Abstract
E2F1 deletion leads to increased mitochondrial number and function, increased body temperature in response to cold and increased resistance to fatigue with exercise. Since E2f1−/− mice show increased muscle performance, we examined the effect of E2f1 genetic inactivation in the mdx background, a mouse model of Duchenne muscular dystrophy (DMD). E2f1−/−;mdx mice demonstrated a strong reduction of physiopathological signs of DMD, including preservation of muscle structure, decreased inflammatory profile, increased utrophin expression, resulting in better endurance and muscle contractile parameters, comparable to normal mdx mice. E2f1 deficiency in the mdx genetic background increased the oxidative metabolic gene program, mitochondrial activity and improved muscle functions. Interestingly, we observed increased E2F1 protein levels in DMD patients, suggesting that E2F1 might represent a promising target for the treatment of DMD.
INTRODUCTION
E2F1 is a transcription factor that participates in the control of cell cycle progression through the regulation of the expression of genes involved in cell proliferation. In addition to its well-known function in cell death and proliferation, an emerging role of E2F1 in metabolic control has been recently described (1,2). It was demonstrated that E2F1 is a transcriptional modulator of the metabolic oxidative response. In particular, it was shown that the glycolytic muscle gastrocnemius (GN) from E2f1−/− mice switched to oxidative metabolism characterized by increased mitochondrial content and activity. E2F1 directly modulated the expression of oxidative genes implicated in mitochondrial activity and biogenesis such as Pgc-1α, Errα and Cpt-1 (3). Consequently, E2f1−/− mice showed increased basal metabolism, resistance to fatigue and increased muscle performance. In our search for potential applications of this ‘oxidative’ phenotype, we found previous studies demonstrating that skeletal muscles of mice lacking dystrophin exhibit abnormalities of mitochondrial function, and more precisely an impaired capacity for oxidative phosphorylation (4). Muscular dystrophies are hereditary diseases involving progressive muscular weakness and degeneration (5). Duchenne muscular dystrophy (DMD), the most prevalent genetically inherited neuromuscular disorder, is caused by mutations or deletions in the gene encoding dystrophin that prevent the synthesis of a full-length dystrophin protein. Microarrays performed in muscles from patients with DMD have confirmed the decrease of the expression of several mitochondrial genes (4). The mdx mouse, an animal model of DMD, was used for the investigation of changes in mitochondrial function associated with dystrophin deficiency (6,7). Interestingly, oxidative fibers seem to be protected with a reduced damage in comparison with glycolytic fibers (8). Moreover, up-regulation of Pgc-1α or activation of AMPK are able to ameliorate DMD through the induction of oxidative program (9,10). Considering the potential advantage of the increase of oxidative program on DMD pathogenesis, we crossbred E2f1−/− and mdx mice to analyze the potential effects of E2F1 in muscle degeneration.
RESULTS AND DISCUSSION
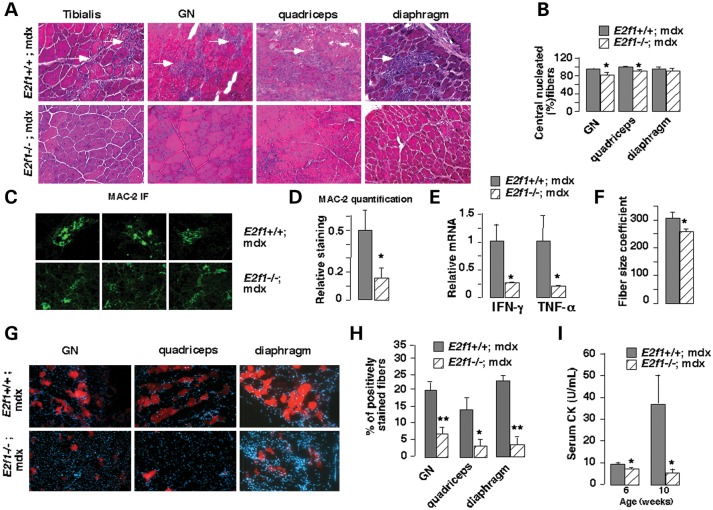
Structural analysis was first performed in order to elucidate any role of E2F1 in muscle pathology. Consistent with previous studies, microscopic analysis of muscle fiber sections of DMD male mice (E2f1+/+;mdx) revealed necrotic lesions, fibrosis and inflammation in GN, tibialis, quadriceps and diaphragm muscles (Fig. 1A). Genetic deletion of E2f1 in the DMD mouse model (E2f1−/−;mdx) resulted in a general improvement of the muscle structure (Fig. 1A). Visualization of muscle fiber cross-sections revealed no signs of lesions in muscles of E2f1−/−;mdx mice, suggesting that E2F1 is involved in muscle function in these mice (Fig. 1A). To further determine whether E2f1 deletion could attenuate the dystrophic phenotype, we examined several morphological parameters known to characterize dystrophic muscle. The number of fibers with central nuclei was reduced in several E2f1−/−;mdx muscles (Fig. 1B), although the high level of fibers with central nuclei strongly suggested that the fibers had been regenerated. The relative area of inflammatory infiltrate was also reduced in E2f1−/−;mdx muscles, as measured by MAC-2 immunofluorescence staining (Fig. 1C and D). This was consistent with decreased expression of inflammatory cytokines in muscles of E2f1−/−;mdx compared with E2f1+/+;mdx mice (Fig. 1E). Assessment of fiber size variability, determined by averaging the standard deviation of the cross-sectional myofiber areas, demonstrated greater homogeneity in E2f1−/−;mdx GN compared with E2f1+/+;mdx muscles (Fig. 1F). No significant differences of fiber size variability were observed in the diaphragm and quadriceps (data not shown). Furthermore, infiltration with Evans blue dye, which stains damaged fibers, was significantly reduced in the quadriceps, GN and diaphragm of the E2F1−/− mdx mice, indicating increased muscle membrane stability (Fig. 1G and H). Finally, circulating creatine kinase (CK) levels were significantly reduced in E2f1−/−;mdx mice aged at 6 and 10 weeks (Fig. 1I). Altogether, these data suggested that genetic deletion of E2f1 in the mdx background improved the dystrophic phenotype.
Figure 1.
Decreased fiber damage in E2f1 −/−;mdx male mice. (A) Hematoxylin staining of different muscles [gastrocnemius (GN)] from E2f1+/+;mdx and E2f1−/−;mdx mice. Necrotic lesions (white arrowhead) are observed only in E2f1+/+;mdx mice. Representative micrographs of one out of four animals are shown. (B) The percentage of centrally located fibers was measured using hematoxylin staining of different muscle sections from three E2f1+/+;mdx and E2f1−/−;mdx mice. All values represent means ± standard error of mean (SEM). *P < 0.05; **P < 0.01 here and in subsequent figures. (C) Inflammatory infiltration in E2f1+/+;mdx and E2f1−/−;mdx mice, as demonstrated by Mac-2 immunofluorescence. Nuclei are stained with Hoechst. (D) Quantification of Mac-2 infiltration using Image J software. Intensity of the staining was corrected by total fiber area. (E) Quantitative mRNA expression of inflammatory cytokines in muscles of E2f1−/−; compared with E2f1+/+;mdx mice. (F) Cross-sectional area of individual fiber was measured on hematoxylin-stained sections of GN muscle and the coefficient of fiber size variability was calculated by averaging the standard deviation of data from three mice for each genotype. (G) Evans blue was injected i.p., and GN and quadriceps tissue sections were analyzed by fluorescence microscopy. (H) Quantification of Evan blue positive-stained fibers in the different E2f1+/+;mdx and E2f1−/−;mdx muscles as indicated. (I) Serum CK in E2f1+/+;mdx and E2f1−/−;mdx mice aged at 6 and 10 weeks.
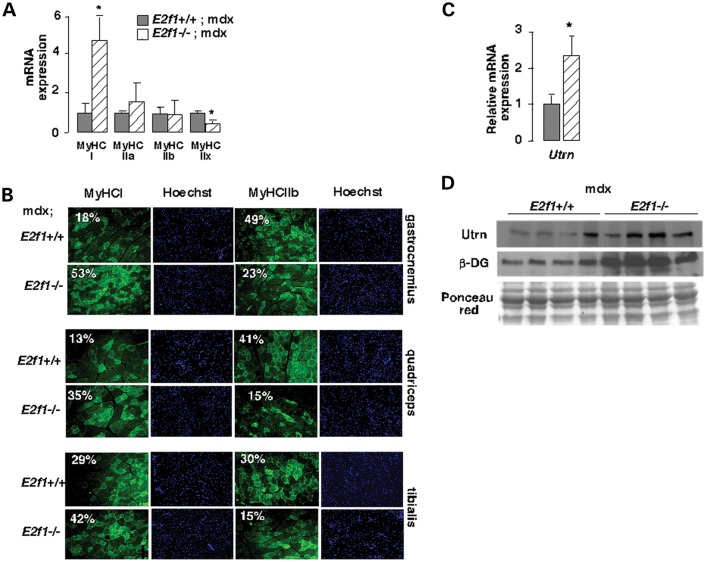
Distinct muscle fibers are differentially affected in DMD. Previous reports demonstrated that fast fibers are preferentially damaged and that slow fibers are protected from degeneration (8). Strikingly, E2f1−/−;mdx muscles showed increased mRNA expression of slow-twitch oxidative myofiber genes (MyHC-I) and a decreased level of fast-twitch glycolytic myofiber genes encoding MyHC-IIb and -IIx (Fig. 2A). Furthermore, immunofluorescence analysis showed a disproportionate increase in MyHC-I staining in sections from E2f1−/−;mdx muscles, and a decrease in MyHC-IIb staining in these sections when compared with sections from muscles of E2f1+/+;mdx mice, indicating a higher proportion of slow fibers in E2f1−/− mice (Fig. 2B). Finally, oxidative muscles express more utrophin in extrasynaptic regions of their fibers than glycolytic fibers (11), which confers resistance to degeneration. Consistently, we observed increased expression of the utrophin mRNA and protein levels in the E2f1−/−;mdx (Fig. 2C and D). We next sought to determine whether the expression of other components of the dystrophin-associated protein complex was also modified. Immunoblot analysis demonstrated an increased accumulation of β-dystroglycan in E2f1−/−;mdx when compared with those in E2f1+/+;mdx GN muscles (Fig. 2D). These results suggested that the particular fiber composition in muscles of E2f1-deficient mice with increased oxidative capacity and utrophin expression protected these mice from muscular dystrophy.
Figure 2.
E2f1 genetic inactivation improves muscle dystrophy in mdx male mice. (A) Relative gene expression of GN myosin heavy chain (MyHC) type I, IIa, IIX and IIb. Results were normalized to the expression of mouse 18S RNA. n = 6 animals/group. (B) Immunofluorescence analysis of distinct muscle types of E2f1+/+;mdx and E2f1−/−;mdx mice showing expression of MyHC type I and IIb (green) in fibers. Nuclei are stained with Hoechst reagent. The percentage of positive-stained fibers is indicated. n = 4 animals per group. (C) Relative mRNA expression of utrophin in E2f1+/+;mdx and E2f1−/−;mdx GN. Results were normalized to the expression of mouse 18S RNA. n = 6 animals/group. Values in A and C represent means ± SEM. *P < 0.05. (D) Western blot showing utrophin and β-dextroglycan in E2f1+/+;mdx and E2f1−/−;mdx GN. Ponceau red staining was used as a loading control.
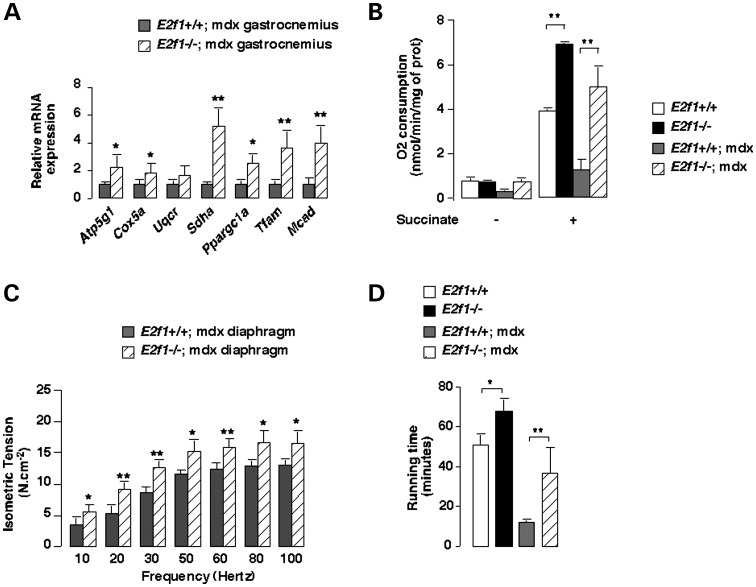
An important feature of slow fibers is increased oxidative metabolism. Indeed, mitochondrial oxidative phosphorylation has been shown to be impaired in muscles of mdx mice. Interestingly, mRNA expression of genes implicated in mitochondrial biogenesis and function, such as Pgc-1α, Tfam or Sdha, was consistently increased in E2f1−/−;mdx compared with those in E2f1+/+;mdx GN muscles (Fig. 3A). Mitochondrial activity studies of the GN muscle further proved the increased oxidative metabolism. Interestingly, deletion of E2f1 in the mdx background (E2f1−/−;mdx) resulted in increased GN mitochondrial oxygen consumption compared with those in E2f1+/+;mdx GN (Fig. 3B).
Figure 3.
Loss of E2f1 in mdx male mice increases oxidative metabolic gene expression, mitochondrial activity, muscle contraction and endurance capacity. (A) Quantification of the expression by Q-PCR of relevant genes involved in mitochondrial biogenesis and function in GN of E2f1+/+;mdx and −/−;mdx mice. Results were normalized by the expression of mouse 18S RNA. n = 6 animals/group. (B) O2 consumption was measured before and after the addition of succinate in isolated mitochondria from E2f1+/+, E2f1−/−, E2f1+/+;mdx and E2f1−/−;mdx GN muscle. n = 4 animals/group. (C) Isometric force production was measured in diaphragm muscle in 12-week-old mice of the indicated genotypes. n = 9 animals/group. (D) Twelve-week-old control E2f1−/−, E2f1+/+;mdx and −/−;mdx mice were run for 1 h on a treadmill with a 15° incline. The average distance run is represented. Values in A to D represent means ± SEM. *P < 0.05; **P < 0.01.
We found that isometric twitch tension of the diaphragm, known to be severely affected in mdx mice, was increased upon deletion of E2F1. Values in E2f1+/+;mdx mice ranged from 3.55 ± 1.0 N/cm2 (10 Hz) to 13.07 ± 0.8 N/cm2 (100 Hz). Tension strength of diaphragm muscle was consistently higher in E2f1−/−;mdx mice [from 5.56 ± 1.1 N/cm2 (20 Hz) to 16.57 ± 1.8 N/cm2 (100 Hz), P < 0.05], which was consistent with the improvement in muscle structure observed in these mice (Fig. 3C). Moreover, E2f1−/−;mdx mice performed similar to wild-type mice in a running test (40 and 50 min running till exhaustion, respectively), whereas E2f1+/+;mdx mice were exhausted after 20 min running (Fig. 3D). Altogether these results suggested that E2f1 deletion preserves muscle function in the mdx model of Duchenne dystrophy.
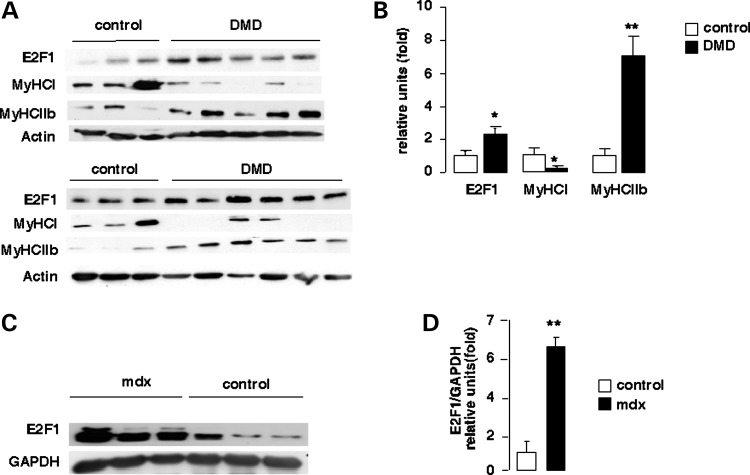
Finally, to assess the potential clinical relevance of our findings, we measured E2F1 protein levels on 6 controls and 11 human subjects with DMD. A robust increase in E2F1 protein level was observed in skeletal muscle from patients with DMD compared with control healthy subjects (Fig. 4A). Moreover, up-regulation of E2F1 protein expression in DMD patients was correlated with increased glycolytic fiber-type expression (MyHCII-b) and a decrease in oxidative fiber-type expression (MyHC-I) (Fig. 4A and B). Expression of E2F1 was also increased in muscles of mdx mice compared with wild-type control mice C57BL/10 (Fig. 4C and D). No correlation was found between E2F1 expression and age of patients, gender, time from loss of ambulation, clinical phenotype, CK levels, presence of dystrophin, or presence and nature of mutations in the gene (Table 1).
Figure 4.
Increased expression of E2F1 in mdx mice and DMD patients. (A) Total proteins were extracted from homogenized skeletal muscle tissues of control and DMD individuals (DMD); 30 µg was subjected to SDS–PAGE followed by immunoblotting with an anti-E2F1, MyHC-I, MyHC-IIb and actin as an internal loading standard. (B) Quantification of the western blot analysis of (c) E2F1, MyHC-I and MyHC-IIb expression in normal (N, n = 6) versus DMD individuals (n = 11). E2F1 protein levels were normalized to GAPDH levels. (C) Total proteins were extracted from GN of control (C57BL/10) and mdx male mice; 30 µg of total proteins were subjected to SDS–PAGE followed by immunoblotting with an anti-E2F1 and anti-GAPDH as an internal loading standard. (D) Quantification of the western blot analysis in (C) of E2F1 expression in normal versus mdx mice. E2F1 protein levels were normalized to GAPDH levels. Values in B and D represent means ± SEM. *P < 0.05; **P < 0.01.
Table 1.
DMD patients and human controls main characteristics
| # | Age at biopsy (y) | Gender | Loss of ambulation (y) | Clinical phenotype | CK level at biopsy (UI/l) | Dystrophin protein, IH/WB | Dystrophin gene mutation | |
|---|---|---|---|---|---|---|---|---|
| DMD patients | 9604 | 2 | M | 7 | Severe DMD | 12 167 | Absence | Deletion |
| 9619 | 3 | M | 18 | Severe BMD | 14 574 | Absence | Deletion | |
| 9631 | 2 | M | 9.5 | DMD | 10 717 | Absence | Deletion | |
| 9908 | 6.5 | M | 8 | Severe DMD | 14 200 | Absence | Deletion | |
| 0007 | 3.7 | M | 11 | DMD | 13 060 | Absence | Duplication | |
| 0008 | 2.1 | M | 11.5 | DMD | 11 218 | Absence | Deletion | |
| W | 6 | M | NA | DMD | 9478 | Quasi-absence | Deletion | |
| 4 | 3.1 | M | NA | DMD | NA | Absence | Punctual mutation | |
| 8 | 4.3 | M | NA | DMD | NA | Quasi-absence | Deletion | |
| 20 | 3.1 | M | NA | DMD | 6629 | Absence | Deletion | |
| 29 | 4 | M | NA | DMD | 11 334 | Quasi-absence | Punctual mutation | |
| Controls | 9613 | 7 | M | NA | Normal | |||
| 9702 | 13.8 | F | 740 | Normal | ||||
| 0408 | 11.9 | F | 197 | Normal | ||||
| 0606 | 2.5 | M | 395 | Normal | ||||
| 27 | 0.8 | M | NA | Normal | ||||
| 30 | 8.5 | F | 6336 | Normal |
DMD, Duchenne muscular dystrophy; BMD, Becker muscular dystrophy; IH, immunohistochemistry; WB, western blot; NA, not applicable.
Muscle degeneration, likely triggered by increased Ca2+ load in muscle cells, involves changes in mitochondrial function (6,7). Several studies have shown that oxidative slow fibers, such as type I, are resistant to degeneration in mdx mice (8,10). Furthermore, oxidative fibers express higher levels of utrophin, which could compensate for dystrophin deficiency (12). Whether mitochondrial activity contributes or is secondary to the muscle degeneration process still remains as an open question. Dystrophic muscles have decreased oxidative metabolism in mice models (7) and in DMD patients (4). In contrast, E2F1 deficiency results in increased oxidative metabolism (3). Consequently, we show now in this study that deletion of E2f1 increases oxidative metabolism in mdx mouse model of DMD. Strikingly, muscles of E2f1−/−;mdx mice contain a higher proportion of type I fibers than the E2f1+/+;mdx myodystrophic mice. Moreover, E2f1−/−;mdx fibers express higher levels of utrophin as well as proteins contained in the dystrophin-associated complex. This correlates with improved muscle performance in these mice. Conversely, it was shown that a fiber-type switch from slow to fast (oxidative to glycolytic) occurred in transgenic mice expressing CaM-binding protein in slow fibers. CaM-binding protein expression interfered with Ca2+/CaM-dependent signaling. Strikingly, when crossed to mdx, transgenic mice showed exacerbated dystrophic phenotype (13). Interestingly, this correlated with the higher presence of fast-type fibers, decreased expression of utrophin and decreased expression of PGC-1α, which is the opposite to what we found in E2f1−/−;mdx mice (13). These results suggest that the effects of E2F1 in this mouse model are mediated by its ability to repress oxidative metabolism. In contrast to E2F1, PPARδ, which is the member of the nuclear receptors family, promotes oxidative metabolism in muscle and facilitates the switch from fast to slow fiber type (14). Consistent with our results, it was shown that activation of PPARδ with specific agonists resulted in enhanced expression of slow oxidative fibers and increased the expression of utrophin, which was proved to be a transcriptional target of PPAR. Moreover, the treatment of mdx mice with PPARδ agonists improved the dystrophic phenotype (15). From these data, it is tempting to speculate that E2F1 regulates the expression of genes, such as CaM-binding protein, or represses the expression of PGC-1α or PPARδ. Further evidence for the implication of metabolic pathways in Duchenne's dystrophy comes from an elegant work in which it was shown through global analyses of published data sets of differential gene expression results comparing normal to DMD muscles that almost all genes downregulated in DMD could be clustered to a single common pathway, which is the fast to slow fiber-type twitch transition (16).
Alternatively, or in complement to the metabolic effects underlying protection to degeneration, we cannot exclude the possibility that E2F1 regulates other physiological processes involved in tissue degeneration in DMD. Interestingly, human patients of DMD express higher levels of E2F1 protein in the muscle, which strongly suggests that this transcription factor contributes also in humans to the physiopathology of DMD. Genetic studies are guaranteed to further demonstrate the relative contribution of E2F1 to DMD.
Finally, our data showing a dramatic increase in muscle performance upon E2f1 deletion in mdx mice, notably in the running test, are very promising for the development of new therapies for DMD treatment.
MATERIALS AND METHODS
Materials and oligonucleotides
Chemicals, unless stated otherwise, and anti-actin were purchased from Sigma (St Louis, MO, USA). Anti-E2F1 (C-20) and anti-GAPDH antibodies were from Santa Cruz. Anti-MyHC-I (BA-F8) and IIb (BF-F3) were purchased from the Developmental Studies Hybridoma Antibody Core at the University of Iowa. Anti-Mac-2 was purchased from eBioscience. Anti-utrophin, anti-β-dystroglycan and anti-γ-sarcoglycan were described elsewhere. Oligonucleotide sequences appear in Supplementary Material, Table S1.
Animal experiments
C57Bl/10J, E2f1+/+, and E2f1−/− and mdx mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and were maintained according to European Union Guidelines for use of laboratory animals. The endurance test was performed on 2 h fasted weight-matched E2f1 +/+, −/−, +/+;mdx and −/−;mdx mice at the beginning speed of 10 cm/s for 5 min. The speed was progressively increased (around 2 cm/s each 5 min) to reach, at the end of the running test, the maximal speed of 25 cm/s. The distance run was recorded all along the test and a mouse was removed from the experiment when considered exhausted. Experiments on E2f1 +/+;mdx/+ and E2f1 −/−;mdx/+ mice were performed on males at the age of 8 weeks unless stated otherwise. Serum CK levels were measured at the Service Phenotypage de l'IFR 150 (Toulouse, France) on mice fed ad libitum.
Oxygen consumption
Measurements of oxygen consumption were performed in mitochondria isolated from muscles. Briefly, tissues were homogenized into isolation buffer [250 mm mannitol, 10 mm ethylenediaminetetraacetic acid (EDTA), 45 mm Tris–HCl, 5 mm Tris-Base, pH 7.4] using a Dounce homogenizer. Nuclei and cell debris were removed by centrifugation at 2000 rpm for 10 min. Mitochondria were isolated from supernatant by spinning twice at 10 000 rpm for 10 min. The mitochondrial pellet was resuspended in isolation buffer and kept on ice. Mitochondrial protein was measured by Bradford method (BioRad). The rate of mitochondrial oxygen consumption was measured at 37°C in an incubation chamber with a Clark-type O2 electrode as described in (17) (Strathkelvin Instruments, Glasgow, UK) filled with 1 ml of respiration medium [15 mm KCl, 30 mm KH2PO4, 25 mm Tris-Base, 45 mm sucrose, 12 mm mannitol, 5 mm MgCl2, EDTA 7, 20 mm glucose, 0.2% bovin serum albumin (BSA)]. The measurements were performed using mitochondria (3 mg/ml) incubated with succinate (5 mm) as substrate in the presence or in the absence of 1 mm adenosin diphosphate. The incubation medium was constantly stirred with a magnetic stirrer. Respiration values were normalized to mitochondrial protein content.
RNA extraction, reverse transcriptase-polymerase chain reaction and quantitative polymerase chain reaction
For gene expression analysis, total RNA extraction from E2f1 +/+;mdx/+ and −/−;mdx/+ tissues and reverse transcription were performed as described (18,19). Quantitative polymerase chain reaction (Q-PCR) was carried out using a LightCycler 480 and the DNA double-strand-specific Power SYBR Green master mix for detection (Applied Biosystems, Foster City, CA, USA). Q-PCR was performed using gene-specific oligonucleotides and the results were then normalized to 18S or Cyclophilin B levels (Supplementary Material, Table S1).
Histology, immunofluorescence and biochemical analysis
Evans blue was injected intra-peritoneally (100 mg/kg) and tissues were harvested 16 h later, embedded in optimal cutting temperature. Evans blue detection was performed on 10 µm frozen sections as previously described (20,21). Hematoxylin, eosin staining and immunofluorescence microscopy (MyHC, 2 µg per section) were performed on 5 µm formalin-fixed paraffin-embedded tissue sections. Briefly, after deparaffinizing and rehydrating the sections, antigen retrieval was performed on muscle sections by incubating tissue preparations in sodium citrate buffer (pH 6.0) at 95°C for 30 min. Sections were then rinsed two times in tris-buffer solution (TBS) plus 0.025% triton X-100 for 5 min and subsequently blocked for 1 h with 10% normal goat serum in TBS containing 1% BSA and then incubated for 16 h with the indicated antibodies in TBS containing 1% BSA. After washing two times in tris-buffer solution (TBS) plus 0.025% triton X-100 for 5 min, immunostainings were revealed using a 488 Alexa-conjugated anti-rabbit or anti-mouse secondary antibodies (Jackson Immunoresearch) and observed under a fluorescence microscope (Leica Microsystemes SAS). Negative controls using rabbit or mouse IgGs were performed and no staining was observed in these conditions.
Morphometric analysis of histological data
The percentage of centrally nucleated fibers was counted on three hematoxylin–eosin (H&E)-stained cross-sections of three different mice from each genotype. The muscle fiber mean cross-sectional area was determined by using ImageScope software on H&E sections. Myofiber size variability, which is a feature of mdx muscles, was determined by multiplying the standard deviation of each individual cross-sectional area by 1000 and dividing it by the mean fiber diameter. For all experiments, approximately 150 fibers/mice were analyzed per genotype, and at least three mice per genotype were used.
Protein extracts and immunoblot analysis
Protein extracts and sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE), electrotransfer and immunoblotting were performed as described (22).
Statistical analysis
Data are presented as means ± standard error of mean. Statistical analyses were performed with unpaired Student's t-test. Differences were considered statistically significant at P < 0.05.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from Agence Nationale pour la Recherche (ANR Genopath/MetabocycleII), Association pour la Recherche sur le Diabète, Association Française des Diabètiques, Société Francophone du Diabète and Ligue Contre le Cancer. E.B. was supported by a grant from the French Ministère de l'Enseignement Supérieur et de la Recherche. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Supplementary Material
Acknowledgments
We thank members of the Fajas’ lab for support and discussions. We are grateful to S. Lehmann for patients biopsies. We acknowledge the RHEM (Réseau d'Histologie Expérimentale de Montpellier) for histology and in particular for tissue and slide preparations. We thank Noemi Taylor's lab, Claude Sardet lab and Valerie Pinet for providing tools and advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Aguilar V., Fajas L. Cycling through metabolism. EMBO Mol. Med. 2010;2:338–348. doi: 10.1002/emmm.201000089. doi:10.1002/emmm.201000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchet E., Annicotte J.S., Fajas L. Cell cycle regulators in the control of metabolism. Cell Cycle. 2009;8:4029–4031. doi: 10.4161/cc.8.24.10110. doi:10.4161/cc.8.24.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchet E., Annicotte J.S., Lagarrigue S., Aguilar V., Clape C., Chavey C., Fritz V., Casas F., Apparailly F., Auwerx J., et al. E2F transcription factor-1 regulates oxidative metabolism. Nat. Cell. Biol. 2011;13:1146–1152. doi: 10.1038/ncb2309. doi:10.1038/ncb2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y.W., Zhao P., Borup R., Hoffman E.P. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell. Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. doi:10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emery A.E. The muscular dystrophies. Lancet. 2002;359:687–695. doi: 10.1016/S0140-6736(02)07815-7. doi:10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 6.Even P.C., Decrouy A., Chinet A. Defective regulation of energy metabolism in mdx-mouse skeletal muscles. Biochem. J. 1994;304:649–654. doi: 10.1042/bj3040649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuznetsov A.V., Winkler K., Wiedemann F.R., von Bossanyi P., Dietzmann K., Kunz W.S. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol. Cell. Biochem. 1998;183:87–96. doi: 10.1023/a:1006868130002. doi:10.1023/A:1006868130002. [DOI] [PubMed] [Google Scholar]
- 8.Webster C., Silberstein L., Hays A.P., Blau H.M. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52:503–513. doi: 10.1016/0092-8674(88)90463-1. doi:10.1016/0092-8674(88)90463-1. [DOI] [PubMed] [Google Scholar]
- 9.Handschin C., Kobayashi Y.M., Chin S., Seale P., Campbell K.P., Spiegelman B.M. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21:770–783. doi: 10.1101/gad.1525107. doi:10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljubicic V., Miura P., Burt M., Boudreault L., Khogali S., Lunde J.A., Renaud J.M., Jasmin B.J. Chronic AMPK activation evokes the slow, oxidative myogenic program and triggers beneficial adaptations in mdx mouse skeletal muscle. Hum. Mol. Genet. 2011;20:3478–3493. doi: 10.1093/hmg/ddr265. doi:10.1093/hmg/ddr265. [DOI] [PubMed] [Google Scholar]
- 11.Chakkalakal J.V., Stocksley M.A., Harrison M.A., Angus L.M., Deschenes-Furry J., St-Pierre S., Megeney L.A., Chin E.R., Michel R.N., Jasmin B.J. Expression of utrophin A mRNA correlates with the oxidative capacity of skeletal muscle fiber types and is regulated by calcineurin/NFAT signaling. Proc. Natl Acad. Sci. USA. 2003;100:7791–7796. doi: 10.1073/pnas.0932671100. doi:10.1073/pnas.0932671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gramolini A.O., Belanger G., Thompson J.M., Chakkalakal J.V., Jasmin B.J. Increased expression of utrophin in a slow vs. a fast muscle involves posttranscriptional events. Am. J. Physiol. Cell. Physiol. 2001;281:C1300–C1309. doi: 10.1152/ajpcell.2001.281.4.C1300. [DOI] [PubMed] [Google Scholar]
- 13.Chakkalakal J.V., Michel S.A., Chin E.R., Michel R.N., Jasmin B.J. Targeted inhibition of Ca2+/calmodulin signaling exacerbates the dystrophic phenotype in mdx mouse muscle. Hum. Mol. Genet. 2006;15:1423–1435. doi: 10.1093/hmg/ddl065. doi:10.1093/hmg/ddl065. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y.X., Zhang C.L., Yu R.T., Cho H.K., Nelson M.C., Bayuga-Ocampo C.R., Ham J., Kang H., Evans R.M. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. doi:10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura P., Chakkalakal J.V., Boudreault L., Belanger G., Hebert R.L., Renaud J.M., Jasmin B.J. Pharmacological activation of PPARbeta/delta stimulates utrophin A expression in skeletal muscle fibers and restores sarcolemmal integrity in mature mdx mice. Hum. Mol. Genet. 2009;18:4640–4649. doi: 10.1093/hmg/ddp431. doi:10.1093/hmg/ddp431. [DOI] [PubMed] [Google Scholar]
- 16.Kotelnikova E., Shkrob M.A., Pyatnitskiy M.A., Ferlini A., Daraselia N. Novel approach to meta-analysis of microarray datasets reveals muscle remodeling-related drug targets and biomarkers in Duchenne muscular dystrophy. PLoS Comput. Biol. 2012;8:e1002365. doi: 10.1371/journal.pcbi.1002365. doi:10.1371/journal.pcbi.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. doi:10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Annicotte J., Fayard E., Swift G.H., Selander L., Edlund H., Tanaka T., Kodama T., Schoonjans K., Auwerx J. Pancreatic–duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol. Cell. Biol. 2003;23:6713–6724. doi: 10.1128/MCB.23.19.6713-6724.2003. doi:10.1128/MCB.23.19.6713-6724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fajas L., Annicotte J.S., Miard S., Sarruf D., Watanabe M., Auwerx J. Impaired pancreatic growth, beta cell mass, and beta cell function in E2F1 (−/−)mice. J. Clin. Invest. 2004;113:1288–1295. doi: 10.1172/JCI18555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mensink M., Hesselink M.K., Russell A.P., Schaart G., Sels J.P., Schrauwen P. Improved skeletal muscle oxidative enzyme activity and restoration of PGC-1 alpha and PPAR beta/delta gene expression upon rosiglitazone treatment in obese patients with type 2 diabetes mellitus. Int. J. Obes. (Lond.) 2007;31:1302–1310. doi: 10.1038/sj.ijo.0803567. doi:10.1038/sj.ijo.0803567. [DOI] [PubMed] [Google Scholar]
- 21.Handschin C., Chin S., Li P., Liu F., Maratos-Flier E., Lebrasseur N.K., Yan Z., Spiegelman B.M. Skeletal muscle fiber-type switching, exercise intolerance and myopathy in PGC-1alpha muscle-specific knockout animals. J. Biol. Chem, 2007;41:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 22.Sarruf D.A., Iankova I., Abella A., Assou S., Miard S., Fajas L. Cyclin D3 promotes adipogenesis through activation of peroxisome proliferator-activated receptor gamma. Mol. Cell. Biol. 2005;25:9985–9995. doi: 10.1128/MCB.25.22.9985-9995.2005. doi:10.1128/MCB.25.22.9985-9995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.