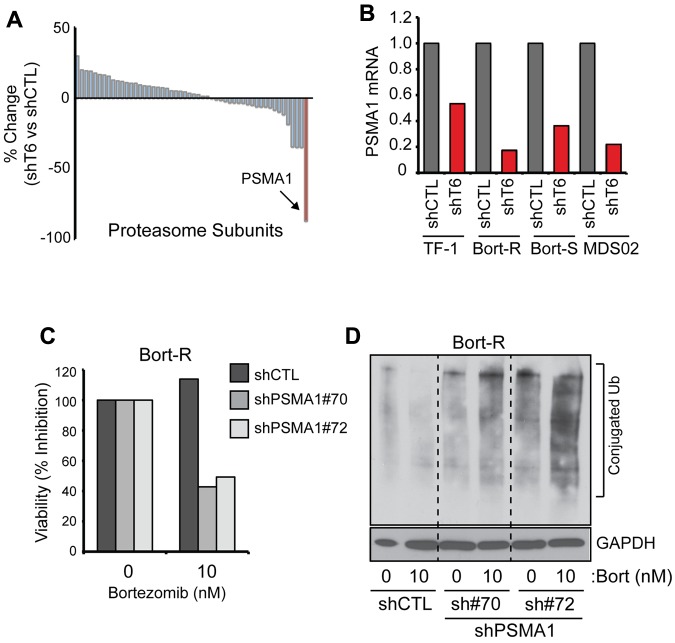
We next performed a gene-expression analysis to explore a potential explanation for the increased sensitivity of Bort-R cells after knockdown of TRAF6 in TF-1, a human AML cell line. Multiple RNAi constructs targeting TRAF6 were evaluated for TRAF6 expression and an effect on cell viability in TF-1 cells (supplemental Figure 5A-C). TF-1 cells isolated 2.5 days after transduction were used for the gene-expression analysis, because this time point precedes measurable effects on cell viability after loss of TRAF6. We identified 185 genes that were expressed differentially after TRAF6 knockdown (greater than 1.5-fold; P < .05). It was reported previously that the stoichiometry of proteasome subunits affects the effectiveness of proteasome inhibition by bortezomib.34,35 Therefore, we searched for differential expression of proteasome subunit genes in TRAF6-depleted cells. Of 52 proteasome subunit genes expressed, only PSMA1 was down-regulated significantly in cells with TRAF6 knockdown (P = .022; Figure 6A and supplemental Table 1). To confirm that TRAF6 regulates PSMA1 expression, Bort-R and Bort-S THP1 cells and MDS patient cells were transduced with shTRAF6 and evaluated for PSMA1 expression (Figure 6B). Consistent with the microarray analysis, PSMA1 expression was down-regulated by 85% and 70% in Bort-R and Bort-S THP1 cells, respectively, and by 80% in MDS patient BM cells after knockdown of TRAF6 (Figure 6B). Analysis of gene-expression data from primary MDS and AML CD34+ cells revealed overexpression of TRAF6 mRNA in a subset of patients and a positive correlation between TRAF6 and PSMA1 mRNA expression, further supporting our findings that TRAF6 regulates PSMA1 expression (supplemental Figure 6).
Figure 6.
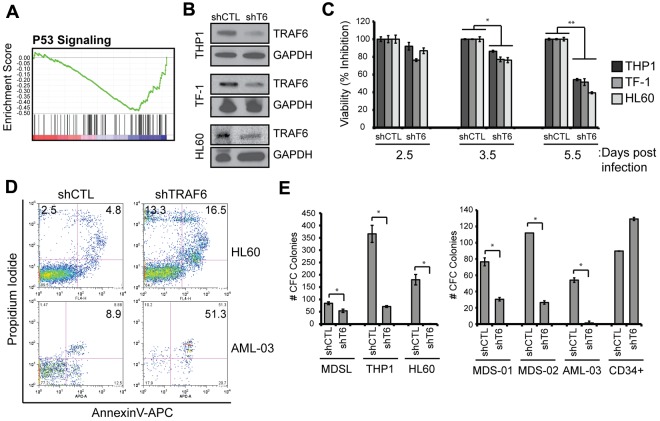
Depletion of TRAF6 impairs MDS and AML cell viability. (A) Microarray analysis was performed on TF-1 cells after knockdown of TRAF6 (as in Figure 6A). Gene set enrichment analysis was performed, and the profile of the P53 signaling gene set is shown. (B) Knockdown of TRAF6 mRNA and protein by shTRAF6 was confirmed by quantitative PCR (supplemental Figure 5) and immunoblot analysis, respectively. (C) The indicated cell lines transduced with shCTL or shTRAF6 were evaluated for cell viability using the MTT assay. Cell viability is shown at the indicated days after transduction. (D) Cell viability of transduced HL60 and AML-03 patient cells was measured by annexin V/propidium iodide staining. Knockdown of TRAF6 mRNA in AML-03 was confirmed by quantitative PCR and showed > 50% reduced expression (not shown). (E) Hematopoietic stem/progenitor cell colony-forming potential of the indicated cell lines and MDS/AML patient BM cells after knockdown of TRAF6 was determined in methylcellulose. Colonies were evaluated after 10 days. *P < .05.
Elevated expression of proteasome subunits has been implicated previously in bortezomib resistance in several hematologic and solid cancers.25,36–38 Therefore, down-regulation of PSMA1 in shTRAF6-expressing cells may reveal a mechanism resulting in increased sensitivity to bortezomib (despite a mutation in the PSMB5 subunit). To test this hypothesis, PSMA1 expression was depleted using 2 shRNAs in Bort-R cells and evaluated for sensitivity to bortezomib and proteasome function (Figure 6C-D). Similar to deletion of TRAF6, knockdown of PSMA1 in Bort-R cells treated with bortezomib resulted in reduced cell viability (Figure 6C) and impaired proteasome function (Figure 6D), as evident by the accumulation of ubiquitinated proteins. These findings suggest that bortezomib cytotoxicity occurs through activation of autophagy and an auto-inhibitory loop involving TRAF6 and PSMA1.
TRAF6 is essential for the survival of MDS/AML cell lines and primary patient cells
To uncover the relevant signaling pathways regulated by TRAF6 in human MDS/AML that may explain its role in bortezomib-induced cytotoxicity of MDS/AML cells, we searched for previously defined expression signatures that overlap genes regulated by TRAF6 using gene set enrichment analysis.27 The gene-expression pattern after TRAF6 knockdown showed a significant enrichment of genes implicated in P53 activation (normalized enrichment score = 0.48; false discover rate q-val = 0.083; Figure 5A). Interestingly, gene sets generated from drug studies that induce autophagy (sirolimus/rapamycin) or inhibit the proteasome (Epigallocatch, ALLN, Disulfiram, and bortezomib) also overlap significantly the gene signature regulated by TRAF6 (supplemental Table 2). To corroborate that TRAF6 regulates cell survival and to investigate the cellular consequences of TRAF6 loss in human MDS/AML, lentiviral vectors encoding a control vector (shCTL) or shTRAF6 were transduced into 3 independent MDS/AML cell lines (THP-1, TF-1, and HL60) and in CD34+ cells from 1 AML patient (Figure 5B). In all MDS/AML cell lines and patient samples examined, loss of TRAF6 correlated with reduced cell viability (as measured by MTT; Figure 5C) and increased apoptosis (as measured by annexin V/propidium iodide staining; Figure 5D, supplemental Figure 5). Despite reduced cell viability, the percentage of viable cells within the G1, S, and G2/M phases at day 4 after transduction were indistinguishable between shCTL- and shTRAF6-transduced cells (supplemental Figure 5D). These observations indicate that TRAF6 primarily affects cell survival/viability and not cell-cycle progression in MDS/AML.
Figure 5.
Depletion of TRF6 sensitizes cells to bortezomib by regulating PSMA1. (A) Microarray analysis was performed on TF-1 cells after knockdown of TRAF6. Shown are differential gene-expression data between shTRAF6 and vector-transduced cells for all proteasome subunit genes (see supplemental Table 1). Expression of PSMA1 (highlighted in red) was most significantly down-regulated in cells with reduced TRAF6 expression (P < .05). (B) TF-1, Bort-S THP-1, Bort-R THP-1, and MDS BM cells (MDS-02) were transduced with shRNA lentiviral vectors targeting TRAF6 or a nontargeting control (shCTL). Down-regulation of PSMA1 in cells with reduced TRAF6 expression was confirmed by quantitative PCR. (C) Bort-R cells were transduced with 2 independent shRNA lentiviral vectors (sh#70 and sh#72) targeting PSMA1 or shCTL. Knockdown of PSMA1 was confirmed by quantitative PCR (sh#70, 89.6% knockdown; sh#72, 85.6% knockdown). Cell viability of Bort-R cells transduced with shPSMA1 or control vector was determined by MTT assay after treatment with 10nM bortezomib for 24 hours. (D) Proteasome inhibition in bortezomib-treated Bort-R cells transduced with 2 independent shPSMA1 or control vector was determined by immunoblotting for ubiquitinated proteins on total cell lysates. *P < .05.
To determine whether TRAF6 is critical for MDS/AML progenitor function, we assayed 3 MDS/AML cell lines, 2 MDS primary samples, and 1 primary AML sample transduced with control vector (shCTL) or shTRAF6 in methylcellulose. MDS/AML cell lines and patient samples with reduced TRAF6 expression exhibited a significant decrease in progenitor function/content (Figure 5E). In contrast, normal CD34+ cells were not sensitive to loss of TRAF6 and formed colonies at the same level as control-transduced CD34+ cells (Figure 5E). We conclude that TRAF6 is necessary for maintaining the survival of MDS/AML progenitors, and its degradation by bortezomib-induced autophagy contributes to cell death (Figure 7).
Figure 7.
Model of TRAF6 degradation by bortezomib-induced autophagy and its role in cell leukemia cell survival. (A) Under normal conditions, TRAF6 induces expression of survival genes. In addition, TRAF6 mediates the expression of PSMA1, an α-subunit of the proteasome. (B) Proteasome inhibition with bortezomib coincides with induction of autophagy, potentially due to ER stress of accumulated nondegraded proteins. TRAF6 is degraded by autophagic lysosomes, which can be blocked with the autophagy inhibitor 3-MA. Depletion of TRAF6 results in apoptotic cell death, in part by down-regulation of survival genes. In addition, loss of TRAF6 results in reduced PSMA1 expression and increased bortezomib sensitivity. α indicates the α-subunit; and β, the β-subunit.
Discussion
Bortezomib has been successful in the treatment of MM and lymphoma. However, comparatively few studies have evaluated the therapeutic efficacy and mechanism of bortezomib in MDS and AML. A case study reported that a del(5q) MDS patient treated with bortezomib achieved a major erythroid response and normalized platelet counts.23 In a larger trial of 19 MDS patients of various subtypes, a cytogenetic and/or hematologic response or stable disease was achieved in the majority of patients who completed the trial.39 More recently, bortezomib combined with low-dose cytarabine in intermediate-2 and high-risk MDS patients (n = 43) improved hematologic response and overall survival compared with cytarabine alone.40 Current and future trials will reveal the efficacy of bortezomib as a single agent and will identify drug combinations showing therapeutic efficacy in MDS and AML. In support of the clinical trials, in vitro treatment of high-risk MDS CD34+ cells or AML blasts with bortezomib results in apoptosis and cell-cycle arrest in part because of inhibition of Spi1/NF-κB–dependent DNA methyltransferase activity.7–9,41,42 Despite ongoing evaluation of bortezomib safety and efficacy in MDS/AML, little is known about its mechanism of action in these related myeloid malignancies.
In the present study, we report that bortezomib-treatment results in degradation of TRAF6 by autophagic lysosomes. In addition, we show that loss of TRAF6 contributes to the inhibitory effects of bortezomib in MDS/AML. To investigate the molecular and cellular consequences resulting from degradation of TRAF6, we performed gene-expression profiling and found that loss of TRAF6 contributes to the cytotoxic effects of bortezomib by 2 parallel mechanisms: (1) by inducing a P53/apoptosis gene program in MDS/AML cells and (2) by inducing an auto-inhibitory loop resulting in further inhibition of PSMA1 expression and proteasome function. TRAF6 expression was elevated significantly in a subset of CD34+ BM cells from MDS and AML patients compared with control CD34+ cells (supplemental Figure 6),43 and overexpression of TRAF6 in primary mouse BM cells results in an MDS-like disease that progresses to AML.15 Although the overexpression of the TRAF6 protein has not been reported in AML, expression of its inhibitor, miR-146a, is frequently reduced in AML patient samples,17 suggesting that the TRAF6 protein may be elevated in certain subtypes of AML. Recent studies have implicated TRAF6 as a critical survival gene in nonhematopoietic human cancers,44,45 so the proposed mechanism of bortezomib action on TRAF6 may extend to other human cancers with elevated TRAF6 expression/signaling. We conclude that inhibiting TRAF6 expression or its function may be therapeutically beneficial in certain subtypes of MDS/AML (or other human cancers with elevated TRAF6 expression) and may enhance the efficacy of bortezomib.
Bortezomib and related proteasome inhibitors have been shown to activate autophagy in various cancers, but with different consequences. Cotreatment with bortezomib and autophagy inhibitors can result in a synergistic cytotoxic effect, indicating that autophagy can be cytoprotective.46 Alternatively, under conditions of persistent bortezomib stress, autophagy can serve as a cell death mechanism.31,47 According to our present findings, bortezomib may induce the autophagy pathway in MDS/AML to initiate cell death. Regulation of TRAF proteins by autophagy/lysosomes has been described previously. TRAF2, a functionally related member and homolog of TRAF6, is sequestered to lysosomes by TNFAIP3/A20, resulting in its degradation.48 Consistent with our present results, a recent study showed that TRAF6 is degraded by autophagy in cells treated with poly(I:C) and that this degradation can be blocked by the addition of 3-MA but not BafA.49 Interestingly, in the present study, we found that 3-MA protected bortezomib-treated cells from cell death and, in contrast, BafA synergized with bortezomib to induce cell death (supplemental Figure 3D). This discrepancy between the effect of 3-MA and BafA on cancer cells has been described previously.50 The use of autophagy modulators in cancer therapy needs to be evaluated carefully.
There are ongoing efforts to uncover the molecular mechanism(s) of bortezomib resistance in hematologic malignancies. Recently, independent groups have associated bortezomib resistance with mutations and/or alterations in proteasome subunit expression.25,32,37 Up-regulated expression or mutations in PSMB5, which results in disrupted binding to bortezomib, have been implicated in drug resistance in lymphoid and myeloid malignancies.25,32,37,38 Additional evidence that proteasome subunits play a role in bortezomib resistance comes from an RNAi screen to identify modulators of proteasome inhibitor sensitivity in MM.51 According to this screen, the strongest bortezomib sensitizers were proteasome subunits (one of which was PSMA1). Lastly, 5-amino-8-hydroxyquinoline, an α subunit 20S proteasome inhibitor, exhibits cytotoxicity in bortezomib-resistant cells.52 These reports are consistent with our observation in the present study that down-regulation of PSMA1 after either knockdown of PSMA1 or TRAF6 by shRNAs sensitizes Bort-R cells to bortezomib (Figures 4 and 6). One potential explanation for the finding that Bort-R cells become resensitized to bortezomib despite its impaired binding to PSMB5 is that reduced PSMA1 protein expression disrupts the stoichiometry of noncatalytic proteasome subunits. Therefore, our present findings suggest that monitoring proteasome subunit expression in MDS/AML patients may predict therapeutic response to bortezomib.
In summary, the findings in the present study implicate TRAF6 in bortezomib-induced cell death in human cancer, reveal a novel mechanism of TRAF6 regulation through bortezomib/autophagy-mediated degradation, and suggest that the expression of certain proteasome subunits affects bortezomib function.
Supplementary Material
Acknowledgments
The authors thank the Mt Auburn Ob-Gyn associates and delivery nursing staff at Christ Hospital (Cincinnati, OH) for collecting cord blood (CD34+) samples from normal deliveries; Dr Kaoru Tohyama for kindly providing MDSL cells; and Dr Jose Cancelas for critical evaluation of the manuscript.
This work was supported by the Cincinnati Children's Hospital Research Foundation, the American Society of Hematology, the National Institutes of Health (RO1HL111103), and the Department of Defense (to D.T.S.). D.T.S. is an American Society of Hematology Scholar. The umbilical cord blood samples were received through the Normal Donor Repository in the Translational Core Laboratory at Cincinnati Children's Research Foundation, which is supported by the National Institute of Diabetes and Digestive and Kidney Diseases Centers of Excellence in Experimental Hematology (P30DK090971).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.F. and D.T.S. designed the research and wrote the manuscript; J.F, G.R., L.B., C. Rasch, and M.V. assisted with the experiments and edited the manuscript; M.W., C. Rigolino, A.C., J.C.M., E.N.O., and M.C. collected and provided patient samples; S.G. helped analyze the AML gene expression data; and G.J. and J.C. provided the Bort-S and Bort-R THP1 cell lines.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel Starczynowski, Division of Experimental Hematology and Cancer Biology, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: daniel.starczynowski@cchmc.org.
References
- 1.Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007;7(2):118–129. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- 2.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111(10):4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 3.Masonic Cancer Center, University of Minnesota. Bortezomib and vorinostat in treating patients with high-risk myelodysplastic syndrome or acute myeloid leukemia. [Accessed April 3, 2012]. Available from: http://clinicaltrials.gov/ct2/show/NCT00818649.
- 4.Virginia Commonwealth University. Belinostat and bortezomib in treating patients with relapsed or refractory acute leukemia or myelodysplastic syndrome. [Accessed April 3, 2012]. Available from: http://clinicaltrials.gov/ct2/show/NCT01075425.
- 5.Children's Oncology Group. Bortezomib and combination chemotherapy in treating younger patients with recurrent, refractory, or secondary acute myeloid leukemia. [Accessed April 3, 2012]. Available from: http://clinicaltrials.gov/ct2/show/NCT00666588.
- 6.Chauhan D, Hideshima T, Mitsiades C, Richardson P, Anderson KC. Proteasome inhibitor therapy in multiple myeloma. Mol Cancer Ther. 2005;4(4):686–692. doi: 10.1158/1535-7163.MCT-04-0338. [DOI] [PubMed] [Google Scholar]
- 7.Braun T, Carvalho G, Coquelle A, et al. NF-kappaB constitutes a potential therapeutic target in high-risk myelodysplastic syndrome. Blood. 2006;107(3):1156–1165. doi: 10.1182/blood-2005-05-1989. [DOI] [PubMed] [Google Scholar]
- 8.Dai Y, Chen S, Wang L, et al. Bortezomib interacts synergistically with belinostat in human acute myeloid leukaemia and acute lymphoblastic leukaemia cells in association with perturbation in NF-kappaB and Bim. Br J Haematol. 2011;153(2):222–235. doi: 10.1111/j.1365-2141.2011.08591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riccioni R, Senese M, Diverio D, et al. M4 and M5 acute myeloid leukaemias display a high sensitivity to bortezomib-mediated apoptosis. Br J Haematol. 2007;139(2):194–205. doi: 10.1111/j.1365-2141.2007.06757.x. [DOI] [PubMed] [Google Scholar]
- 10.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr., Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107(12):4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fribley A, Zeng Q, Wang CY. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24(22):9695–9704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milani M, Rzymski T, Mellor HR, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with bortezomib. Cancer Res. 2009;69(10):4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 13.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 14.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17(1):1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 16.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26(1):13–22. doi: 10.1038/leu.2011.221. [DOI] [PubMed] [Google Scholar]
- 17.Starczynowski DT, Morin R, McPherson A, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117(2):595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 18.Starczynowski DT, Kuchenbauer F, Wegrzyn J, et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39(2):167–178. doi: 10.1016/j.exphem.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011;108(22):9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boldin MP, Taganov KD, Rao DS, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hongming H, Jian H. Bortezomib inhibits maturation and function of osteoclasts from PBMCs of patients with multiple myeloma by downregulating TRAF6. Leuk Res. 2009;33(1):115–122. doi: 10.1016/j.leukres.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 23.Terpos E, Verrou E, Banti A, Kaloutsi V, Lazaridou A, Zervas K. Bortezomib is an effective agent for MDS/MPD syndrome with 5q- anomaly and thrombocytosis. Leuk Res. 2007;31(4):559–562. doi: 10.1016/j.leukres.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka A, Tochigi A, Kishimoto M, et al. Lenalidomide induces cell death in an MDS-derived cell line with deletion of chromosome 5q by inhibition of cytokinesis. Leukemia. 2010;24(4):748–755. doi: 10.1038/leu.2009.296. [DOI] [PubMed] [Google Scholar]
- 25.Oerlemans R, Franke NE, Assaraf YG, et al. Molecular basis of bortezomib resistance: proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112(6):2489–2499. doi: 10.1182/blood-2007-08-104950. [DOI] [PubMed] [Google Scholar]
- 26.Ng KP, Ebrahem Q, Negrotto S, et al. p53 independent epigenetic-differentiation treatment in xenotransplant models of acute myeloid leukemia. Leukemia. 2011;25(11):1739–1750. doi: 10.1038/leu.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellagatti A, Cazzola M, Giagounidis AA, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108(1):337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- 29.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 30.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 31.Belloni D, Veschini L, Foglieni C, et al. Bortezomib induces autophagic death in proliferating human endothelial cells. Exp Cell Res. 2010;316(6):1010–1018. doi: 10.1016/j.yexcr.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Franke NE, Niewerth D, Assaraf YG, et al. Impaired bortezomib binding to mutant beta5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cells. Leukemia. 2012;26(4):757–768. doi: 10.1038/leu.2011.256. [DOI] [PubMed] [Google Scholar]
- 33.Wu YT, Tan HL, Shui G, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285(14):10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs D, Berges C, Opelz G, Daniel V, Naujokat C. Increased expression and altered subunit composition of proteasomes induced by continuous proteasome inhibition establish apoptosis resistance and hyperproliferation of Burkitt lymphoma cells. J Cell Biochem. 2008;103(1):270–283. doi: 10.1002/jcb.21405. [DOI] [PubMed] [Google Scholar]
- 35.Rückrich T, Kraus M, Gogel J, et al. Characterization of the ubiquitin-proteasome system in bortezomib-adapted cells. Leukemia. 2009;23(6):1098–1105. doi: 10.1038/leu.2009.8. [DOI] [PubMed] [Google Scholar]
- 36.Skov V, Larsen ST, Thomassen M, et al. Increased expression of proteasome-related genes in patients with primary myelofibrosis [abstract]. Blood (ASH Annual Meeting Abstracts) 2010 Abstract 4117. [Google Scholar]
- 37.Lü S, Chen Z, Yang J, et al. Overexpression of the PSMB5 gene contributes to bortezomib resistance in T-lymphoblastic lymphoma/leukemia cells derived from Jurkat line. Exp Hematol. 2008;36(10):1278–1284. doi: 10.1016/j.exphem.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Balsas P, Galan-Malo P, Marzo I, Naval J. Bortezomib resistance in a myeloma cell line is associated to PSMbeta5 overexpression and polyploidy. Leuk Res. 2012;36(2):212–218. doi: 10.1016/j.leukres.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Alimena G, Breccia M, Musto P, et al. Erythroid response and decrease of WT1 expression after proteasome inhibition by bortezomib in myelodysplastic syndromes. Leuk Res. 2011;35(4):504–507. doi: 10.1016/j.leukres.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Natarajan-Ame S, Park S, Ades L, et al. Bortezomib combined with low-dose cytarabine in Intermediate-2 and high risk myelodysplastic syndromes. A phase I/II study by the GFM. Br J Haematol. 2012;158(2):232–237. doi: 10.1111/j.1365-2141.2012.09153.x. [DOI] [PubMed] [Google Scholar]
- 41.Colado E, Alvarez-Fernandez S, Maiso P, et al. The effect of the proteasome inhibitor bortezomib on acute myeloid leukemia cells and drug resistance associated with the CD34+ immature phenotype. Haematologica. 2008;93(1):57–66. doi: 10.3324/haematol.11666. [DOI] [PubMed] [Google Scholar]
- 42.Liu S, Liu Z, Xie Z, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008;111(4):2364–2373. doi: 10.1182/blood-2007-08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofmann WK, de Vos S, Komor M, Hoelzer D, Wachsman W, Koeffler HP. Characterization of gene expression of CD34+ cells from normal and myelodysplastic bone marrow. Blood. 2002;100(10):3553–3560. doi: 10.1182/blood.V100.10.3553. [DOI] [PubMed] [Google Scholar]
- 44.Starczynowski DT, Lockwood WW, Delehouzee S, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-kappaB pathways in human lung cancer. J Clin Invest. 2011;121(10):4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang WL, Wang J, Chan CH, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawaguchi T, Miyazawa K, Moriya S, et al. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol. 2011;38(3):643–654. doi: 10.3892/ijo.2010.882. [DOI] [PubMed] [Google Scholar]
- 47.Wu WK, Wu YC, Yu L, Li ZJ, Sung JJ, Cho CH. Induction of autophagy by proteasome inhibitor is associated with proliferative arrest in colon cancer cells. Biochem Biophys Res Commun. 2008;374(2):258–263. doi: 10.1016/j.bbrc.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Soetandyo N, Wang Q, Ye Y. The zinc finger protein A20 targets TRAF2 to the lysosomes for degradation. Biochim Biophys Acta. 2009;1793(2):346–353. doi: 10.1016/j.bbamcr.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inomata M, Niida S, Shibata KI, Into T. Regulation of Toll-like receptor signaling by NDP52-mediated selective autophagy is normally inactivated by A20. Cell Mol Life Sci. 2012;69(6):963–979. doi: 10.1007/s00018-011-0819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11(4):448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 51.Zhu YX, Tiedemann R, Shi CX, et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117(14):3847–3857. doi: 10.1182/blood-2010-08-304022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Wood TE, Sprangers R, et al. Effect of noncompetitive proteasome inhibition on bortezomib resistance. J Natl Cancer Inst. 2010;102(14):1069–1082. doi: 10.1093/jnci/djq198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.