Abstract
In the oral cavity, mucosal keratinocytes resist bacterial infection, in part, by producing broad-spectrum antimicrobial peptides (AMPs) including defensin, adrenomedullin and calprotectin. Epidermal keratinocyte expression of many AMPs increases in response to interleukin-1α (IL-1α). IL-1α is produced by epidermal keratinocytes and regulates cell differentiation. To better understand innate immunity in the oral cavity, we sought to determine how IL-1α might regulate expression of AMPs by human gingival keratinocytes (HGKs) using DNA microarray and western blot analyses. HGKs from three subjects expressed eleven AMPs, including S100A7, S100A8, S100A9, S100A12, secretory leukocyte protease inhibitor, lipocalin 2 (LCN2), cystatin C and β-defensin 2. Of the expressed AMPs, S100A7, S100A12 and LCN2 were up-regulated by IL-1α (inducible AMPs); the other AMPs were considered to be constitutive. Human gingival keratinocytes, therefore, express constitutive and IL-1α-inducible AMPs to provide a rapid and robust innate response to microbial infection.
Keywords: antimicrobial peptide, gingival keratinocyte, interleukin-1α, microarray analysis, analysis
1. Introduction
In the oral cavity, the epithelium functions as a barrier against many microbes. In addition to physically separating the mucosal microflora from the connective tissues and circulation, the oral epithelium produces innate immune effector molecules, providing a more direct attack against infecting microbes. Oral keratinocyte innate immune effector molecules include the antimicrobial peptides (AMPs) defensin, cathelicidin, adrenomedullin and calprotectin.1 Human β-defensin 2 and β-defensin 3 have broad-spectrum antimicrobial activities against oral anaerobic bacteria including Porphyromonas gingivalis (P. gingivalis) and Aggregatibacter (Actinobacillus) actinomycetemcomitans, facultative anaerobes such as Streptococcus sanguinis, S. gordonii and S. mutans, and the opportunistic fungal pathogen, Candida.2,3 Human cathelicidin shows antimicrobial activity against periodontopathic bacteria and other bacteria.3 Calprotectin, a heterocomplex of S100 calcium binding protein (S100) A8 (S100A8) and S100A9, is also expressed in human oral and gingival keratinocytes.4,5 Calprotectin inhibits growth of P. gingivalis and adhesion and invasion into epithelial cells.4,6,unpublished data Functioning in defense against periodontopathic bacteria, calprotectin also appears to increase resistance to invasion by transient enteric bacteria.7,8
Interleukin-1α (IL-1α), which is produced by and signals epithelial cells9–11, regulates expression of selected AMPs including lipocalin 2, S100A7, S100A8, S100A9 and secretory leukocyte protease inhibitor (SLPI) in epidermal keratinocytes.12 Extracellular IL-1α signals through the IL-1α receptor (IL-1R1) to regulate epidermal keratinocyte differentiation in an autocrine manner and modulate keratinocyte growth factor production by fibroblasts in a paracrine manner.9–11 In human gingival keratinocytes, IL-1α induces cell differentiation and up-regulates calprotectin expression.13 To better understand the potential of gingival mucosal keratinocytes to contribute to innate immunity, we sought to develop a more comprehensive analysis of the constitutive and regulated AMPs. In the present study, AMPs expressed in human gingival keratinocytes (HGKs) were analyzed using DNA microarray and western blots. Constitutive and IL-1α-inducible AMPs were also distinguished.
2. Material and methods
2.1. Culture of human gingival keratinocytes
Human gingival fragments were obtained from three subjects (No.1: age 23 years, female; No.2: 15 years, female; No. 3: 26 years, male) during extraction of impacted mandibular third teeth. The Ethics Committee of Tokushima University Hospital approved the protocol (Approval No. 570) and informed consent was obtained from gingival tissue donors. Normal HGKs were isolated from excised gingival tissues as reported by Matsuyama et al.14 Briefly, gingival tissue was cut into 2–3 mm fragments, immobilized as explants on collagen I-coated dishes (IWAKI, Tokyo, Japan), and cultured for 1–2 days in Dulbecco’s Modified Eagle’s Minimal Essential Medium (NISSUI PHARMACEUTICAL Co., Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS; HyClone, Utah, USA), 100 U/ml penicillin and 100 µg/ml streptomycin. HGK explants were further cultured for 20 days in Keratinocyte-SFM (Gibco Invitrogen Co., Carlsbad, CA, USA) containing 1 ng/ml epidermal growth factor (Gibco Invitrogen) and 30 µg/ml bovine pituitary extract (Gibco Invitrogen). After out-growth from gingival explants, cells were cultured until the third passage. HGKs (4.8 × 104 cells/cm2) were seeded on collagen I-coated dishes, cultured for 6 days, and some cultures were then supplemented with 30 ng/ml IL-1α (Wako, Osaka, Japan) for 36 (microarray and RT-PCR) or 48 h (Western blot).
2.2. RNA isolation and microarray analysis
Total RNA was isolated from IL-1α-treated or non-treated HGKs using an RNeasy kit (Qiagen, Valencia, CA, USA). Microarray analysis was performed by the modification of previous method12, using GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA). Gene expression data were analyzed using Affymetrix® Expression Console™ Software (Affymetrix). Genes showing fluorescence intensity of less than 100 in any HGK sample were excluded from further analysis. HGK genes up-regulated more than 2-fold or down-regulated 0.5-fold by IL-1α treatment are listed in Table 1; AMP genes expressed in HGKs are shown (Table 2).
Table 1.
Genes differentially expressed in human gingival keratinocytes in response to IL-1α
| Gene | Accession No. | Fold |
|---|---|---|
| Upregulated genes | ||
| Defensin, beta 4 | NM_004942 | 52.44 |
| Chromosome 15 open reading frame 48 | AF228422 | 8.43 |
| Small proline-rich protein 2G | AA456642 | 8.32 |
| S100 calcium binding protein A7 | NM_002963 | 8.13 |
| Serpin peptidase inhibitor, clade B, member 4 | AB046400 | 7.00 |
| Late cornified envelope 3D | AB048288 | 6.08 |
| Small proline-rich protein 2C | NM_006518 | 5.57 |
| Similar to FRAS1 related extracellular matrix protein 2 | BE669806 | 5.45 |
| Dehydrogenase/ Reductase member 9 | AF240698 (NM_005771) |
3.10 (2.36) |
| Ribonuclease, RNase A family, 7 | AJ131212 | 2.60 |
| Down-regulated genes | ||
| Chemokine ligand 14 | AF144103 | 0.29 |
Table 2.
Antimicrobial peptide genes expressed in human gingival keratinocytes in response to IL-1α
| Gene | Accession No. | Cell No. | Signal value |
Fold |
||
|---|---|---|---|---|---|---|
| control | IL-1α | cont/IL-1α | average | |||
| S100 calcium binding protein A8 (S100A8) | NM_00296 | 1. | 10630.17 | 10563.37 | 0.99 | 1.07 |
| 2. | 11569.83 | 12344.92 | 1.07 | |||
| 3. | 11944.66 | 13893.91 | 1.16 | |||
| Secretory leukocyte peptidase inhibitor (SLPI) | NM_003064 | 1. | 6405.07 | 8624.71 | 1.35 | 1.20 |
| 2. | 7128.82 | 8310.68 | 1.17 | |||
| 3. | 10780.03 | 11686.36 | 1.08 | |||
| S100 calcium binding protein A9 (S100A9) | NM_002965 | 1. | 5591.75 | 8264.38 | 1.48 | 1.25 |
| 2. | 8429.60 | 9411.56 | 1.12 | |||
| 3. | 9996.13 | 11462.43 | 1.15 | |||
| Lipocalin 2 (LCN2) | NM_005564 | 1. | 1116.49 | 3549.59 | 3.18 | 2.31 |
| 2. | 1579.89 | 3443.55 | 2.18 | |||
| 3. | 3144.53 | 4960.89 | 1.58 | |||
| Cystatin C (CST3) | NM_000099 | 1. | 1324.43 | 1239.03 | 0.94 | 0.89 |
| 2. | 1693.14 | 1456.90 | 0.86 | |||
| 3. | 1525.31 | 1311.21 | 0.86 | |||
| Adrenomedullin (ADM) | NM_001124 | 1. | 1580.59 | 1029.43 | 0.65 | 0.97 |
| 2. | 1417.43 | 1520.63 | 1.07 | |||
| 3. | 1257.57 | 1505.86 | 1.20 | |||
| S100 calcium binding protein A12 (S100A12) | NM_005621 | 1. | 77.87 | 557.08 | 7.15 | 4.34 |
| 2. | 316.69 | 1271.19 | 4.01 | |||
| 3. | 1218.05 | 2276.57 | 1.87 | |||
| Defensin, beta 1 (DEFB1) | U73945 | 1. | 297.14 | 259.76 | 0.87 | 1.05 |
| 2. | 304.96 | 335.80 | 1.10 | |||
| 3. | 424.97 | 495.50 | 1.17 | |||
| S100 calcium binding protein A7 (S100A7) | NM_002963 | 1. | 33.18 | 327.11 | 9.86 | 8.13 |
| 2. | 125.74 | 1209.30 | 9.62 | |||
| 3. | 655.44 | 3213.26 | 4.90 | |||
| Ribonuclease, RNase A family, 7 (RNASE7) | AK023343 | 1. | 166.55 | 370.08 | 2.22 | 1.90 |
| 2. | 255.15 | 474.85 | 1.86 | |||
| 3. | 294.09 | 480.62 | 1.63 | |||
| AJ131212 | 1. | 122.52 | 443.54 | 3.62 | 2.60 | |
| 2. | 200.69 | 407.02 | 2.03 | |||
| 3. | 324.88 | 698.45 | 2.15 | |||
| Defensin, beta 4 (DEFB4) | NM_004942 | 1. | 5.25 | 609.47 | 116 | 52.44 |
| 2. | 76.44 | 2713.46 | 35.5 | |||
| 3. | 340.70 | 1981.65 | 5.82 | |||
2.3. Reverse transcription-polymerase chain reaction (RT-PCR)
As we reported previously6,12, total RNA was isolated from cultured cells using RNeasy kit (Qiagen), cDNA was synthesized from 1 µg of the RNA sample using ReverTra Ace -α-® (TOYOBO, Osaka, Japan). The cDNA was added to the PCR mixture, which contained primers (Table 3), dNTPs, TaKaRa Taq™ HS (TaKaRa Bio, Otsu, Japan) and PCR Buffer, and amplified for 30–40 cycles under the following conditions: denature 94°C for 1 min, anneal at 50–60°C for 1 min, and extension at 72°C for 1 min. The PCR products were analyzed by electrophoresis on a 1.5% agarose gels containing 0.1 µg/ml ethidium bromide. The expression of AMPs and IL-1R1 genes was verified by RT-PCR.
Table 3.
RT-PCR primers
| Gene | Primer | PCR product size (bp) |
|---|---|---|
| DEFB4 | For: 5’-CCAGCCATCAGCCATGAGGGT-3’ Rev: 5’-GGAGCCCTTTCTGAATCCGCA-3’ |
255 |
| GAPDH | For: 5’-TCCACCACCCTGTTGCTGTA-3’ Rev: 5’-ACCACAGTCCATGCCATCAC-3’ |
451 |
| IL-1R1 | For: 5’-TGCCGCTCTTCTGTCATCCCGCTC-3’ Rev: 5’-GGGGGGACCGTTATTGACCTGAAA-3’ |
716 |
| LCN2 | For: 5’-TGTCACCTCCGTCCTGTTTAG-3’ Rev: 5’-TCTCCCGTAGAGGGTGATCTT-3’ |
226 |
| RNASE7 | For: 5’-TTTGGCTGACCTTCAATTCC-3’ Rev: 5’-TCTTGGGGATAAGCATCTGG-3’ |
199 |
| S100A7 | For: 5’-TGCTGACGATGATGAAGGAG-3’ Rev: 5’-ATTCTCCCAGCAAGGACAG-3’ |
151 |
| S100A12 | For: 5’-TTGAAGAGCATCTGGAGGG-3’ Rev: 5’-CTACTCTTTGTGGGTGTGG-3’ |
269 |
2.4. Western blotting
HGKs were cultured for 6 days and then for 48 h with or without IL-1α (30 ng/ml), harvested, and suspended in 10 mM Tris-HCl (pH 7.4) with a protease inhibitor cocktail including phenylmethylsulfonyl fluoride (174 µg/ml), leupeptin (1 µg/ml), pepstatin (1 µg/ml), N-p-tosyl-l-phenylalanine chloromethyl ketone (1 µg/ml) and N-α-p-tosyl-l-lysine chloromethyl ketone hydrochloride (1 µg/ml). After sonication in an ice water bath, the suspended cells were analyzed for production of AMPs by western blotting using a modification of a method described previously.15 Briefly, 25 µg of cellular protein was electrophoretically separated on 15% polyacrylamide gels and electrically transferred to Hybond-P (GE-Healthcare Life Sciences, Amersham, Buckinghamshire, UK). After blocking with Starting Block™ blocking buffers (PIERCE, Rockford, IL, USA), membranes were separately incubated at 4°C overnight with 1 µg/ml anti-human β-defensin 2 (Santa Cruz Biotechnology, INC; Santa Cruz, CA, USA), 0.2 µg/ml anti-lipocalin 2 (R&D Systems Inc; Minneapolis, MN, USA), 1 µg/ml anti-S100A7 (Acris Antibodies GmbH; Hiddenhausen, Germany) or 0.2 µg/ml anti-S100A12 (R&D Systems Inc; Minneapolis, MN, USA) antibody. Bound primary antibodies were then incubated with horseradish peroxidase-conjugated anti-goat or mouse IgG (1/5000 dilution, Cell Signaling Technology) for 1.5 h at room temperature. The AMP protein antigens were detected using an ECL Western Blotting Detection System (GE-Healthcare Life Sciences Co., Piscataway, NJ, USA) and resolved by exposure to Hyperfilm-ECL (GE-Healthcare Life Sciences Co.).
3. Results
3.1. Genes expressed in human gingival keratinocytes in response to IL-1α
In RNA samples from three subjects, the average expression of 10 genes was increased after incubation with IL-1α more than two-fold compared to un-treated controls; IL-1α decreased expression of one gene to less than 0.5-fold of control (Table 1). Of the ten genes found to be up-regulated by IL-1α, three are putative AMPs: β-defensin 2 (Defensin, beta 4 [DEFB4]; 52.4-fold), S100A7 (8.1-fold), and ribonuclease 7 (ribonuclease, RNase A family, 7 [RNASE7]; 2.6-fold). Other seven are as follows; Chromosome 15 open reading frame 48 (8.4-fold), Small proline-rich protein 2G (8.3-fold), Serpin peptidase inhibitor, clade B, member 4 (7.0-fold), Late cornified envelope 3D (6.1-fold), Small proline-rich protein 2C (5.6-fold), Similar to FRAS1 related extracellular matrix protein 2 (5.5-fold) and Dehydrogenase/Reductase member 9 (3.1-fold). IL-1α decreased the expression of chemokine ligand 14 (0.29-fold).
3.2. AMP expression in human gingival keratinocytes
The three HGK samples expressed eleven AMP genes with signal intensity of more than 100 (Table 2). The S100A8, SLPI and S100A9 genes appeared to be expressed in greatest abundance, showing the highest signal intensity among 11 AMP genes. Lipocalin 2 (LCN2), cystatin C (CST3) and adrenomedullin (ADM) showed moderate gene expression levels, whereas S100A12, β-defensin 1 (DEFB1), S100A7, RNASE7 and DEFB4 were expressed at low levels.
In the conditions of our experiments, however, the expression of the S100A8 (1.07-fold), SLPI (1.20-fold) and S100A9 (1.25-fold) appeared unaffected by IL-1α. Similarly, the expression of DEFB1 (1.05-fold), CST3 (0.89-fold) and ADM (0.97-fold) were not up-regulated by IL-1α. In contrast, IL-1α up-regulated the expression of LCN2 (2.3-fold), S100A12 (4.3-fold), S100A7 (8.1-fold), RNASE7 (2.6-fold) and DEFB4 (52.4-fold). AMPs such as azurocidin 1, cathelicidin, dermcidin, β-defensin 3 (DEFB103A), hepcidin, mucin 5B and seminal plasmin (PYY2) showed signal intensities less than 100 in IL-1α-stimulated and non-stimulated HGKs.
3.3. Validation of AMP gene expression up-regulated by IL-1α
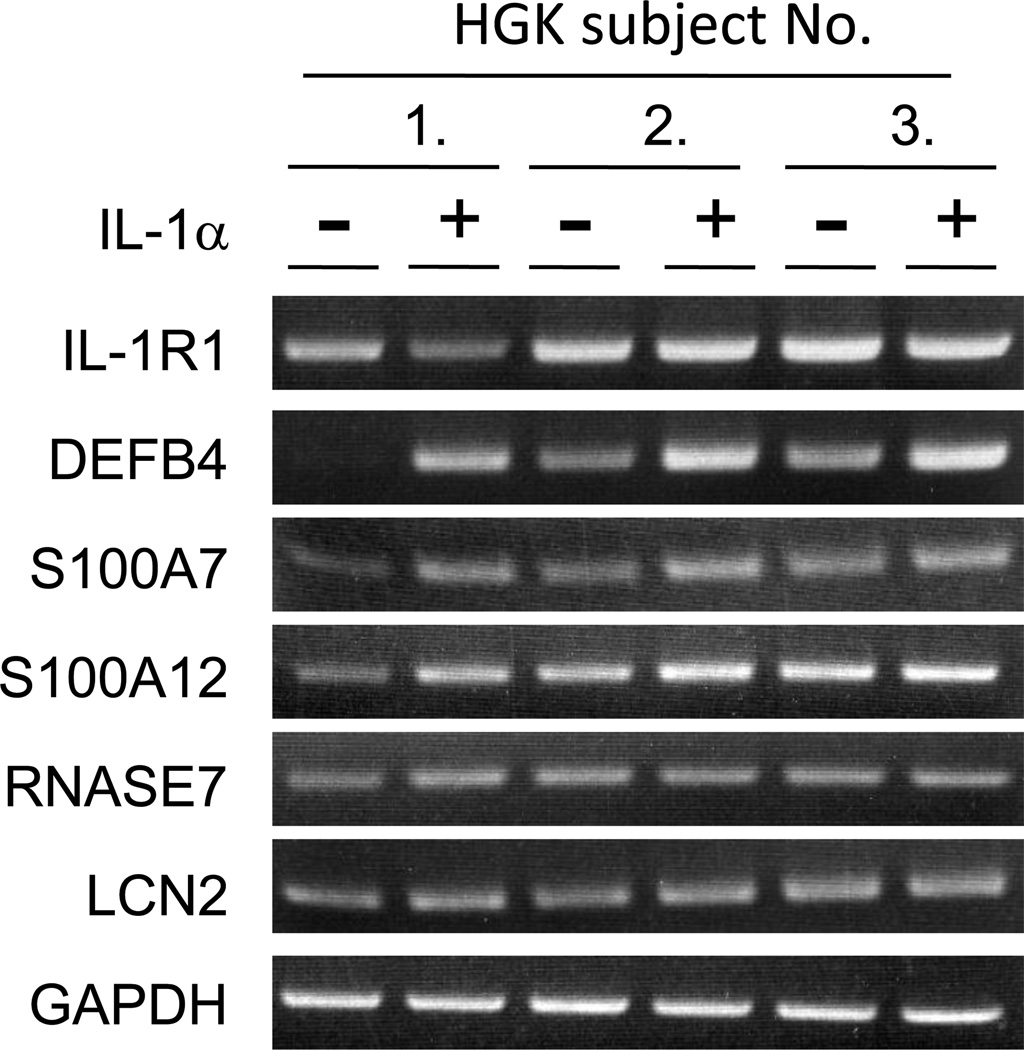
HGKs from three subjects expressed IL-1R1 gene; expression was not affected by IL-1α (Figure 1). In microarray analysis, expression of AMP genes, including DEFB4, S100A7, S100A12, RNASE7 and LCN2, increased at least two-fold in response to IL-1α. To confirm the results of the microarray analysis, RT-PCR analysis of the five AMP genes was performed using RNA samples from HGKs of the three subjects. Expression of DEFB4, S100A7, S100A12, RNASE7 and LCN2 RNA was confirmed in HGKs using RT-PCR analysis and expression of DEFB4, S100A7 and S100A12 were confirmed to be up-regulated by IL-1α; IL-1α-dependent changes in expression of RNASE7 and LCN2 were not apparent (Figure 1).
Figure 1.
Verification of AMP mRNA expression in IL-1α-stimulated HGKs by RT-PCR. HGKs were cultured for 6 days and incubated with IL-1α (30 ng/ml) for 36 h. After RNA isolation, three separate tissue samples were analyzed for expression of DEFB4, S100A7, S100A12, RNASE7, LCN2, IL-1R1 and GAPDH mRNAs using RT-PCR.
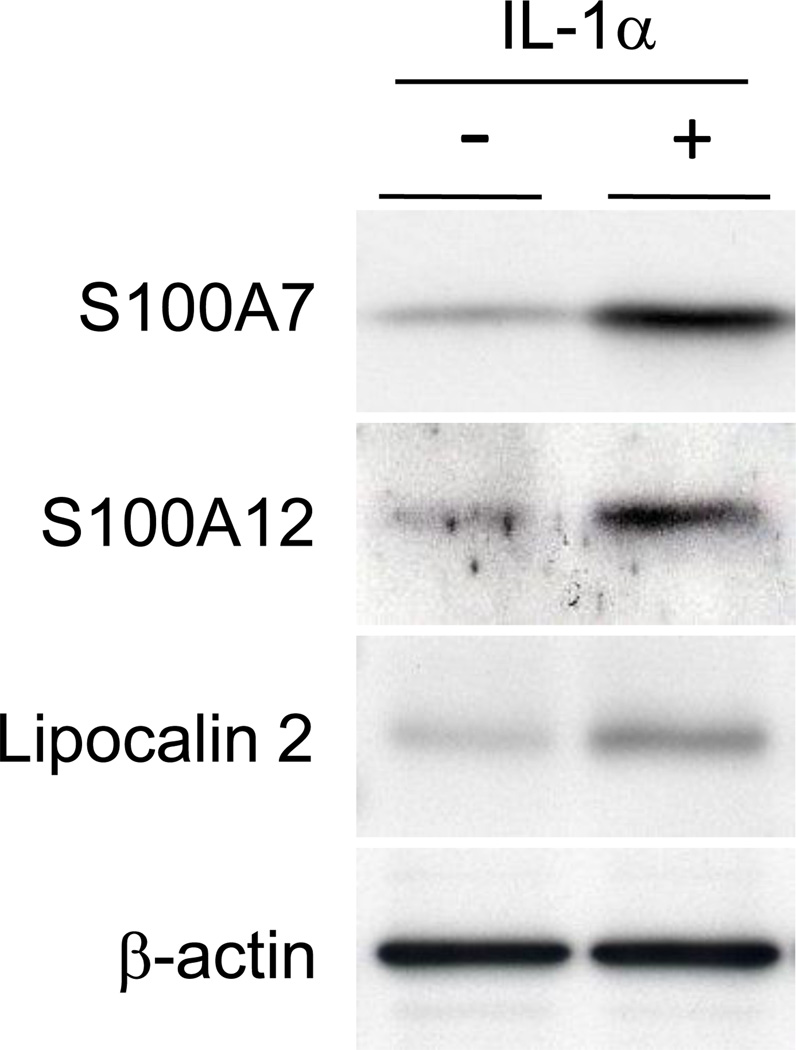
3.4. Up-regulation of AMP proteins by IL-1α
The effect of IL-1α on AMP expression in HGKs was investigated at the protein level by western blot analysis (Figure 2). Incubation with IL-1α for 48 h increased S100A7, S100A12 and lipocalin 2 protein antigens in HGKs. Using several different anti-human β-defensin 2 antibodies, β-defensin 2 protein was below the limits of detection (data not shown). S100A7, S100A12 and lipocalin 2 protein antigen level reflected the up-regulation in gene expression in response to IL-1α.
Figure 2.
AMP proteins up-regulated by IL-1α HGKs were cultured for 6 days and incubated with IL-1α (30 ng/ml) for 48 h. HGKs were sonicated to disrupt the cells, and 30 µg of soluble protein was analyzed for S100A7, S100A12, lipocalin 2 and β-actin by Western blotting. A typical result for protein isolated from three separate HGK tissue samples is shown.
4. Discussion
In the present study, HGKs derived from three subjects each expressed genes encoding eleven putative AMPs. HGKs appeared to express constitutive and IL-1α-inducible AMPs. In the conditions of these experiments, IL-1α did not induce S100A8, S100A9, SLPI, CST3, ADM and DEFB1. Yet, these AMPs showed high or moderate gene array signal intensity in non-stimulated HGKs and appear to be more tightly regulated in response to IL-1α than AMP genes that are expressed at lower levels. The relative importance of each of the AMPs to providing innate resistance against infection of HGKs is unclear.
Calprotectin (S100A8/S100A9) is expressed in human oral and gingival keratinocytes, and its expression is increased by pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α, and stimulators of keratinocyte differentiation.12,13,16–18 IL-1α also increases expression of calprotectin in keratinocytes, with a narrow dose-response range that differs with the cell lineage.11–13,Sorenson et al., 2010, in preparation In the conditions of our experiments, we were unable to see a regulatory effect of IL-1α on calprotectin expression. SLPI, a potent serine protease inhibitor, is constitutively expressed in human keratinocytes, and has been reported to be up-regulated by IL-1α, IL-1β, TNF-αand epidermal growth factor12,19–22 but SLPI was minimally up-regulated by IL-1α in our experiments. CST3 has been detected in gingival crevicular fluid, tears and cerebrospinal fluid, and inhibits the replication of viruses and bacteria.23–25 ADM, a potent vasoactive peptide, is actively secreted from human oral and skin keratinocytes, and is increased by IL-1α, IL-1β, TNF-α and lipopolysaccharide.26 Human β-defensins including β-defensin 1, 2 and 3 are major cationic AMPs and expressed in epidermal and epithelial keratinocytes, and have antimicrobial activity against Gram-positive and -negative bacteria, and Candida.1,27–29 DEFB1 was not induced by IL-1α but showed moderate expression in untreated HGKs as reported previously.30,31 Constitutive AMPs are likely to contribute rapidly to innate oral immunity, assuming that like calprotectin, the protein products are stored and provide intracellular protection against invading microbes7,8,Sorenson et al. 2010, in preparation or are released to add to the antimicrobial content of the mucosal surface fluids.
In contrast, IL-1α increased expression of five AMP genes including DEFB4, S100A7, S100A12, RNASE7 and LCN2. These inducible AMPs showed low signal intensity in non-stimulated HGKs, but highly responded to IL-1α. DEFB4 expression was most strongly up-regulated by IL-1α among the eleven HGK AMPs and up-regulation by IL-1α mirrored what we have reported in human skin keratinocytes.32 S100A7 and S100A12 are members of the S100 family of calcium binding proteins. S100A7 is expressed in healthy skin and mucosal epithelial cells, and increased in response to pro-inflammatory cytokines and stimulators of keratinocyte differentiation.12,33,34 S100A12 is expressed in monocytes, neutrophils and epithelial cells, and its expression markedly increased in response to infection and inflammation.35,36 Ribonuclease 7 has ribonuclease activity and antimicrobial activity with broad spectrum and is expressed in various epithelial cells.37,38 RNASE7 expression is up-regulated by pro-inflammatory cytokines and bacterial infection.37 Lipocalin 2 is an abundant protein in specific granules of human neutrophils and is also secreted from lacrimal gland.39 As a major component in tears, lipocalin 2 inhibits bacterial growth through binding to iron siderophores.40 LCN2 expression is up-regulated by IL-1α, IL-1β and TGF-α in human keratinocytes.12,21 Inducible AMPs are produced in mucosal and gingival tissues after microbial infection or exposure to pro-inflammatory cytokines. When compared to the constitutive AMPs, the inducible AMPs would appear to function as a secondary antimicrobial barrier in the human oral cavity. Constitutive AMPs are suggested to contribute to oral epithelial health in the presence of the endogenous mucosal flora and transient pathogens. The inducible AMPs appear to function as an early response to infection or inflammatory injury. The dichotomous regulation of AMP expression in HGKs may contribute to a robust innate immune response in health and during initial infections of the oral cavity.
HGKs derived from three subjects showed individual variations in AMP expression and responses to IL-1α. For example, the inducible AMPs, DEFB4 (range, 5.8 ~ 116-fold) and S100A12 (1.9 ~ 7.2-fold), showed a wide range of responses to IL-1α. Similar inter-individual variability also occurs in primary human skin keratinocytes, whereby IL-1α up-regulated DEFB4 (1.6 ~ 45.5-fold) and S100A12 (1.1 ~ 9.4-fold).36 While it is tempting to speculate that the range of response reflects individual differences in innate immunity in the gingiva and skin, we cannot rule out cell selection that occurs whenever primary cultures are established from explants.
The response to IL-1α by keratinocytes is more generalized than an antimicrobial response. IL-1α also regulates keratinocyte differentiation, proliferation and growth factor production.13,41–44 In the present study, IL-1α increased the expression of small proline-rich protein (SPRR) family including SPRR2G (8.3-fold) and SPRR2C (5.6-fold). The SPRR genes are conserved in the human epidermal differentiation complex45, and expressed in stratified squamous epithelia, keratinized and non-keratinized mucosal epithelia.46,47 The expression of SPRR genes increases during normal keratinocyte differentiation48, 49, suggesting that IL-1α may regulate the expression of AMP genes coordinately with keratinocyte differentiation.
Acknowledgements
The authors are very grateful to Dr. Masaru Nagayama, Dr. Youji Miyamoto and Dr. Kazuhito Satomura (The University of Tokushima Graduate School) for the help to supply of gingival fragments. This study was supported in part by Grants-in Aid (19592388 and 21592625) for Scientific Research from the Japan Society for the Promotion and by NIH grants RO1DE11831 and RO1DE15503 to MCH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dale BA, Fredericks LP. Antimicrobial peptides in the oral environment: expression and function in health and disease. Curr Issues Mol Biol. 2005;7:119–133. doi: 10.1093/jac/dki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joly S, Maze C, McCray PB, Jr, Guthmiller JM. Human β-defensin 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–1029. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji S, Hyun J, Park E, Lee BL, Kim KK, Choi Y. Susceptibility of various oral bacteria to antimicrobial peptides and to phagocytosis by neutrophils. J Periodont Res. 2007;42:410–419. doi: 10.1111/j.1600-0765.2006.00962.x. [DOI] [PubMed] [Google Scholar]
- 4.Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infec Immun. 2001;69:4242–4247. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross KF, Herzberg MC. Calprotectin expression by gingival epithelial cells. Infec Immun. 2001;69:3248–3254. doi: 10.1128/IAI.69.5.3248-3254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiroshima Y, Bando M, Kataoka M, Shinohara Y, Herzberg MC, Ross KF, et al. Shosaikoto increases calprotectin expression in human oral epithelial cells. J Periodont Res. 2010;45:79–86. doi: 10.1111/j.1600-0765.2009.01203.x. [DOI] [PubMed] [Google Scholar]
- 7.Champaiboon C, Sappington KJ, Guenther BD, Ross KF, Herzberg MC. Calprotectin S100A9 calcium-binding loops I and II are essential for keratinocyte resistance to bacterial invasion. J Biol Chem. 2009;284:7078–7090. doi: 10.1074/jbc.M806605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaia AA, Sappington KJ, Nisapakultorn K, Chazin WJ, Dietrich EA, Ross KF, et al. Subversion of antimicrobial calprotectin (S100A8/S100A9 complex) in the cytoplasm of TR146 epithelial cells after invasion by Listeria monocytogenes. Mucoal Immunol. 2009;2:43–53. doi: 10.1038/mi.2008.63. [DOI] [PubMed] [Google Scholar]
- 9.Sims JE, March CJ, Cosman D, Widmer MB, MacDonald HR, McMahan CJ, et al. cDNA expression cloning of the IL-1 receptor, a member of the immunoglobulin superfamily. Science. 1988;241:585–589. doi: 10.1126/science.2969618. [DOI] [PubMed] [Google Scholar]
- 10.Sims JE, Gayle MA, Slack JL, Alderson MR, Bird TA, Giri JG, et al. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc Natl Acad Sci USA. 1993;90:6155–6159. doi: 10.1073/pnas.90.13.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bando M, Hiroshima Y, Kataoka M, Herzberg MC, Ross KF, Shinohara Y, et al. Modulation of calprotectin in human keratinocytes by keratinocyte growth factor and interleukin-1α. Immunol Cell Biol. 2010;88:328–333. doi: 10.1038/icb.2009.104. [DOI] [PubMed] [Google Scholar]
- 12.Bando M, Hiroshima Y, Kataoka M, Shinohara Y, Herzberg MC, Ross KF, at al. Interleukin-1α regulates antimicrobial peptide expression in human keratinocytes. Immunol Cell Biol. 2007;85:532–537. doi: 10.1038/sj.icb.7100078. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi N, Kido J, Suryono, Kido R, Wada C, Kataoka M, et al. Regulation of calprotectin expression by interleukin-1α and transforming growth factor- β in human gingival keratinocytes. J Periodont Res. 2007;42:1–7. doi: 10.1111/j.1600-0765.2005.00857.x. [DOI] [PubMed] [Google Scholar]
- 14.Matsuyama T, Izumi Y, Sueda T. Culture and characterization of human junctional epithelial cells. J Periodontol. 1997;68:229–239. doi: 10.1902/jop.1997.68.3.229. [DOI] [PubMed] [Google Scholar]
- 15.Kido J, Kido R, Suryono, Kataoka M, Fagerhol MK, Nagata T. Induction of calprotectin release by Porphyromonas gingivalis lipopolysaccharide in human neutrophils. Oral Microbiol Immunol. 2004;19:182–187. doi: 10.1111/j.0902-0055.2004.00139.x. [DOI] [PubMed] [Google Scholar]
- 16.Eversole LR, Miyasaki KT, Christensen RE. The distribution of the antimicrobial protein, calprotectin, in normal oral keratinocytes. Archs oral Biol. 1992;37:963–968. doi: 10.1016/0003-9969(92)90068-j. [DOI] [PubMed] [Google Scholar]
- 17.Suryono, Kido J, Hayashi N, Kataoka M, Nagata T. Effect of Porphyromonas gingivalis lipopolysaccharide, tumor necrosis factor-α, and interleukin-1β on calprotectin release in human monocytes. J Periodontol. 2003;74:1719–1724. doi: 10.1902/jop.2003.74.12.1719. [DOI] [PubMed] [Google Scholar]
- 18.Kido J. The role of calprotectin in periodontal diseases and regulation of its expression. J Jpn Soc Periodontol. 2007;49:13–19. [Google Scholar]
- 19.Furutani Y, Kato A, Yasue H, Alexander LJ, Beattie CW, Hirose S. Evolution of the trappin multigene family in the Suidae. J Biochem. 1998;124:491–502. doi: 10.1093/oxfordjournals.jbchem.a022140. [DOI] [PubMed] [Google Scholar]
- 20.Doumas S, Kolokotronis A, Stefanopoulos P. Anti-inflammatory and antimicrobial roles of secretory leukocyte protease inhibitor. Infec Immun. 2005;73:1271–1274. doi: 10.1128/IAI.73.3.1271-1274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sørensen OE, Cowland JB, Theilgaard-Mönch K, Liu L, Ganz T, Borregaard N. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J Immunol. 2003;170:5583–5589. doi: 10.4049/jimmunol.170.11.5583. [DOI] [PubMed] [Google Scholar]
- 22.Lai JY, Borson ND, Strausbauch MA, Pittelkow MR. Mitosis increases levels of secretory leukocyte protease inhibitor in keratinocytes. Biochem Biophys Res Commun. 2004;316:407–410. doi: 10.1016/j.bbrc.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 23.Jasir A, Kasprzykowski F, Lindström V, Schalén C, Grubb A. New antimicrobial peptide active against Gram-positive pathogens. Indian J Med Res. 2004;119:74–76. [PubMed] [Google Scholar]
- 24.Wassélius J, Håkansson K, Abrahamson M, Ehinger B. Cystatin C in the anterior segment of rat and mouse eyes. Acta Ophthalmol Scand. 2004;82:68–75. doi: 10.1046/j.1600-0420.2003.00178.x. [DOI] [PubMed] [Google Scholar]
- 25.Ülker AE, Tulunoglu Ö, Özmeric N, Can M, Demirtas S. The evaluation of cystatin C, IL-1β, and TNF-α levels in total saliva and gingival crevicular fluid from 11- to 16-year-old children. J Periodontol. 2008;79:854–860. doi: 10.1902/jop.2008.070422. [DOI] [PubMed] [Google Scholar]
- 26.Kapas S, Tenchini ML, Farthing PM. Regulation of adrenomedullin secretion in cultured human skin and oral keratinocytes. J Inves Dermatol. 2001;117:353–359. doi: 10.1046/j.0022-202x.2001.01426.x. [DOI] [PubMed] [Google Scholar]
- 27.Harder J, Bartels J, Christophers E, Schröder JM. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J Bio Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 28.Harder J, Schröder JM. Antimicrobial peptides in human skin. Chem Immunol Allergy. 2005;86:22–41. doi: 10.1159/000086650. [DOI] [PubMed] [Google Scholar]
- 29.Sørensen OE, Thapa DR, Rosenthal A, Liu L, Roberts AA, Ganz T. Differential regulation of β-defensin expression in human skin by microbial stimuli. J Immunol. 2005;174:4870–4879. doi: 10.4049/jimmunol.174.8.4870. [DOI] [PubMed] [Google Scholar]
- 30.Krisanaprakornkit S, Weinberg A, Perez CN, Dale BA. Expression of the peptide antibiotic human β-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infec Immun. 1998;66:4222–4228. doi: 10.1128/iai.66.9.4222-4228.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, et al. Production of β-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infec Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu AY, Destoumieux D, Wong AV, Park CH, Valore EV, Liu L, et al. Human β-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J Invest Dermatol. 2002;118:275–281. doi: 10.1046/j.0022-202x.2001.01651.x. [DOI] [PubMed] [Google Scholar]
- 33.Madsen P, Rasmussen HH, Leffers H, Honoré B, Dejgaard K, Olsen E, et al. Molecular cloning, occurrence, and expression of a novel partially secreted protein “Psoriasin” that is highly up-regulated in psoriatic skin. J Invest Dermatol. 1991;97:701–712. doi: 10.1111/1523-1747.ep12484041. [DOI] [PubMed] [Google Scholar]
- 34.Gläser R, Harder J, Lange H, Bartels J, Christophers E, Schröder JM. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 35.Robinson MJ, Hogg N. A comparison of human S100A12 with MRP-14 (S100A9) Biochem Biophys Res Commun. 2000;275:865–870. doi: 10.1006/bbrc.2000.3407. [DOI] [PubMed] [Google Scholar]
- 36.Mee JB, Johnson CM, Morar N, Burslem F, Groves RW. The psoriatic transcriptome closely resembles that induced by interleukin-1 cultured keratinocytes. Am J Pathol. 2007;171:32–42. doi: 10.2353/ajpath.2007.061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harder J, Schröder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J Bio Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 38.Eberhard J, Menzel N, Dommisch H, Winter J, Jepsen S, Mutters R. The stage of native biofilm formation determines the gene expression of human β-defensin-2, psoriasin, ribonuclease 7 and inflammatory mediators: a novel approach for stimulation of keratinocytes with in situ formed biofilms. Oral Microbiol Immunol. 2008;23:21–28. doi: 10.1111/j.1399-302X.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- 39.Kjeldsen L, Bainton DF, Sengeløv H, Borregaard N. Identification of neutrophil gelatinase-associated lipocalin as a novel matrix protein of specific granules in human neutrophils. Blood. 1994;83:799–807. [PubMed] [Google Scholar]
- 40.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 41.Sauder DN, Stanulis-Praeger BM, Gilchrest BA. Autocrine growth stimulation of human keratinocytes by epidermal cell-derived thymocyte-activating factor: implications for skin aging. Arch Dermatol Res. 1988;280:71–76. doi: 10.1007/BF00417707. [DOI] [PubMed] [Google Scholar]
- 42.Dinarello CA, Wolff SM. The role of interleukin-1 in disease. N Engl J Med. 1993;328:106–113. doi: 10.1056/NEJM199301143280207. [DOI] [PubMed] [Google Scholar]
- 43.Eller MS, Yaar M, Ostrom K, Harkness DD, Gilchrest BA. A role for interleukin-1 in epidermal differentiation: regulation by expression of functional versus decoy receptors. J Cell Sci. 1995;108:2741–2746. doi: 10.1242/jcs.108.8.2741. [DOI] [PubMed] [Google Scholar]
- 44.Feliciani C. Keratinocytes and cytokine/growth factors. Crit Rev Oral Biol Med. 1996;7:300–318. doi: 10.1177/10454411960070040101. [DOI] [PubMed] [Google Scholar]
- 45.Lohman FP, Medema JK, Gibbs S, Ponec M, van de Putte P, Backendorf C. Expression of the SPRR cornification genes is differentially affected by carcinogenic transformation. Exp Cell Res. 1997;231:141–148. doi: 10.1006/excr.1996.3458. [DOI] [PubMed] [Google Scholar]
- 46.Hohl D, de Viragh PA, Amiguet-Barras F, Gibbs S, Backendorf C, Huber M. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursor proteins. J Invest Dermatol. 1995;104:902–909. doi: 10.1111/1523-1747.ep12606176. [DOI] [PubMed] [Google Scholar]
- 47.Katou F, Shirai N, Kamakura S, Tagami H, Nagura H, Motegi K. Differential expression of cornified cell envelope precursors in normal skin, intraorally transplanted skin and normal oral mucosa. Br J Dermatol. 2003;148:898–905. doi: 10.1046/j.1365-2133.2003.05288.x. [DOI] [PubMed] [Google Scholar]
- 48.Kartasova T, van Muijen GNP, van Pelt-Heerschap H, van de Putte P. Novel protein in human epidermal keratinocytes: regulation of expression during differentiation. Mol Cell Biol. 1988;8:2204–2210. doi: 10.1128/mcb.8.5.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibbs S, Lohman F, Teubel W, van de Putte P, Backendorf C. Characterization of the human spr2 promoter: induction after UV irradiation or TPA treatment and regulation during differentiation of cultured primary keratinocytes. Nucleic Acids Res. 1990;18:4401–4407. doi: 10.1093/nar/18.15.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]