Abstract
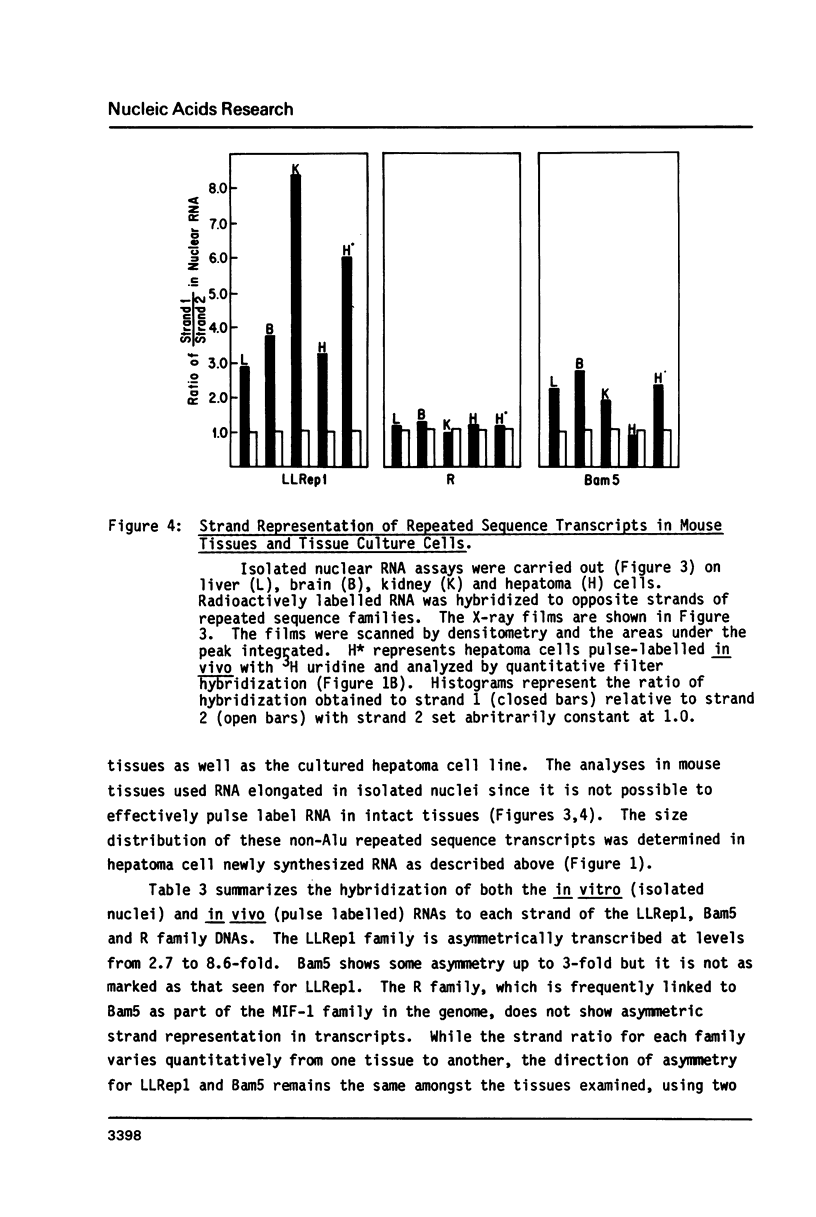
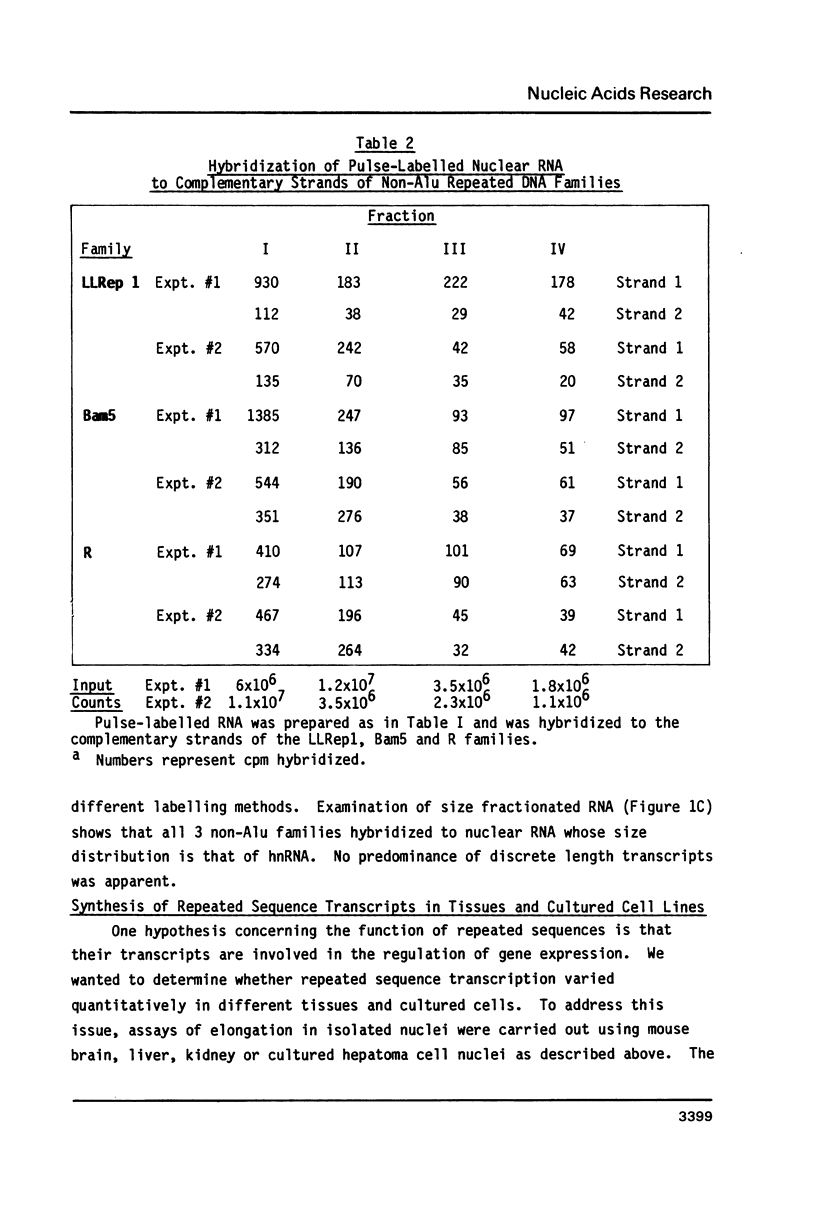
We have carried out transcriptional measurements on several families of repeated sequences to define their expression in mouse cells. The majority of Alu family transcripts result from read-through from adjacent structural gene promoters while 20% are discrete RNA polymerase III products. Alu repeat members show preferential orientation within RNA polymerase II transcription units as evidenced by asymmetric representation of the complementary strands of the Alu family in hnRNA. We assessed whether 3 non-Alu repeated sequence families had their own promoters by strand symmetry measurements and size distribution analysis of repeat-homologous newly synthesized nuclear RNA. Transcription homologous to the R family is totally symmetric and is likely due to read-through from adjacent structural gene promoters. LLRep1 and Bam5 repeats, in contrast, exhibit consistent strand asymmetry which is suggestive that at least some members may be transcribed by their own promoters. Among 3 mouse tissues and 1 cultured cell line analyzed, no quantitative variation in the expression of any of these sequences was observed.
Full text
PDF



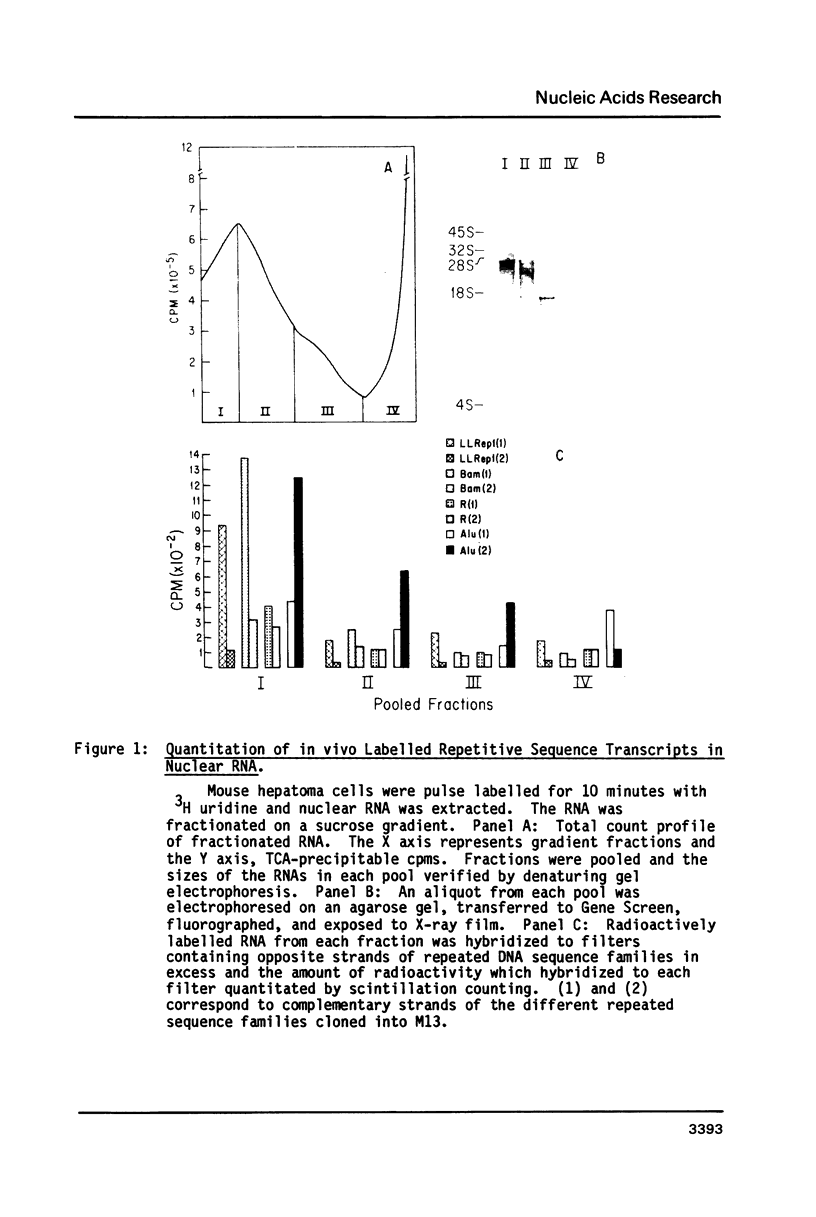

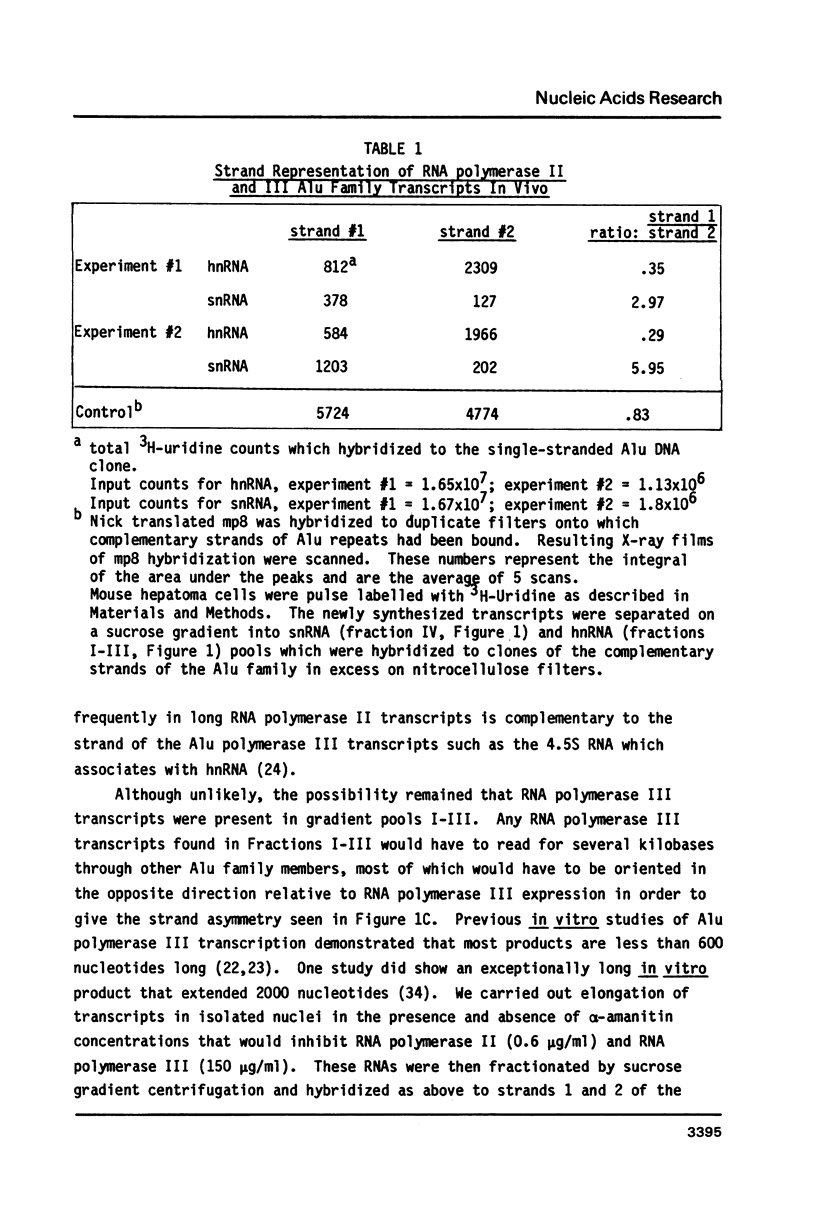






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. W., Kaufman R. E., Kretschmer P. J., Harrison M., Nienhuis A. W. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980 Dec 20;8(24):6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Maxson R., Kedes L. H. Histone gene expression during sea urchin embryogenesis: isolation and characterization of early and late messenger RNAs of Strongylocentrotus purpuratus by gene-specific hybridization and template activity. Dev Biol. 1979 Nov;73(1):153–173. doi: 10.1016/0012-1606(79)90144-1. [DOI] [PubMed] [Google Scholar]
- Costantini F. D., Scheller R. H., Britten R. J., Davidson E. H. Repetitive sequence transcripts in the mature sea urchin oocyte. Cell. 1978 Sep;15(1):173–187. doi: 10.1016/0092-8674(78)90093-4. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Duncan C., Biro P. A., Choudary P. V., Elder J. T., Wang R. R., Forget B. G., de Riel J. K., Weissman S. M. RNA polymerase III transcriptional units are interspersed among human non-alpha-globin genes. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5095–5099. doi: 10.1073/pnas.76.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Characterization of a highly repetitive family of DNA sequences in the mouse. Nucleic Acids Res. 1982 Aug 25;10(16):5003–5013. doi: 10.1093/nar/10.16.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard W., Meitinger T., Höchtl J., Zachau H. G. A new family of interspersed repetitive DNA sequences in the mouse genome. J Mol Biol. 1982 May 25;157(3):453–471. doi: 10.1016/0022-2836(82)90471-5. [DOI] [PubMed] [Google Scholar]
- Groudine M., Peretz M., Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981 Mar;1(3):281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Kato N. Nucleotide sequences of 4.5S RNAs associated with poly(A)-containing RNAs of mouse and hamster cells. Nucleic Acids Res. 1980 Mar 25;8(6):1273–1285. doi: 10.1093/nar/8.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Jelinek W. R. Low molecular weight RNAs transcribed in vitro by RNA polymerase III from Alu-type dispersed repeats in Chinese hamster DNA are also found in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6130–6134. doi: 10.1073/pnas.78.10.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller D., Jackson M., Leinwand L. Organization and expression of non-Alu family interspersed repetitive DNA sequences in the mouse genome. J Mol Biol. 1984 Mar 15;173(4):419–436. doi: 10.1016/0022-2836(84)90389-9. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R. Specific nucleotide sequences in HeLa cell inverted repeated DNA: enrichment for sequences found in double-stranded regions of heterogeneous nuclear RNA. J Mol Biol. 1977 Oct 5;115(4):591–601. doi: 10.1016/0022-2836(77)90104-8. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Evans R., Wilson M., Salditt-Georgieff M., Darnell J. E. Oligonucleotides in heterogeneous nuclear RNA: similarity of inverted repeats and RNA from repetitious DNA sites. Biochemistry. 1978 Jul 11;17(14):2776–2783. doi: 10.1021/bi00607a012. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Leinwand L. Low molecular weight RNAs hydrogen-bonded to nuclear and cytoplasmic poly(A)-terminated RNA from cultured Chinese hamster ovary cells. Cell. 1978 Sep;15(1):205–214. doi: 10.1016/0092-8674(78)90095-8. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. A family of short, interspersed repeat sequences at the 5' end of a set of Dictyostelium single-copy mRNAs. Cell. 1979 Apr;16(4):787–796. doi: 10.1016/0092-8674(79)90094-1. [DOI] [PubMed] [Google Scholar]
- Klein W. H., Thomas T. L., Lai C., Scheller R. H., Britten R. J., Davidson E. H. Characteristics of individual repetitive sequence families in the sea urchin genome studied with cloned repeats. Cell. 1978 Aug;14(4):889–900. doi: 10.1016/0092-8674(78)90344-6. [DOI] [PubMed] [Google Scholar]
- Laird C. D. Chromatid structure: relationship between DNA content and nucleotide sequence diversity. Chromosoma. 1971 Mar 16;32(4):378–406. doi: 10.1007/BF00285251. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Colozzo M. T. Synthesis in vitro of an exceptionally long RNA transcript promoted by an AluI sequence. Nature. 1982 Nov 25;300(5890):376–379. doi: 10.1038/300376a0. [DOI] [PubMed] [Google Scholar]
- Meunier-Rotival M., Soriano P., Cuny G., Strauss F., Bernardi G. Sequence organization and genomic distribution of the major family of interspersed repeats of mouse DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):355–359. doi: 10.1073/pnas.79.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolella G., Lucero M. A., Murphy M. H., Baralle F. E. The Alu family repeat promoter has a tRNA-like bipartite structure. EMBO J. 1983;2(5):691–696. doi: 10.1002/j.1460-2075.1983.tb01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. R., Westphal K. H., Rigby P. W. Activation of mouse genes in transformed cells. Cell. 1983 Sep;34(2):557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Maio J. J., Brown F. L. KpnI families of long, interspersed repetitive DNAs in human and other primate genomes. Nucleic Acids Res. 1982 May 25;10(10):3175–3193. doi: 10.1093/nar/10.10.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Paulson K. E., Schmid C. W., Kadyk L., Leinwand L. Non-Alu family interspersed repeats in human DNA and their transcriptional activity. Nucleic Acids Res. 1984 Mar 26;12(6):2669–2690. doi: 10.1093/nar/12.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Bloom F. E., Lerner R. A. Common 82-nucleotide sequence unique to brain RNA. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4942–4946. doi: 10.1073/pnas.79.16.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Gottesfeld J. M., Lerner R. A. Identifier sequences are transcribed specifically in brain. Nature. 1984 Mar 15;308(5956):237–241. doi: 10.1038/308237a0. [DOI] [PubMed] [Google Scholar]
- Van Arsdell S. W., Denison R. A., Bernstein L. B., Weiner A. M., Manser T., Gesteland R. F. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981 Oct;26(1 Pt 1):11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Voliva C. F., Jahn C. L., Comer M. B., Hutchison C. A., 3rd, Edgell M. H. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res. 1983 Dec 20;11(24):8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]