Abstract
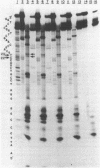
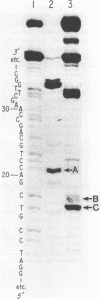
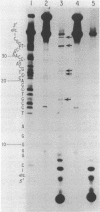
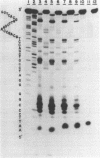
Ultraviolet light irradiation of DNA results in the formation of two major types of photoproducts, cyclobutane dimers and 6-4' [pyrimidin-2'-one] -pyrimidine photoproducts. The enzyme T4 DNA polymerase possesses a 3' to 5' exonuclease activity and hydrolyzes both single and double stranded DNA in the absence of deoxynucleotide triphosphate substrates. Here we describe the use of T4 DNA polymerase associated exonuclease for the detection and quantitation of UV light-induced damage on both single and double stranded DNA. Hydrolysis of UV-irradiated single or double stranded DNA by the DNA polymerase associated exonuclease is quantitatively blocked by both cyclobutane dimers and (6-4) photoproducts. The enzyme terminates digestion of UV-irradiated DNA at the 3' pyrimidine of both cyclobutane dimers and (6-4) photoproducts. For a given photoproduct site, the induction of cyclobutane dimers was the same for both single and double stranded DNA. A similar relationship was also found for the induction of (6-4) photoproducts. These results suggest that the T4 DNA polymerase proofreading activity alone cannot remove these UV photoproducts present on DNA templates, but instead must function together with enzymes such as the T4 pyrimidine dimer-specific endonuclease in the repair of DNA photoproducts. The T4 DNA polymerase associated exonuclease should be useful for the analysis of a wide variety of bulky, stable DNA adducts.
Full text
PDF




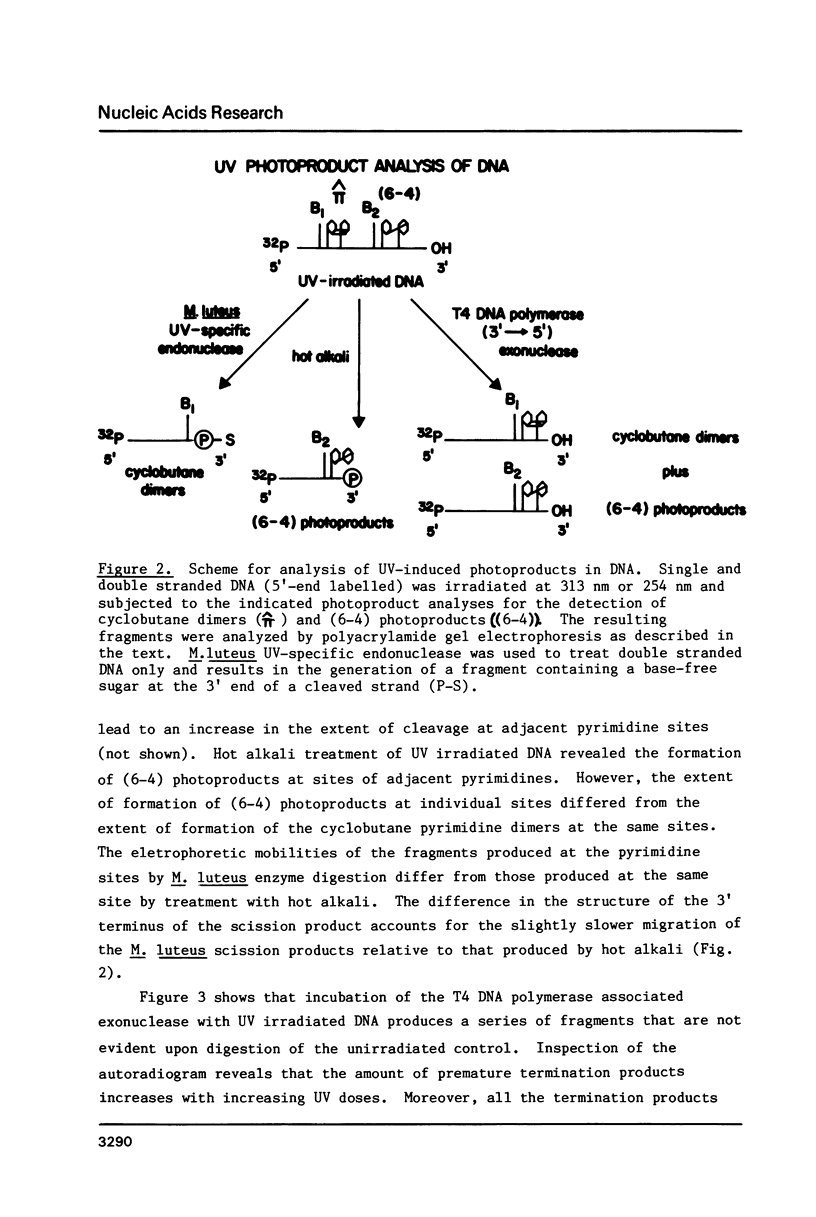

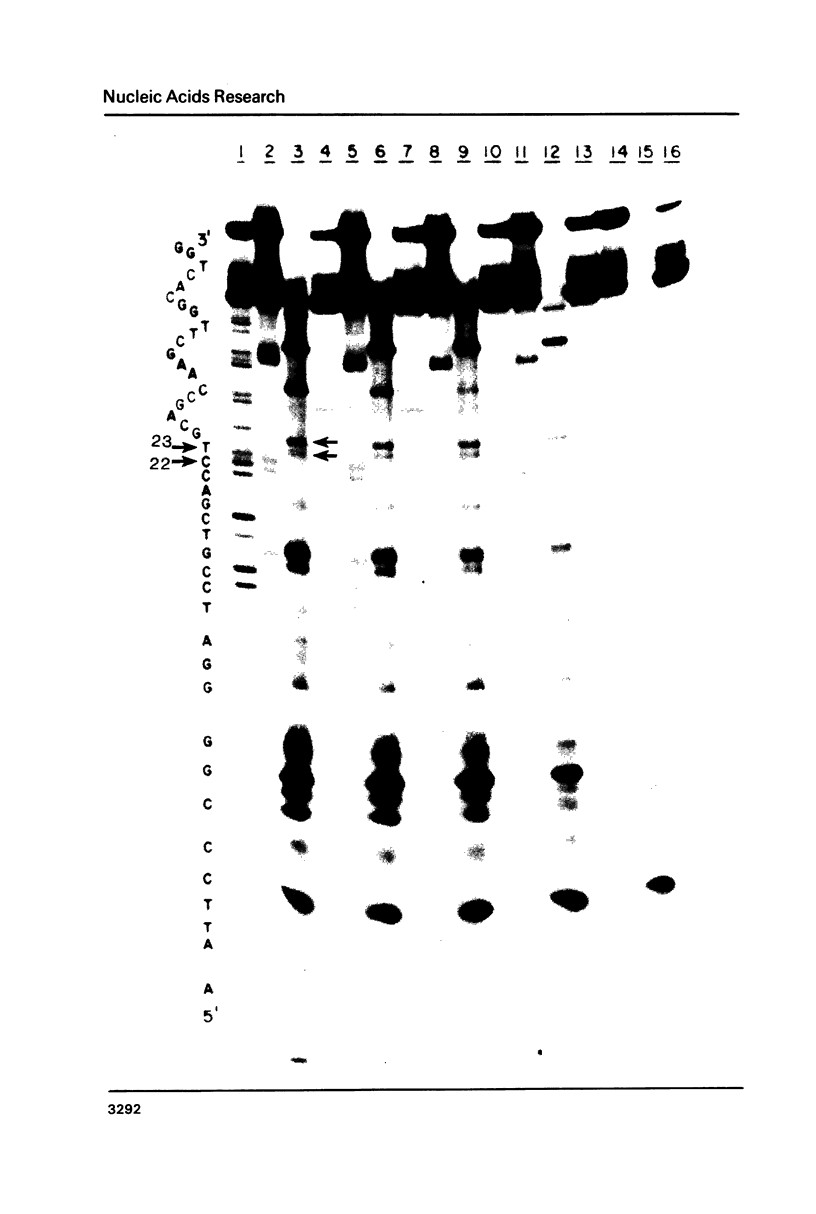

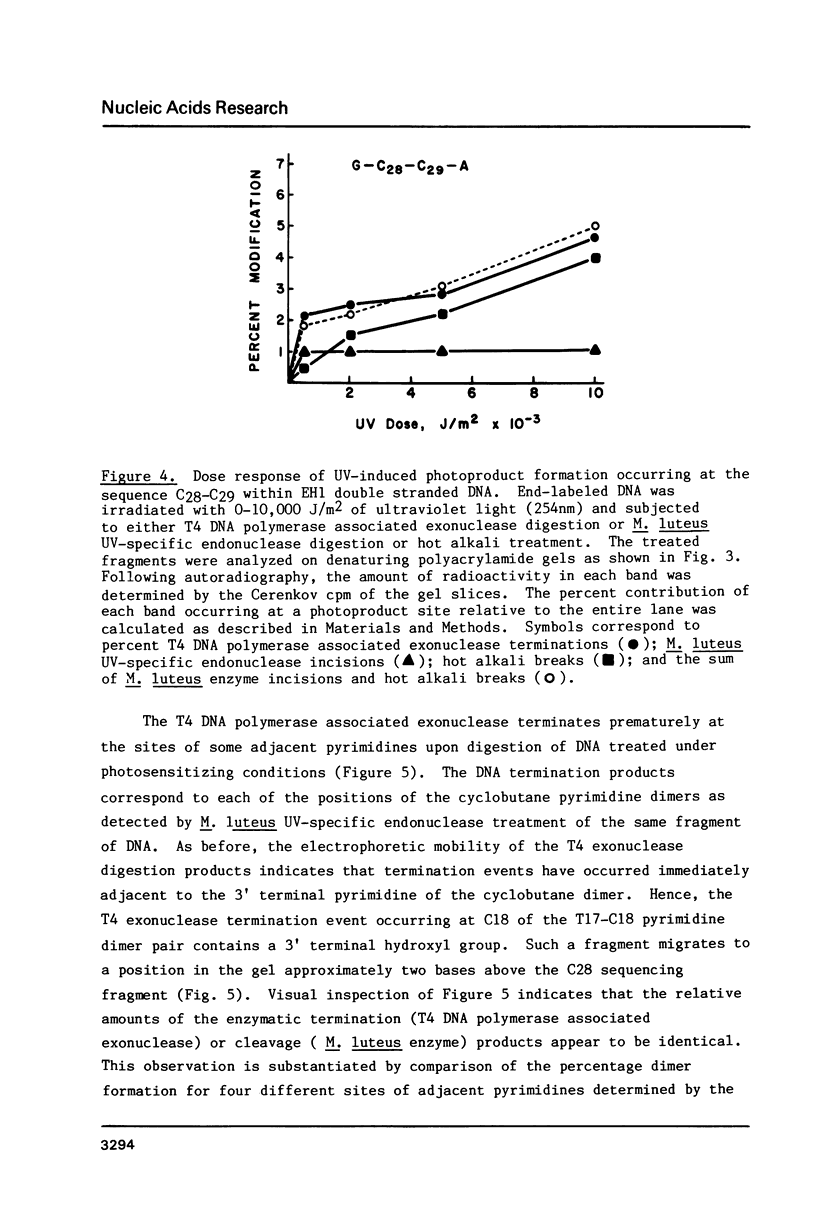

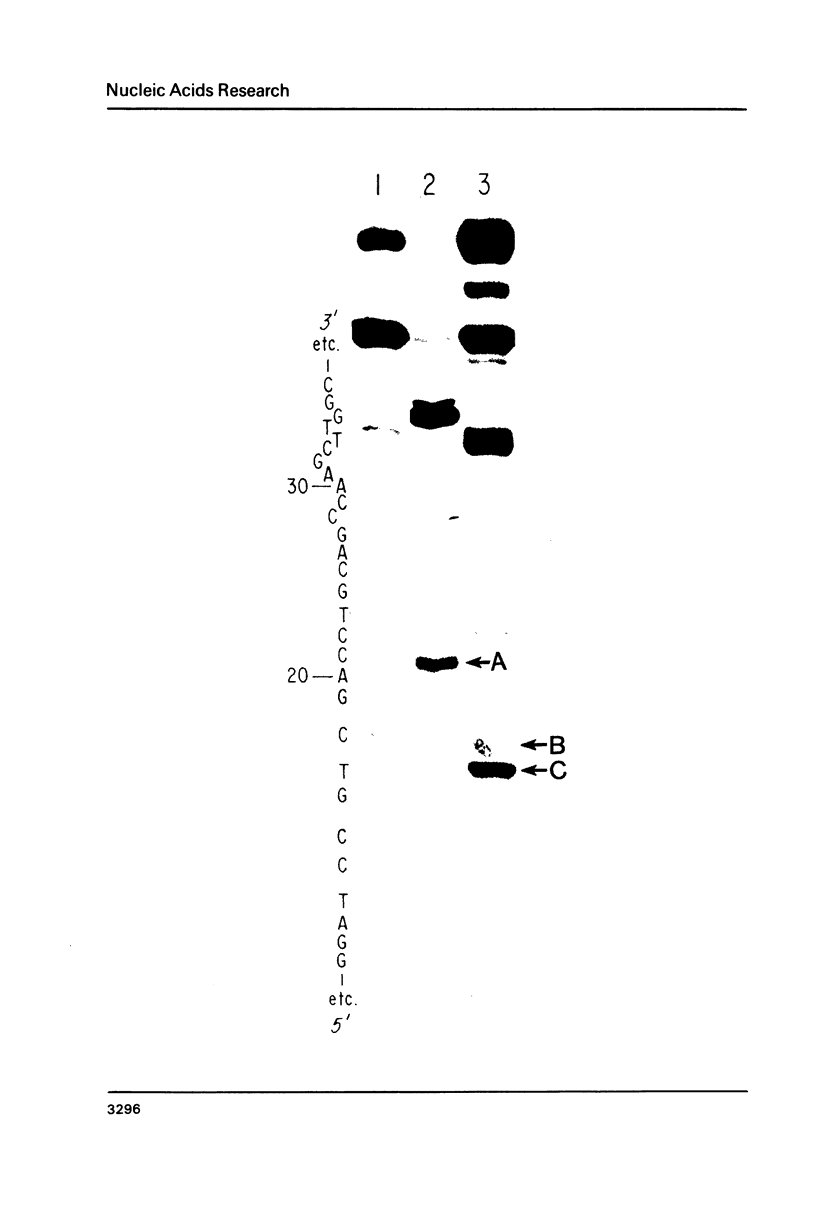

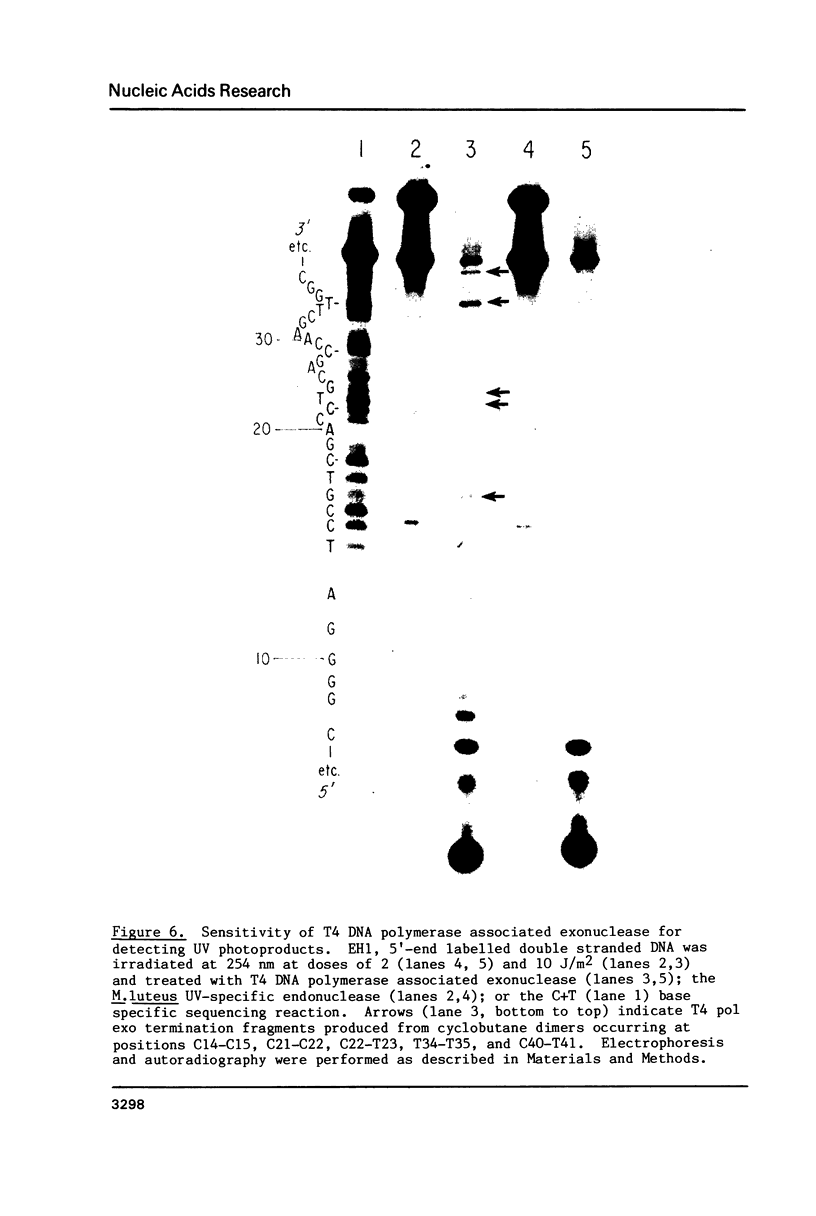
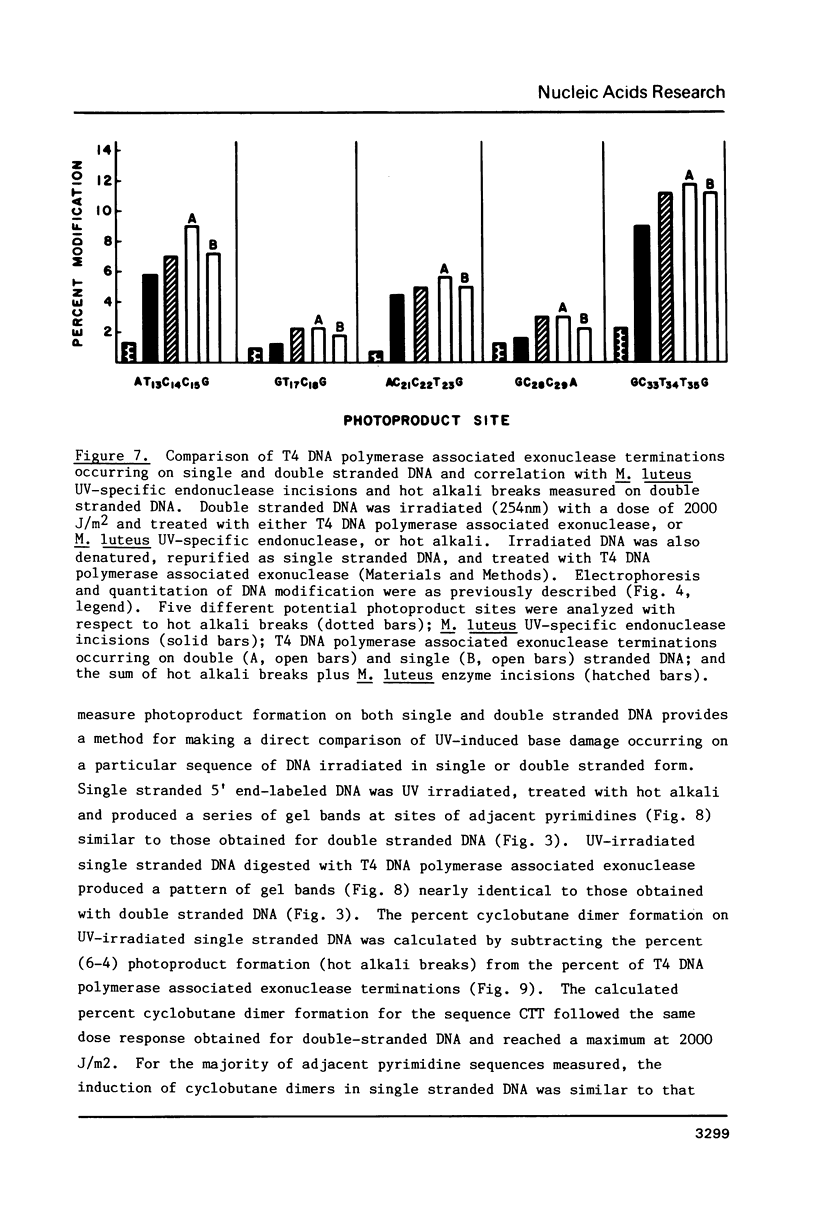
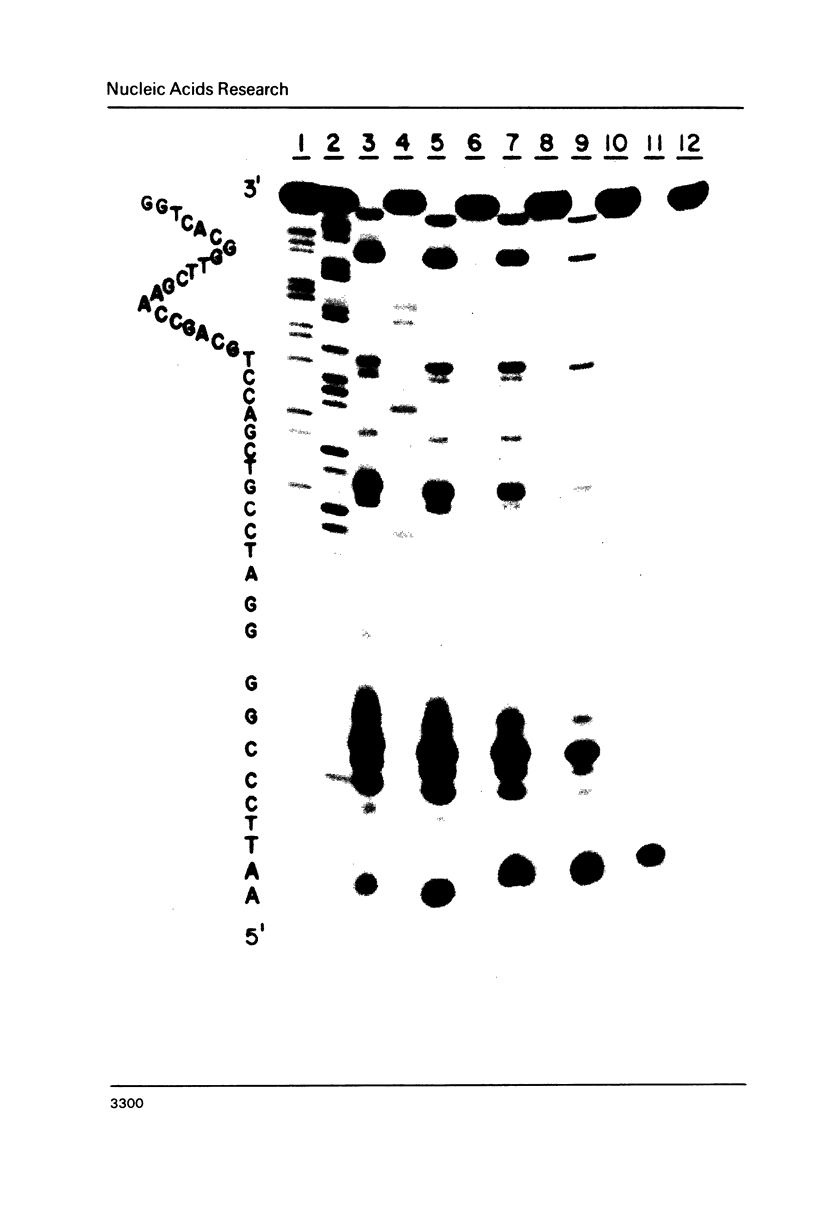

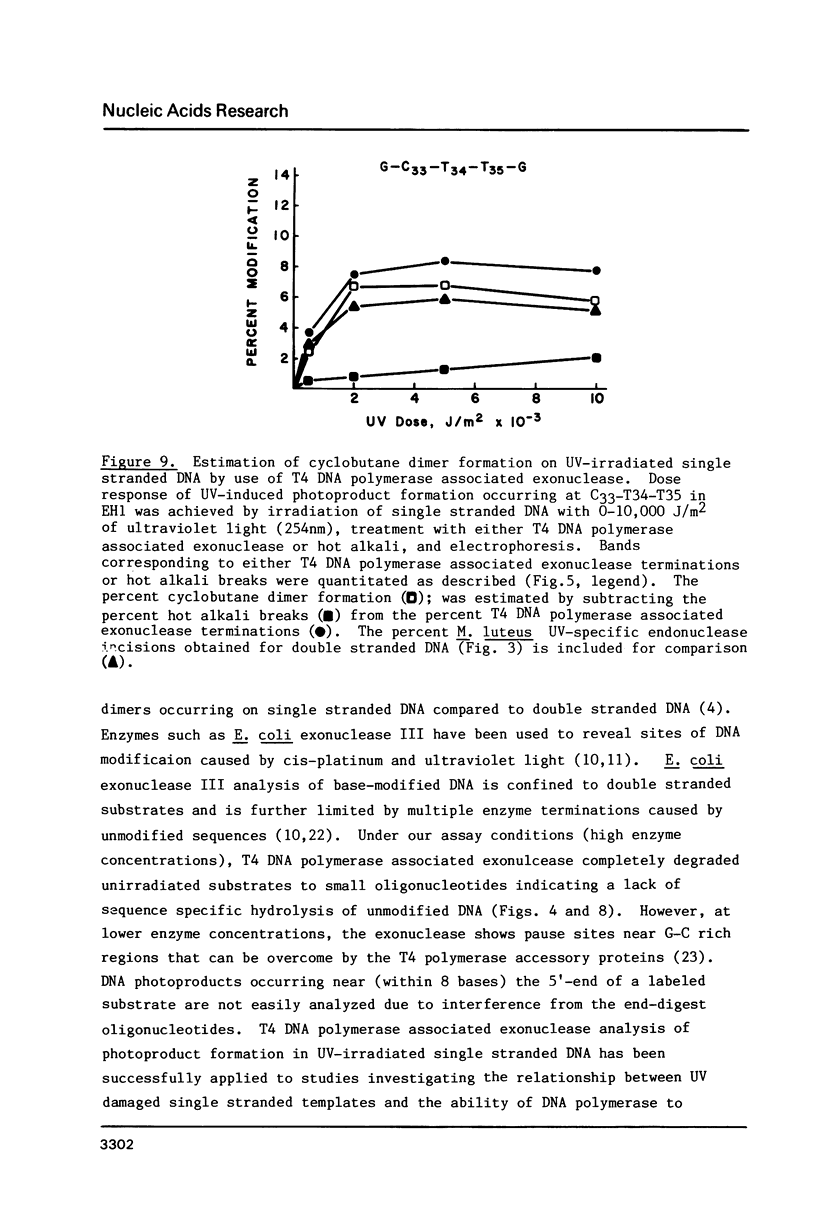


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bessman M. J., Muzyczka N., Goodman M. F., Schnaar R. L. Studies on the biochemical basis of spontaneous mutation. II. The incorporation of a base and its analogue into DNA by wild-type, mutator and antimutator DNA polymerases. J Mol Biol. 1974 Sep 15;88(2):409–421. doi: 10.1016/0022-2836(74)90491-4. [DOI] [PubMed] [Google Scholar]
- Brash D. E., Haseltine W. A. UV-induced mutation hotspots occur at DNA damage hotspots. Nature. 1982 Jul 8;298(5870):189–192. doi: 10.1038/298189a0. [DOI] [PubMed] [Google Scholar]
- Clayton L. K., Goodman M. F., Branscomb E. W., Galas D. J. Error induction and correction by mutant and wild type T4 DNA polymerases. Kinetic error discrimination mechanisms. J Biol Chem. 1979 Mar 25;254(6):1902–1912. [PubMed] [Google Scholar]
- D'Andrea A. D., Haseltine W. A. Modification of DNA by aflatoxin B1 creates alkali-labile lesions in DNA at positions of guanine and adenine. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4120–4124. doi: 10.1073/pnas.75.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin W. A., Lo K. M., Haseltine W. A. Alkaline lability of fluorescent photoproducts produced in ultraviolet light-irradiated DNA. J Biol Chem. 1982 Nov 25;257(22):13535–13543. [PubMed] [Google Scholar]
- Fuchs R. P. DNA binding spectrum of the carcinogen N-acetoxy-N-2-acetylaminofluorene significantly differs from the mutation spectrum. J Mol Biol. 1984 Jul 25;177(1):173–180. doi: 10.1016/0022-2836(84)90063-9. [DOI] [PubMed] [Google Scholar]
- Gordon L. K., Haseltine W. A. Comparison of the cleavage of pyrimidine dimers by the bacteriophage T4 and Micrococcus luteus UV-specific endonucleases. J Biol Chem. 1980 Dec 25;255(24):12047–12050. [PubMed] [Google Scholar]
- Gordon L. K., Haseltine W. A. Quantitation of cyclobutane pyrimidine dimer formation in double- and single-stranded DNA fragments of defined sequence. Radiat Res. 1982 Jan;89(1):99–112. [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Grunberg S. M., Haseltine W. A. Use of an indicator sequence of human DNA to study DNA damage by methylbis(2-chloroethyl)amine. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6546–6550. doi: 10.1073/pnas.77.11.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Gordon L. K., Lindan C. P., Grafstrom R. H., Shaper N. L., Grossman L. Cleavage of pyrimidine dimers in specific DNA sequences by a pyrimidine dimer DNA-glycosylase of M. luteus. Nature. 1980 Jun 26;285(5767):634–641. doi: 10.1038/285634a0. [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Lo K. M., D'Andrea A. D. Preferred sites of strand scission in DNA modified by andi-diol epoxide of benzo[a]pyrene. Science. 1980 Aug 22;209(4459):929–931. doi: 10.1126/science.7403858. [DOI] [PubMed] [Google Scholar]
- Hershfield M. S., Nossal N. G. Hydrolysis of template and newly synthesized deoxyribonucleic acid by the 3' to 5' exonuclease activity of the T4 deoxyribonucleic acid polymerase. J Biol Chem. 1972 Jun 10;247(11):3393–3404. [PubMed] [Google Scholar]
- Hershfield M. S. On the role of deoxyribonucleic acid polymerase in determining mutation rates. Characterization of the defect in the T4 deoxyribonucleic acid polymerase caused by the ts L88 mutation. J Biol Chem. 1973 Feb 25;248(4):1417–1423. [PubMed] [Google Scholar]
- Huang W. M., Lehman I. R. On the exonuclease activity of phage T4 deoxyribonucleic acid polymerase. J Biol Chem. 1972 May 25;247(10):3139–3146. [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A., Goodman M. F. On the fidelity of DNA replication. The accuracy of T4 DNA polymerases in copying phi X174 DNA in vitro. J Biol Chem. 1984 Feb 10;259(3):1539–1545. [PubMed] [Google Scholar]
- Linxweiler W., Hörz W. Sequence specificity of exonuclease III from E. coli. Nucleic Acids Res. 1982 Aug 25;10(16):4845–4859. doi: 10.1093/nar/10.16.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippke J. A., Gordon L. K., Brash D. E., Haseltine W. A. Distribution of UV light-induced damage in a defined sequence of human DNA: detection of alkaline-sensitive lesions at pyrimidine nucleoside-cytidine sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3388–3392. doi: 10.1073/pnas.78.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Muench K. F., Misra R. P., Humayun M. Z. Sequence specificity in aflatoxin B1--DNA interactions. Proc Natl Acad Sci U S A. 1983 Jan;80(1):6–10. doi: 10.1073/pnas.80.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzyczka N., Poland R. L., Bessman M. J. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972 Nov 25;247(22):7116–7122. [PubMed] [Google Scholar]
- Nossal N. G., Hershfield M. S. Nuclease activity in a fragment of bacteriophage T4 deoxyribonucleic acid polymerase induced by the amber mutant am B22. J Biol Chem. 1971 Sep 10;246(17):5414–5426. [PubMed] [Google Scholar]
- Rahn R. O., Landry L. C. Ultraviolet irradiation of nucleic acids complexed with heavy atoms. II. Phosphorescence and photodimerization of DNA complexed with Ag. Photochem Photobiol. 1973 Jul;18(1):29–38. doi: 10.1111/j.1751-1097.1973.tb06389.x. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Bessman M. J. Studeis on the biochemical basis of mutation. IV. Effect of amino acid substitution on the enzymatic and biological properties of bacteriophage T4 DNA polymerase. J Mol Biol. 1977 Oct 15;116(1):99–113. doi: 10.1016/0022-2836(77)90121-8. [DOI] [PubMed] [Google Scholar]
- Riazuddin S., Grossman L. Micrococcus luteus correndonucleases. I. resolution and purification of two endonucleases specific for DNA containing pyrimidine dimers. J Biol Chem. 1977 Sep 25;252(18):6280–6286. [PubMed] [Google Scholar]
- Royer-Pokora B., Gordon L. K., Haseltine W. A. Use of exonuclease III to determine the site of stable lesions in defined sequences of DNA: the cyclobutane pyrimidine dimer and cis and trans dichlorodiammine platinum II examples. Nucleic Acids Res. 1981 Sep 25;9(18):4595–4609. doi: 10.1093/nar/9.18.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Olivera B. M. Processivity of DNA exonucleases. J Biol Chem. 1978 Jan 25;253(2):424–429. [PubMed] [Google Scholar]
- Venkatesan M., Nossal N. G. Bacteriophage T4 gene 44/62 and gene 45 polymerase accessory proteins stimulate hydrolysis of duplex DNA by T4 DNA polymerase. J Biol Chem. 1982 Oct 25;257(20):12435–12443. [PubMed] [Google Scholar]