SUMMARY
DNA methylation has been implicated as an epigenetic component of mechanisms that stabilize cell-fate decisions. Here, we have characterized the methylomes of human female hematopoietic stem/progenitor cells (HSPCs) and mature cells from the myeloid and lymphoid lineages. Hypomethylated regions (HMRs) associated with lineage-specific genes were often methylated in the opposing lineage. In HSPCs, these sites tended to show intermediate, complex patterns that resolve to uniformity upon differentiation, by increased or decreased methylation. Promoter HMRs shared across diverse cell types typically display a constitutive core that expands and contracts in a lineage-specific manner to fine-tune the expression of associated genes. Many newly identified intergenic HMRs, both constitutive and lineage specific, were enriched for factor binding sites with an implied role in genome organization and regulation of gene expression, respectively. Overall, our studies represent an important reference data set and provide insights into directional changes in DNA methylation as cells adopt terminal fates.
INTRODUCTION
Development and tissue homeostasis rely on the balance between faithful stem-cell self-renewal and the ordered, sequential execution of programs essential for lineage commitment. Under normal circumstances, commitment is thought to be unidirectional with repressive epigenetic marks stabilizing loss of plasticity (De Carvalho et al., 2010). However, certain differentiated mammalian cells can be reverted to an induced pluripotent state (iPSCs) through exogenous transduction of specific transcription factors (Takahashi and Yamanaka, 2006). Yet, even these reprogrammed cells retain a residual “memory” of their former fate, displaying DNA methylation signatures specific to their tissue of origin (Kim et al., 2010).
DNA methylation is critical for the self-renewal and normal differentiation of somatic stem cells. For example, within the hematopoietic compartment, impaired DNA methyltransferase function disrupts stem cell maintenance (Maunakea et al., 2010; Trowbridge and Orkin, 2010), and loss of DNMT1 leads to defective differentiation and unbalanced commitment to the myeloid and lymphoid lineages (Bröske et al., 2009; Trowbridge et al., 2009). These studies highlight the well-characterized hematopoietic compartment as a context in which to study the link between DNA methylation patterns and cell-fate specification.
Toward this end, DNA methylation profiles of murine hematopoietic progenitors through early stages of lineage commitment were recently compared with CHARM (Irizarry et al., 2008; Ji et al., 2010), which profiles a predefined set of CpG-dense intervals. Overall, CHARM revealed that early lymphopoeisis involves more global acquisition of DNA methylation than myelopoiesis and that DNMT1 inhibition skews progenitors toward the myeloid state. These data support earlier reports that DNMT1 hypomorphic hematopoietic stem and progenitor cells (HSPCs) show reduced lymphoid differentiation potential (Bröske et al., 2009). Importantly, regions identified to have differential methylation through sequential stages of differentiation most often did not correspond to CpG islands (CGIs) but instead lay adjacent in areas referred to as “shores.”
Higher-resolution maps of DNA methylation with shotgun bisulfite sequencing have mainly been produced from cultured cells (Laurent et al., 2010; Lister et al., 2009) or mixed cell types (Li et al., 2010). Several unexpected findings emerged from these early studies including significant frequencies of cytosines methylated in a non-CpG context in human embryonic stem cells (ESCs), a characteristic previously thought to be restricted to plants. Other genome-wide studies have implicated DNA methylation in the regulation of alternative promoters and even RNA splicing patterns (Maunakea et al., 2010). These observations emphasize the need for complete, unbiased, and quantitative assessment of cytosine methylation and the establishment of reference methylomes from purified populations of primary cells.
Here, we performed whole-genome shotgun bisulfite sequencing on female human HSPCs, B cells, and neutrophils to examine the relationships between the methylation states of multipotent blood-forming stem cells and two divergent derived lineages. This enabled us to probe directional changes in DNA methylation associated with cell-fate specification. Comparison of the three reference methylomes revealed a number of important principles of epigenetic regulation, in addition to providing insights into the dynamics of epigenetic changes during development.
RESULTS AND DISCUSSION
Lineage-Specific Hypomethylated Regions Extend beyond Annotated CGIs
We sought to generate reference, single nucleotide-resolution methylation profiles for several nodes within the human hematopoietic lineage using whole-genome bisulfite sequencing (see the Experimental Procedures). Therefore, we examined CD34+ CD38–Lin– HSPCs, CD19+ B cells, and granulocytic neutrophils from peripheral blood of pooled human female donors. These cell types represent one of the earliest self-renewing, multipotent populations, and two derived, mature cell types from the lymphoid and myeloid lineages, respectively. For comparison, we generated methylomes from HSPCs from male umbilical cord blood (CD133+CD34+CD38–Lin–) and compared to data sets created from primate sperm (Molaro et al., 2011) and embryonic stem cells (Laurent et al., 2010). In all cases, we achieved a median of 10× independent sequence coverage, sufficient to interrogate 96% of genomic CpG sites (Figure S1A and Table S1A available online). While this level of coverage is still subject to sampling error at individual sites (see discussion in Hodges et al., 2009), features such as transitions from high to low levels of methylation can still be identified with a resolution of the boundaries to within a few CpG sites.
In the genome as a whole, CpG dinucleotides have a strong tendency to be methylated (70%–80%) (Lister et al., 2009). Coincidently, CpGs are also underrepresented, perhaps because of their vulnerability to methylation-induced deamination and consequent loss over evolutionary time (Cooper and Krawczak, 1989; Gardiner-Garden and Frommer, 1987). Areas of increased CpG density, called CpG islands (CGIs) have a lower probability of being methylated and these or their adjacent regions (CGI shores) have been implicated as potential regulatory domains (Gardiner-Garden and Frommer, 1987; Irizarry et al., 2009a; Wu et al., 2010). Though CGIs have been defined computationally (Irizarry et al., 2009b), we developed an algorithm to identify hypomethylated regions (HMRs) empirically in bisulfite sequencing data sets, based on their methylation state alone (see Figures 1A and 1B).
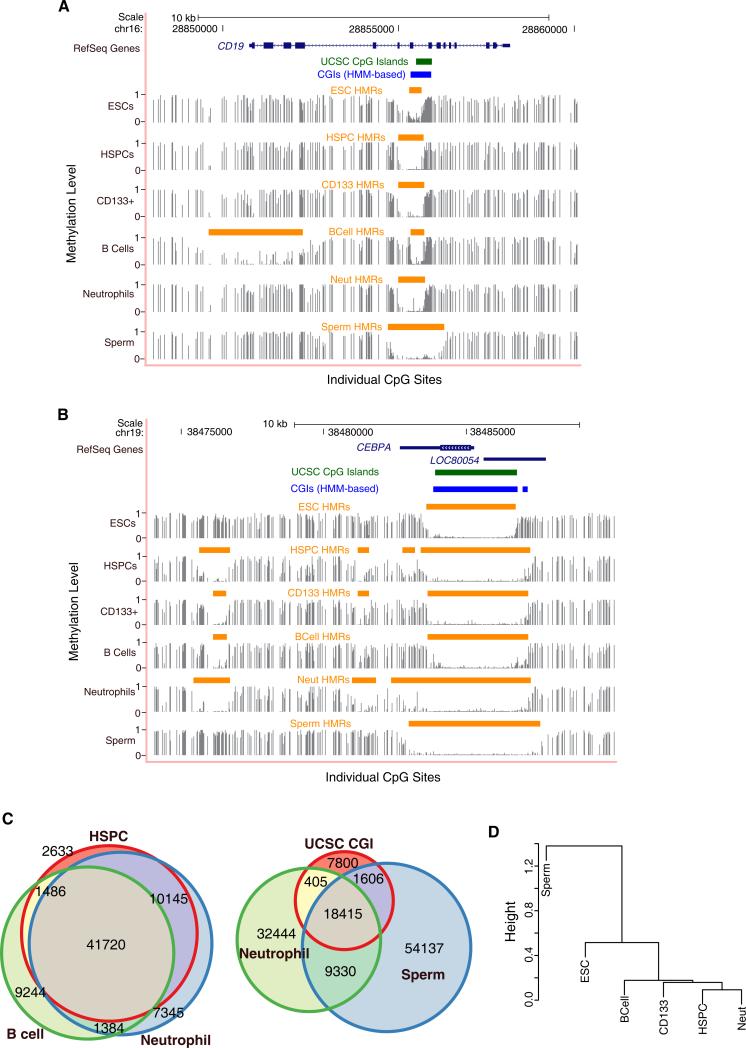
Figure 1. Features of Methylomes in Hematopoietic Cells.
(A and B) Genome browser tracks depict methylation profiles across a lymphoid (A) and myeloid (B) specific locus in blood cells, ESCs, and sperm. Methylation frequencies, ranging between 0 and 1, of unique reads covering individual CpG sites are shown in gray with identified hypomethylated regions (HMRs) indicated by orange bars. UCSC predicted/annotated CpG islands (green bars) and HMM-based CpG islands (blue bars) (Irizarry et al., 2009b) are also displayed. Numbers (top) indicate base position along the chromosome.
(C) Venn diagrams depict the intersection between HMRs identified in blood as well as the overlap between blood-derived cells, sperm, and UCSC CpG islands. The size of the circles and the proportion of circle overlap reflect the relative number of HMRs identified as well as the degree of intersection between each set of HMRs.
(D) Dendrogram clusters cell-types according to their pearson correlations of individual CpG methylation levels within HMRs, both overlapping and nonover-lapping, across all tissues examined.
See also Figures S1 and S2 and Table S1.
Between 50,000 and 60,000 HMRs were identified from each hematopoietic profile (Table S1B), with neutrophils displaying the greatest number (~60,000), followed by HSPCs (~55,000) and B lymphocytes (~53,000) (Figure 1C). Interestingly, this was lower than the number in male germ cells (~80,000), perhaps because of the extensive repeat hypomethylation observed in sperm as compared to somatic cells.
Certainly, many annotated CGIs were contained within our set of functionally defined HMRs; however, CGIs appeared to fall short as a benchmark by which to define all HMRs with probable regulatory significance. Annotated CGIs accounted for fewer than half of the HMRs identified in any cell type (Figure 1C and Figure S1B). Moreover, many HMRs whose biological relevance is supported by lineage-specific methylation failed to meet the conservative CGI criteria.
Sequence tracks showing methylation levels for a lymphoid-(Figure 1A) or myeloid- (Figure 1B) specific gene illustrate several characteristics of HMRs. The locus for the B cell marker CD19 displays a broad, cell type-specific HMR at its transcriptional start site (TSS), which does not overlap a predicted CGI. In contrast, “tidal” methylation at CGI shores characterizes several HMRs surrounding the myeloid transcription factor, CEBPA. The cores of these HMRs are shared among blood forming cells, but their widths differ, with neutrophils demonstrating the most expansive hypomethylation. Infact, shared HMRsoften show variablewidths, suggesting that the boundaries of HMRs fluctuate in a cell type-dependent manner. Due to the dynamic behavior of the HMRs, we were motivated to seek further validation of these characteristics as biological phenomena, rather than as technical artifacts of the methodology. Therefore, we focused on an independent data-set derived from chimpanzee. We reasoned that genic relationships to methylation dynamics should be preserved in closely related species. Indeed, HMRs show significant overlap between human and chimp, with chimp HMRs following very similar patterns of boundary fluctuations (Table S1C and Figure S2).
While a high proportion of identified HMRs (≥70%) intersected all blood cell types studied, ~10-fold more HMRs were shared only between HSPCs and neutrophils than exclusively between HSPCs and B cells (Figure 1C). In contrast, ~45%–50% of HMRs identified in blood cells overlap sperm HMRs. Interestingly, the diversity of differentially expressed genes within the hematopoietic lineage has been reported to be similar to the complexity observed across human tissues (Novershtern et al., 2011). However, at the epigenetic level, HMR profiles easily distinguished closely related cell types (blood forming) from distantly related ones (Figure 1D), indicating that patterns of DNA methylation are strongly correlated within a lineage.
HMR Expansion Correlates with Differential Expression
Differentially methylated regions (DMRs) at promoters have been ascribed regulatory roles, with differential methylation being linked to tissue-specific expression. Yet, HSPCs, B cells, and neutrophils mainly share promoter-associated HMRs at differentially expressed genes. Prior studies have associated changes in gene expression with changes in methylation states adjacent to constitutively hypomethylated CGIs, in so-called “CGI shores” (Irizarry et al., 2009a). Therefore, we examined correlations between the geography of promoter HMRs and changes in lineage-specific expression, focusing on a comparison of B cells and neutrophils.
Differential methylation often manifested as a broadening of TSS-associated HMRs in a specific lineage (Table S2A). The changes were asymmetric, with the greatest loss of methylation on the gene-ward side (Wilcoxon ranks sum: p < 5e-60, both DMR sets). Globally, these HMRs were broadest in sperm and constricted in ESCs (Figure 2A) (see also Molaro et al., 2011), widening again in a tissue-specific fashion. Thus, our analyses provide global support for “tidal” methylation changes at CGI shores.
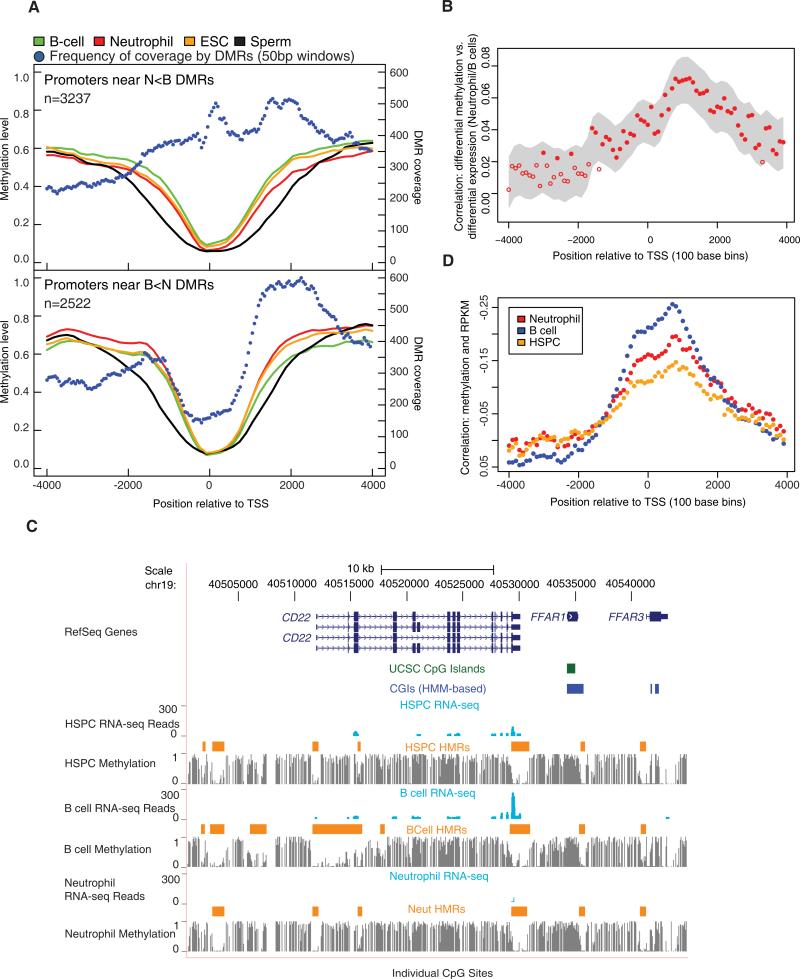
Figure 2. Promoter Differential Methylation and Gene Expression.
(A) Average methylation levels across promoters of genes having a DMR within 4 kb of the TSS are shown. Two separate graphs display neutrophil hypomethylated promoter DMRs relative to B cells (N < B, top) and B cell hypomethylated promoter DMRs relative to neutrophils (B < N, bottom). The number of DMRs covering nonoverlapping 50 bp windows across the promoter is also shown.
(B) Correlations between differential methylation and differential expression between neutrophils and B cells as a function of position relative to the TSS are shown. The correlations were obtained by comparing log odds of differential methylation and log of RPKM. The probability for differential methylation at a given CpG is described in the Supplemental Experimental Procedures. The gray area displays the smoothed 95% confidence interval. The closed circles indicate correlation coefficients that are significantly different from 0.
(C) The browser image shows gene expression for CD22 in the form of mapped read profiles from RNA-seq data. Methylation profiles are also shown (as in Figure 1A) along with HMRs.
(D) Correlations between methylation levels and expression levels represented by RPKM values are shown as a function of position relative to the TSS. Correlation coefficients were averaged in 100 bp bins across regions between 4 kb upstream and downstream of the TSS. Y axis labels were reversed.
See also Figure S3 and Table S2.
For deeper analysis of these tidal patterns, we measured differential methylation in 50 base windows surrounding TSSs (Figure 2A). Moving 3′ toward B cell hypomethylated promoters (B < N), coverage by DMRs peaked between 1.5 Kbp and 2 Kbp downstream of the TSS. A slightly different pattern was observed for neutrophil hypomethylated promoters (N < B), with DMRs rising to a peak directly at the TSS. In both data sets, the greatest concentration of differential methylation occurred ~1–2 Kb downstream of the TSS, consistent with overall methylation being selectively reduced in the transcribed regions of genes with tissue-specific DMRs.
We next asked whether any element of DMR geography correlated with tissue-specific gene expression. We carried out RNA-seq and computed RPKM values for each cell type (Table S2B). We then computed the correlation between differential expression and differential methylation in 100 base windows surrounding the TSS (see the Experimental Procedures). This correlation was strongly asymmetric, peaking ~1,000 bases downstream of the TSS. Notably, this corresponded with the expansion of HMRs that contributes to tissue-specific promoter hypomethylation (Figure 2B).
CD22 provides a specific example of the general phenomena that we observed (Figure 2C). CD22 is expressed in B cells, but not neutrophils. In each cell type its TSS is covered by an HMR, which in HSPCs and neutrophils extends ~500 bp and centered on the TSS. In B cells, the HMR begins at the same position upstream of the CD22 TSS, but extends more than 4,300 bp into the transcribed region.
The properties noted for differentially expressed genes were extensible to the entire set of REFSEQ genes. Though hypomethylation was largely symmetric around REFSEQ TSSs, a strong correlation could be seen between RPKM and lower methylation levels peaking 1.0 Kb downstream of the TSS (Figure 2D). This was true of all cell types examined, though the magnitude of the effect was lowest in HSPCs.
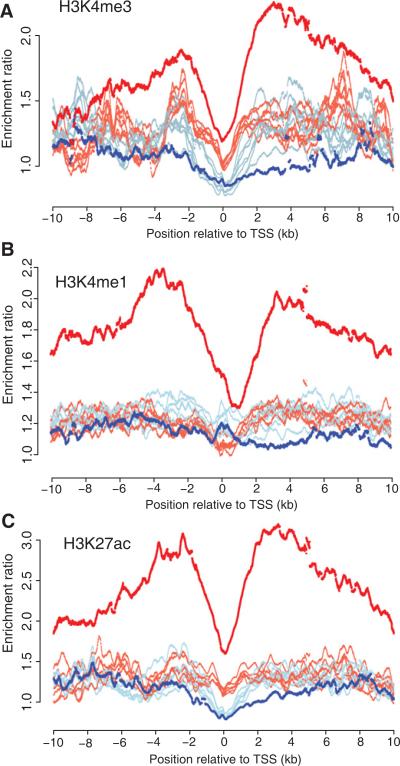
Our results are in accord with a recent study that revealed a unique chromatin signature surrounding the TSS of tissue-specific loci. Spreading of H3K4me2 into the 5′ untranslated region (UTR) was observed at tissue-specific genes, whereas it remained as a discrete peak at the TSS of ubiquitously expressed genes (Pekowska et al., 2010). To look for similar relationships between histone profiles and expanding promoter HMRs, we analyzed chromatin immunoprecipitation sequencing (ChIP-seq) data for H3K4me3, H3K4me1, and H3K27ac enrichment across eight different ENCODE cell lines (Bernstein et al., 2005; Birney et al., 2007). The ENCODE cell lines are derived from a variety of tissues and include GM12878, which is a lymphoblastoid cell line. First, we observe a strong enrichment for these histone marks at B cell promoters containing expanded HMRs. In addition, the greatest difference between the lymphoid cell line and the other cell lines appears upstream and downstream of the TSS compared to all promoters. Interestingly, the H3K4me3 differential enrichment is biased on the 3′ side of the TSS (Figure 3).
Figure 3. Histone Enrichment across Expanded HMRs.
Read count enrichment ratios per 25 bp bins located 10 kb upstream and 10 kb downstream of the TSS were calculated for promoters overlapping HMRs included in Figure 2A for B cell HMRs (red lines) or neutrophil HMRs (blue lines) for H3K4me3 (A), H3K4me1 (B), and H3K27ac (C) by comparison of read counts across all REFSEQ annotated promoters. Data were obtained from ENCODE and include histone profiles for eight different cell lines. The lymphoblastoid cell line GM12878 is highlighted in darker shaded colors.
It has also been noted that for a subset of CGI-associated promoters, high CpG density extends downstream of the TSS and hypomethylation of the extended region is required for RNA polymerase II binding (Appanah et al., 2007). In fact, analysis of existing lymphoid ChIP-seq data of RNA polymerase II revealed a 33 enrichment in B cell expanded HMR regions compared to neutrophil-expanded regions (Table S2C) (Barski et al., 2010). This suggests that while core CGI promoters remain hypomethylated by default, expansion downstream of the TSS may be important for productive transcription.
Features of Shared and Lineage-Specific Intergenic HMRs
While REFSEQ gene promoters were often associated with an HMR, the majority of HMRs were not found at promoters (Figure S3). Nearly half of all identified HMRs were located in gene bodies. An additional quarter lay >10 Kb from the nearest annotated genes, and we defined this class as “intergenic HMRs.”
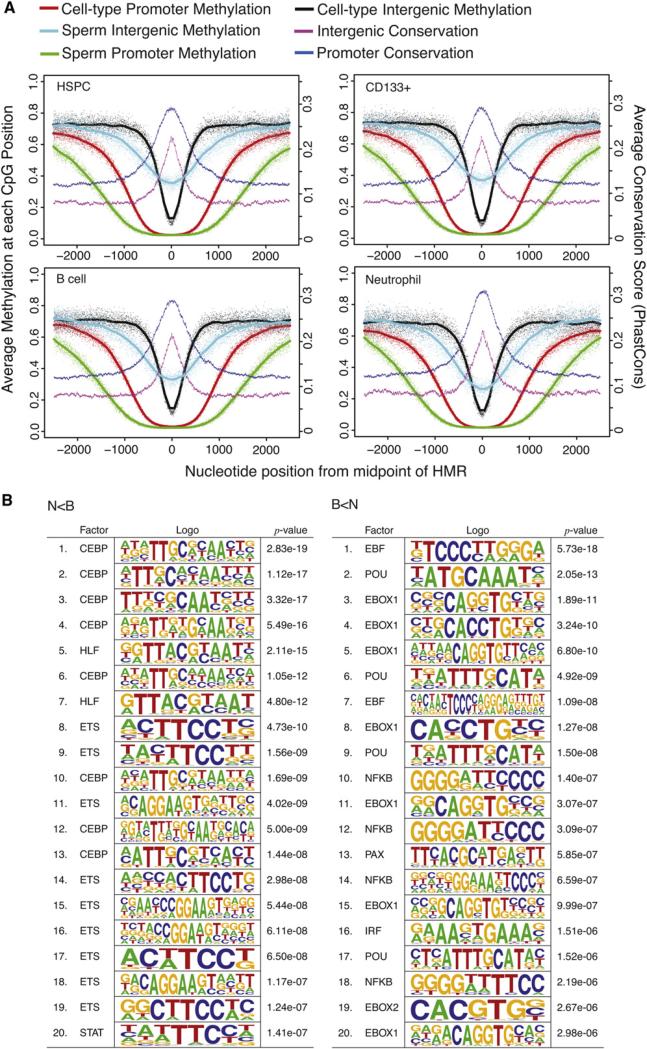
Like promoter-associated HMRs, intergenic HMRs showed sequence conservation, suggesting that these are functional elements (Figure 4A). In fact, genome-wide comparisons of methylation states of orthologous sites in the corresponding cell types of chimpanzee supported concomitant conservation of constitutive and cell type-specific patterns of intergenic methylation (data not shown). Intergenic HMRs tended to be narrower than those found at promoters and were less likely to be shared among cell types. When they were shared, they displayed patterns of expansion and contraction very similar to what was observed for promoter-associated regions (Figure 4A), with their overall extent being widest in sperm.
Figure 4. Features of Intergenic HMRs and DMRs.
(A) Composite methylation profiles are plotted for individual CpG sites within HMRs. The x axes of the plots indicate genomic position centered on the midpoint of HMRs in the reference cell type labeled for each plot. Methylation profiles are given for the reference cell and sperm, separately for regions where the reference cell HMR spans a TSS and intergenic region (>10 Kbp from any RefSeq transcript; not overlapping a repeat). Average cross-species conservation scores from PhyloP probabilities derived from 44-way multiple alignments are plotted separately for promoter and intergenic HMRs.
(B) Transcription factor binding site motifs enriched in DMRs between neutrophils and B cells are shown. The top 20 most enriched motifs are shown separately for N < B and B < N DMRs, based on the motifclass tool in the CREAD package. See the Supplemental Experimental Procedures for details of enrichment calculations.
See also Figures S3 and S4.
An early, pervasive view of DNA methylation proposed that germ cell profiles should represent a default state of hypomethylation in all potential regulatory regions (Gardiner-Garden and Frommer, 1987). This was based on the idea that hypomethylation in germ cells would prevent CpG erosion over evolutionary time spans. The high number of nonoverlapping HMRs in the adult somatic cell strongly argues against both of these notions (Figure 1C). However, the width of both genic and intergenic HMRs in sperm compared to somatic cells suggests that germ cells can define the ultimate boundaries of somatic HMRs.
Guided by the strong general enrichment for potential transcription factor binding sites in all HMRs (see Table 1), we searched for motifs in intergenic DMRs specific to neutrophils or B cells (Figure 4B). The strongest scoring motifs in the neutrophil-specific intergenic DMRs included those associated with C/EBP and ETS families, along with HLF and STAT motifs. This striking enrichment for C/EBP and ETS family binding sites is consistent with the functions of ETS factor PU.1 and several C/EBP factors as multipotent progenitors commit to become myeloblasts, which ultimately give rise to neutrophils (Nerlov and Graf, 1998). Because the ETS family contains a large number of transcription factors, we sought experimental support for their binding at HMRs. Therefore we probed existing ChIP-seq data of PU.1 from human HSPCs (Novershtern et al., 2011). We find numerous examples PU.1 enrichment in HMRs, several of which are provided in Figure S4. In contrast, the strongest scoring motifs in B cell-specific intergenic DMRs included the EBF motif, POU family motifs, E-boxes, a PAX motif, and those associated with NFκB and IRF. The simultaneous enrichment of EBF, E-box, and PAX motifs is consistent with the interacting roles of EBF, E2A (which binds E-boxes) and PAX5 as common lymphoid progenitors progress along the B cell lineage (Lin et al., 2010; Medina et al., 2004; Sigvardsson et al., 2002). The enrichment of NFκB and IRF motifs is consistent with the known roles for these factors in both activation and differentiation of lymphocytes (Hayden et al., 2006). Considered together, these analyses strongly suggest that at least a subset of intergenic DMRs can be engaged by tissue-specific transcription factors, leading to changes in chromatin organization that might have long-distance impacts on annotated genes or more local impacts on as yet unidentified ncRNAs. In fact, we do find evidence of transcriptional activity surrounding intergenic DMRs in our RNA-seq data sets, but we have not yet pursued this observation further (data not shown). Irrespective of the model, our results strongly support the biological relevance of tissue-specific intergenic HMRs.
Table 1.
TFBS Enrichment in HMRs across Intergenic and Promoter Regions
| Cell | Region | CGI? | HMRa | TFBS | Expected | Enrichment |
|---|---|---|---|---|---|---|
| N/A | promoter | 34,257 | 244,998 | 91,570.8 | 2.7 | |
| promoter | cgi | 24,601 | 191,452 | 65,760.9 | 2.9 | |
| promoter | nocgi | 9,656 | 53,852 | 25,810 | 2.1 | |
| intergenic | cgi | 10,630 | 13,608 | 4,603.76 | 3.0 | |
| B Cell | all | 53,834 | 339,943 | 76,196.1 | 4.5 | |
| intergenic | 5,849 | 16,150 | 3,779 | 4.3 | ||
| intergenic | cgi | 1,670 | 4,802 | 1,194.97 | 4.0 | |
| intergenic | nocgi | 4,179 | 11,348 | 2,584.01 | 4.4 | |
| promoter | 13,650 | 212,644 | 36,548.3 | 5.8 | ||
| promoter | cgi | 12,828 | 206,556 | 35,080 | 5.9 | |
| promoter | nocgi | 822 | 6,088 | 1,468.27 | 4.1 | |
| CD133 | all | 49,593 | 339,191 | 67,778.2 | 5.0 | |
| intergenic | 6,494 | 17,708 | 3,816.73 | 4.6 | ||
| intergenic | cgi | 1,630 | 4,817 | 1,207.45 | 4.0 | |
| intergenic | nocgi | 4,864 | 12,891 | 2,609.26 | 4.9 | |
| promoter | 13,745 | 224,955 | 37,395.1 | 6.0 | ||
| promoter | cgi | 12,965 | 219,407 | 36,309.9 | 6.0 | |
| promoter | nocgi | 780 | 5,548 | 1,085.18 | 5.1 | |
| ESC | all | 40,476 | 318,377 | 65,062.3 | 4.9 | |
| intergenic | 3,768 | 11,220 | 2,404.28 | 4.7 | ||
| intergenic | cgi | 1,151 | 3,295 | 882.802 | 3.7 | |
| intergenic | nocgi | 2,617 | 7,925 | 1,521.45 | 5.2 | |
| promoter | 13,098 | 222,654 | 36,332.4 | 6.1 | ||
| promoter | cgi | 12,661 | 218,765 | 35,769.4 | 6.1 | |
| promoter | nocgi | 437 | 3,889 | 562.951 | 6.9 | |
| HSPC | all | 55,984 | 352,574 | 77,671.2 | 4.5 | |
| intergenic | 6,154 | 17,619 | 3,972.1 | 4.4 | ||
| intergenic | cgi | 1,663 | 4,775 | 1,222.27 | 3.9 | |
| intergenic | nocgi | 4,491 | 12,844 | 2,749.81 | 4.7 | |
| promoter | 13,820 | 222,635 | 37,830.8 | 5.9 | ||
| promoter | cgi | 12,948 | 216,433 | 36,461.3 | 5.9 | |
| promoter | nocgi | 872 | 6,202 | 1,369.4 | 4.5 | |
| Neut. | all | 60,594 | 362,074 | 82,427.7 | 4.4 | |
| intergenic | 6,422 | 18,515 | 4,212.75 | 4.4 | ||
| intergenic | cgi | 1,626 | 4,760 | 1,243.88 | 3.8 | |
| intergenic | nocgi | 4,796 | 13,755 | 2,968.85 | 4.6 | |
| promoter | 13,862 | 224,621 | 38,503.6 | 5.8 | ||
| promoter | cgi | 12,950 | 218,281 | 37,060.6 | 5.9 | |
| promoter | nocgi | 912 | 6,340 | 1,442.93 | 4.4 | |
| Sperm | all | 81,446 | 440,856 | 201,006 | 2.2 | |
| intergenic | 2,616 | 14,903 | 3,158.15 | 4.7 | ||
| intergenic | cgi | 865 | 6,181 | 1,307.11 | 4.7 | |
| intergenic | nocgi | 1,751 | 8,722 | 1,851.02 | 4.7 | |
| promoter | 14,051 | 270,798 | 63,641.3 | 4.3 | ||
| promoter | cgi | 13,588 | 266,658 | 62,357.8 | 4.3 | |
| promoter | nocgi | 463 | 4,140 | 1,283.49 | 3.2 |
Enrichment of predicted transcription factor binding sites (TFBSs) in intergenic HMRs and HMRs that overlap promoters. For each set of HMRs, corresponding to a cell type, the TFBS enrichment (observed/expected site counts) is given for all HMRs, those overlapping promoters, those that are intergenic, separately according to whether the HMRs overlap CGIs. Data are presented for each of the following cell types: B cells, CD133 cord blood, HSPCs, ESCs, neutrophils, and sperm. For comparison, the TFBS enrichment in the full set of promoters (including those overlapping CGIs) is given, along with enrichment in the full set of intergenic CGIs.
For the “N/A” group, the HMRs are simply the number of promoters or CGIs.
We also probed the possible functions of shared intergenic HMRs. Prior studies had experimentally identified binding sites for the insulator protein, CTCF, by chromatin immunoprecipitation (Kim et al., 2007). These sites are strongly enriched (155-fold) in nonrepeat intergenic HMRs that are common to all cell types examined. In fact, ~90% (>500) of the nonrepeat, shared intergenic HMRs contain a CTCF site. This correlates with the known propensity of CTCF to bind unmethylated regions and suggests that many of the shared intergenic HMRs that we detect may function in the structural organization of chromosomes and nuclear domains.
Myeloid-Biased, Poised Methylation States Characterize HSPC Methylomes
For loci whose differential expression characterizes the lymphoid and myeloid lineages, we set out with a simple general expectation. Low methylation levels in stem and progenitor cells would be permissive for expression in either lineage, and an accumulation of methylation during differentiation would correlate with silencing of loci in the lineage in which they are not expressed.
To test this hypothesis, we selected lineage-specific HMRs arising from a comparison of neutrophils and B cells and examined their status in HSPCs. Both at the level of individual CpGs (Figure 5A) and at the level of overall methylation (Figure 5B), HSPCs showed intermediate methylation states at sites where B cells and neutrophils show opposing methylation patterns. This suggests that differentiation involves both gains and losses of DNA methylation at lineage-specific HMRs, an observation consistent with recent studies using other methodologies (Attema et al., 2007; Claus et al., 2005; Ji et al., 2010).
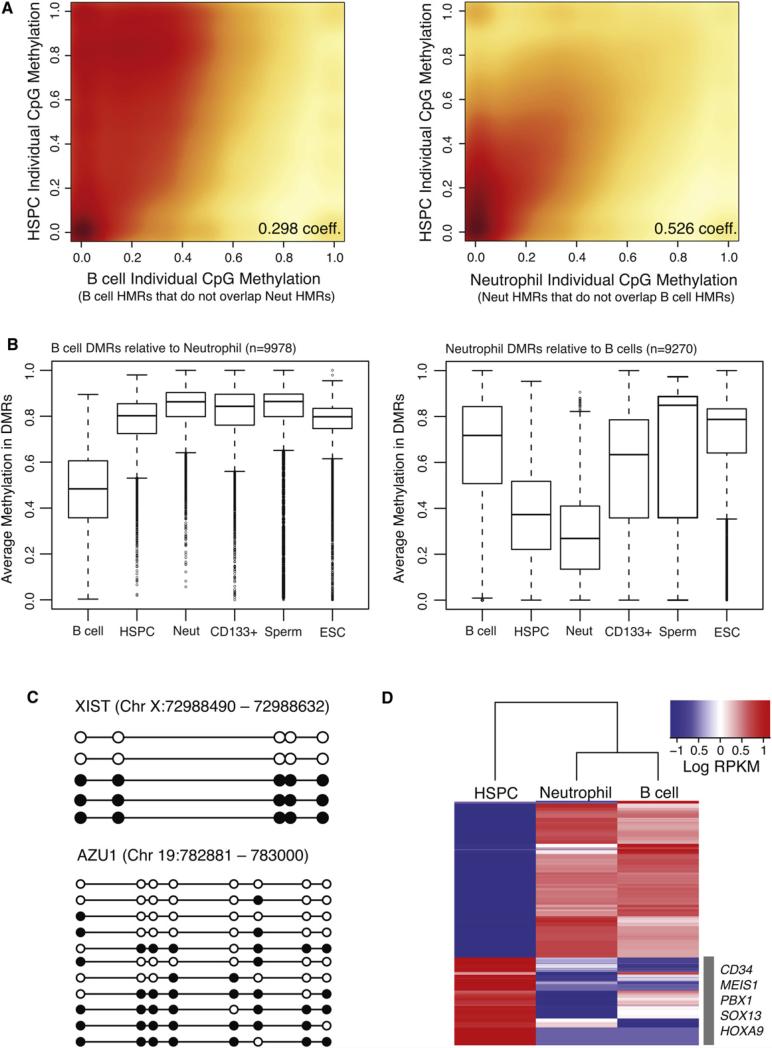
Figure 5. Methylation Dynamics during Lineage Selection.
(A) Smoothed scatter plot heat maps showing the correlation between individual CpG methylation levels in HSPCs versus B cells (left) and HSPCs versus neutrophils (right) within B cell- and neutrophil-specific HMRs, respectively. Darker shading (red) indicates greater density of data points, while lighter (yellow) shading reflects lower density. Positive correlations between HSPCs and both B cells and neutrophils indicate an intermediate state for HSPCs.
(B) Box plots show the distribution of average methylation levels in regions of differential methylation (DMRs) between B cells and neutrophils. Whiskers represent minimum and maximum values, while boxes depict the interquartile range, with horizontal lines indicating the median value. Outliers are shown as open circles.
(C) Lollipop diagrams display the methylation status of HSPC-derived clones sequenced by conventional methods following bisulfite conversion and site-specific PCR amplification across an interval near the XIST gene (top) and the AZU1 gene (bottom). Filled and open circles represent methylated and unmethylated CpG sites, respectively.
(D) Heat map of log RPKM values show expression levels for the top 100 differentially expressed genes (rows), selected for high expression in one cell type compared to the other, in each cell population (columns). Signature marker genes found within the HSPC cluster are listed.
See also Figures S3, S5, and S6 and Table S3.
At the level of individual CpGs, HSPC patterns correlated better with those seen in neutrophils at myeloid HMRs than they did with B cell methylation patterns at nonoverlapping lymphoid HMRs (Figure 5A). Moreover, the median methylation level for B cells at B cell DMRs was more than twice as high as the median level at neutrophil specific DMRs (Figure 5B). This finding, along with the fact that B cells exhibited fewer total HMRs than either HSPCs or neutrophils, supported an earlier observation that lymphoid commitment in mice involves globally increased DNA methylation (Ji et al., 2010). As a whole, our results indicate that the HSPC methylome has more myeloid than lymphoid character. Many fewer DMRs were identified in comparisons of HSPC and neutrophil methylation profiles than of HSPCs and B cells (Figure S3). Such a myeloid bias is also consistent with prior studies, which point to the myeloid lineage as a default differentiation path for HSPCs (Månsson et al., 2007).
Regions that exhibit intermediate methylation occurred in two forms. The well-documented mode is allelic methylation that is characteristic of dosage compensated and imprinted genes. We detected such loci abundantly in our data sets, and these encompassed both known monoallelic genes and new candidates for monoallelic expression (data not shown). More prevalent were regions of intermediate methylation wherein each chromosome displayed different patterns of CpG modification with little correlation between the states of adjacent CpGs. Partially methylated regions were previously noted in ESCs (Lister et al., 2009), though they did not investigate whether these presented allelic versus stochastic and complex patterns.
To discriminate between allelic and complex patterns, we performed targeted conventional bisulfite PCR sequencing of individual clones from HSPCs across a selected set of myeloid loci and a known locus with allele-specific methylation (Figure 5C, Figure S5, and Table S3). This allowed detailed analysis of adjacent CpG methylation on individual molecules. As expected, for the allelic XIST locus on chromosome X, we observed uniform methylation profiles of adjacent CpG sites within individual clones representing two states that contributed nearly equally to the partial methylation observed. In contrast, the myeloid AZU1 locus exemplified a stochastic pattern of methylation in HSPC. We cannot determine whether the complex states that we observed were in dynamic equilibrium or whether they were fixed in each chromosome that contributed to our analysis.
While the mechanisms underlying complex, partial methylation patterns in HSPCs are unclear, they are reminiscent of bivalent promoters that contain both repressive and active histone marks (Bernstein et al., 2006). Both during embryonic development and during stem cell differentiation, such poised promoters are converted to a determinate chromatin state by shifting the balance of histone marks. This has already been noted for lineage-specific genes in HSPCs (Attema et al., 2007), and our data indicate that this well-established property of chromatin may also extend to DNA methylation patterns.
Alternative explanations for our results must also be considered. Since we have used pooled individuals, each of the observed patterns could be specific to one donor, giving rise to a complex pool of clones; however, this seems unlikely as we also detect lower correlations between neighboring CpGs within single clones. Alternatively, complex states could represent heterogeneity within the isolated HSPC population (see Figure S6), with our data coming from a mixture of self-renewing and more committed cell types. To investigate this possibility, we searched within our RNA-seq data for expression patterns characteristic of each purified cell population. Transcriptional profiles revealed the top differentially expressed genes within the HSPC compartment to be highly enriched for signature gene markers associated with self-renewing hematopoietic stem cells (Figure 5D) and depleted for genes associated with committed progenitors. Collectively, these data suggest that the observed methylation patterns are likely derived from a highly enriched stem cell population, and indicate that those populations may naturally adopt complex, potentially dynamic, methylation patterns at lineage-specific HMRs.
Both the general trends of methylation loss along a lineage and the possibility of dynamic poised methylation states imply that demethylation, either passive or active, is a common event. In mammals, factors capable of promoting active demethylation have remained somewhat elusive (Ooi and Bestor, 2008). In vitro studies have demonstrated that MBD2, a methyl-CpG binding protein, can specifically demethylate cytosines, and components of the elongator complex and the cytidine deaminase, AID, have been implicated in demethylation during early development (Bhattacharya et al., 1999; Okada et al., 2010; Popp et al., 2010). Furthermore, in zebrafish, the coordinated activities of glycosylases, deaminases, and DNA repair proteins have been reported to cause differentiation defects when disrupted, and this has been posited as an effect of improper DNA methylation (Rai et al., 2010). Alternatively, demethylation could potentially be achieved through the action of hydroxymethylases (e.g., TET1-3), which have been proposed to execute an intermediate step toward methylation loss (Ito et al., 2010; Tahiliani et al., 2009; Zhang et al., 2010). Additional information will be necessary to resolve the relevance of any of these pathways to the transition in methylation states between HSPCs and mature neutrophils and B cells.
As a whole, our data not only provide insights into the global behavior of DNA methylation, both in individual cell types and along a well-characterized lineage, but also provide a critical reference data set to enable detailed future studies of both the mechanisms that set somatic DNA methylation patterns and the consequences of those patterns for gene expression and genome organization.
EXPERIMENTAL PROCEDURES
Flow Cytometry and DNA Extraction
Peripheral blood was collected from six healthy female donors ages 25–35 and pooled. After isolation by Ficoll gradient, mononuclear cells were fixed in 1% paraformaldehyde (PFA) and stained with antibodies against the following human cell surface markers (eBiosciences): anti-CD34 (mucosialin) conjugated to PE-Cy7, anti-CD38 conjugated to APC, anti-CD45 conjugated to PE, anti-CD19 conjugated to PE, and anti-CD235a (Glycophorin) conjugated to PE. For lineage depletion, either a combination of PE-conjugated antibodies against CD45, CD19, and CD235a or a commercially available human hematopoietic lineage cocktail was used. CD34+CD38–Lin– hematopoietic stem cells and CD19+ B cells were purified with the FACSAriaII (Becton Dickinson). Neutrophils were purified according to their forward and side-scatter profile. FACS profiles are provided in Figure S6. Umbilical cord blood was collected from a single donor, and CD133+ cells were selected via magnetic separation on CD133+ microbeads (Milteny Biotec) according to instructions supplied by the manufacturer. Two column separations were performed for additional purity. All cells were collected in cell lysis buffer (50 mM Tris, 10 mM EDTA and 1% SDS), and PFA induced crosslinks were reversed with RNase A and a 65° C incubation overnight, after which residual proteins were digested with Proteinase K for 3 hr at 42° C. DNA was extracted with an equal volume of phenol:chloroform, followed by a single extraction with chloroform and ethanol precipitation. Human sperm was purified and sequenced according to methods described in Molaro et al. (2011).
Illumina Library Preparation for Bisulfite Sequencing
Bisulfite sequencing libraries were generated by previously described methods (Hodges et al., 2009) and on the manufacturer's instructions (Illumina) but with several additional modifications. In brief, after each enzymatic step, genomic DNA was recovered by phenol:chloroform extraction and ethanol precipitation. Adenylated fragments were ligated to Illumina-compatible paired-end adaptors synthesized with 5′-methyl-cytosine, and, when necessary, adaptors were diluted 100×–1000× to compensate for low-input libraries and maintain an approximate 10-fold excess of adaptor oligonucleo-tides. After ligation, DNA fragments were purified and concentrated on MinElute columns (QIAGEN). The standard gel purification step for size selection was excluded from the protocol. Fragments were denatured and treated with sodium bisulfite with the EZ DNA Methylation Gold kit according to the manufacturer's instructions (Zymo). Lastly, the sample was desulfonated and the converted, adaptor-ligated fragments were PCR enriched with paired-end adaptor-compatible primers 1.0 and 2.0 (Illumina) and the Expand High Fidelity Plus PCR system (Roche). Paired-end Illumina sequencing was performed on bisulfite converted libraries for 76–100 cycles each end.
RNA-Seq
For isolation of RNA from target cell populations, unfixed (live) cells were sorted as described above into Trizol-LS (Invitrogen), and RNA was purified according to the manufacturer's recommendations. Double-stranded complementary DNA (cDNA) libraries were generated with the Ovation RNA-seq system (Nugen). After reverse transcription and cDNA amplification, double-stranded cDNA fragments were phosphorylated, adenylated, and ligated to Illumina paired-end adaptors followed by 15 cycles of PCR amplification with Phusion HF PCR master mix (Finnzymes) according to the standard Illumina protocol for genomic libraries. Single-end sequencing was performed for 36 cycles.
Conventional Bisulfite Cloning and Sanger Sequencing
Genomic DNA isolated from pooled human HSPCs was bisulfite converted with the EZ DNA Methylation Gold kit (Zymo). For selection of specific regions for amplification, forward and reverse primers were designed with Methprimer (Li and Dahiya, 2002). Primer sequences are provided in the Table S3. The following PCR reaction components were combined in a total volume of 25 μl: 5 μl 5× Expand High Fidelity Plus buffer without MgCl2, 1 μl 10 mM dNTPs, 1 μl 10 mM each forward and reverse primers, 2.5 μl 25 mM MgCl2, 2 μl DNA template, and 11.5 μl nuclease-free water. Thermal cycling was performed as follows: 35 cycles each of denaturation at 94° C for 2 min, annealing at 60° C or 53° C for 1 min, and extension at 72° C for 30 s followed by 7 min at 72° C. The PCR products were purified on columns with a PCR purification kit (QIAGEN). PCR products were adenylated with Klenow exo– and purified. Purified amplicons were cloned and sequenced according to previously described methods (Hodges et al., 2009).
Computational Methods Summary
The Supplemental Experimental Procedures contain a detailed description of computational methods. Mapping bisulfite treated reads was done with methods described by Smith et al. (2009) with tools from the RMAP package (Smith et al., 2009). Hypomethylated regions (HMRs) were identified with a hidden Markov model as described in Molaro et al. (2011). DMRs were identified by (1) computation of probabilities of differential methylation at individual CpGs based on number of reads and frequencies of methylation, and (2) identification of peaks in these profiles after kernel smoothing. Cross-species conservation information was taken from UCSC MULTIZ 44-way vertebrate alignments and PhyloP profiles from these alignments.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the McCombie lab and Michelle Rooks for help with experimental procedures, and Assaf Gordon, Luigi Manna, and the Cold Spring Harbor Laboratory and University of Southern California High Performance Computing Centers for computational support. Chimp blood was supplied by the New Iberia Research Center and the Southwest National Primate Center. This work was supported in part by grants from the National Institutes of Health and by a kind gift from Kathryn W. Davis (A.S., G.J.H.). The ENCODE ChIP-seq data were generated at the Broad Institute and in the Bradley E. Bernstein lab at the Massachusetts General Hospital/Harvard Medical School. Data generation and analysis was supported by funds from the National Human Genome Research Institute, the Burroughs Wellcome Fund, Massachusetts General Hospital, and the Broad Institute.
Footnotes
ACCESSION NUMBERS
Data analyzed herein have been deposited in GEO with accession number GSE31971.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and three tables and can be found with this article online at doi:10.1016/j.molcel.2011.08.026.
REFERENCES
- Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3:e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl. Acad. Sci. USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center; Broad Institute. Children's Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Claus R, Almstedt M, Lübbert M. Epigenetic treatment of hematopoietic malignancies: in vivo targets of demethylating agents. Semin. Oncol. 2005;32:511–520. doi: 10.1053/j.seminoncol.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009a;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Wu H, Feinberg AP. A species-generalized probabilistic model-based definition of CpG islands. Mamm. Genome. 2009b;20:674–680. doi: 10.1007/s00335-009-9222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thorén L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Ferrier P, Spicuglia S. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010;20:1493–1502. doi: 10.1101/gr.109389.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Sarkar S, Broadbent TJ, Voas M, Grossmann KF, Nadauld LD, Dehghanizadeh S, Hagos FT, Li Y, Toth RK, et al. DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell. 2010;142:930–942. doi: 10.1016/j.cell.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M, Clark DR, Fitzsimmons D, Doyle M, Akerblad P, Breslin T, Bilke S, Li R, Yeamans C, Zhang G, Hagman J. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol. Cell. Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Orkin SH. DNA methylation in adult stem cells: New insights into self-renewal. Epigenetics. 2010;5:189–193. doi: 10.4161/epi.5.3.11374. [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11:499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.