Abstract
Cell-penetrating peptides (CPPs) are routinely used for intracellular delivery of a variety of cargo, including drugs, genes, and short interfering RNA (siRNA). Most CPPs are active only upon exposure to acidic environments inside of late endosomes, thereby facilitating the endosomal escape of internalized vectors. Here, we describe the generation of a synthetic polypeptide—PVBLGn-8—that is able to adopt a helical structure independent of pH. Like other CPPs, the helical structure of PVBLGn-8 allows the polypeptide to destabilize membranes. However, since the helix is stable at all physiologically relevant pH values between pH 2 and pH 7.4, the membrane permeation properties of PVBLGn-8 are irreversible. Given its pH-insensitive activity, our results suggest that PVBLGn-8 is able to facilitate efficient siRNA delivery by causing pore formation in the cell membranes through which either free or complexed siRNA is able to diffuse. This nonspecific form of entry into the cell cytosol may prove useful when trying to deliver siRNA to cells which have proven to be difficult to transfect.
Introduction
The past 20 years have witnessed intense interest in the treatment of human disease using genetic information. For a large portion of the time, the focus was on using DNA to treat disease—so called gene therapy. According to the principles of gene therapy, the genetic information added would replace an absent gene, supplement an existing gene that was inadequately produced or impart a completely new functionality to cells. Unfortunately, the clinical translation of gene therapy has been wrought with concerns over its safety and thus remains an elusive goal.1,2 As an alternative to DNA-based therapies, the pioneering work of Fire et al. in the field of RNA interference has spurred tremendous interest in the treatment of disease using short interfering RNA (siRNA).3 Counter to gene therapy's attempts to treat disease through the addition of a gene, RNA interference attempts to treat disease by silencing the expression of a gene in a very precise and specific fashion. Several reports have revealed that RNA interference can be applied to inhibit targets implicated in a variety of diseases ranging from cancer to viral infections to genetic disorders.4,5 However, as of yet, the successful clinical application of RNA interference has been hampered by the lack of an efficient means to deliver siRNA to cells.6,7
Owing to its size, hydrophilicity and negative charge, siRNA delivery to mammalian cells must be achieved through the use of a carrier system. A variety of materials have been explored as RNA delivery vectors, including lipids, polymers, proteins/antibodies, peptides, aptamers, and even small molecules.8,9,10,11,12,13,14,15 Furthermore, both covalent and noncovalent strategies have been used to drive the association of siRNA with the carrier system. Akinc et al., for example, used the electrostatic attraction between the anionic phosphate backbone of siRNA and cationic lipid-like materials to drive the formation of siRNA-containing lipid nanoparticles.8 Alternatively, Rozema et al. covalently conjugated siRNA to an endosomolytic polymer (PBAVE) via disulfide linkages to enable siRNA interaction with the delivery vector.16 Regardless of whether the association of siRNA with its delivery vehicle occurs via covalent or noncovalent means, the vector serves to prolong siRNA circulation in the blood by increasing the siRNA complex diameter from ~2.5 nm to >5 nm.17 With a larger size and effective molecular weight, complexed siRNA is not subject to renal filtration and rapid clearance from the circulatory system. As an added benefit, association with a delivery vehicle can also provide protection from nucleolytic degradation, even if the siRNA is not fully encapsulated in the vehicle and instead is exposed to the surrounding environment.18
Cell-penetrating peptides (CPPs) have been used previously to improve intracellular siRNA delivery efficiency. For example, penetratin covalently conjugated to siRNA has been shown to effectively silence the expression of luciferase, green fluorescent protein, superoxide dismutase 1 and other genes.19,20 Likewise, noncovalent interactions between siRNA and oligoargine-cholesterol conjugates have been used to form nanoparticles for vascular endothelial growth factor knockdown.21 In both of these examples, the inclusion of CPPs is intended to promote the endosomal escape of internalized siRNA-delivery vehicles. One limitation to the use of CPPs like penetratin and oligoarginine, however, is that their activity is dependent upon the formation of an acidic pH environment.22,23 Thus, if cargo is internalized via a process that avoids rapid acidification (i.e., caveolae-mediated update) or is trafficked outside of the endolysosomal system, the CPPs will be ineffective. It has recently been suggested that internalization routes which avoid rapid acidification may result in more efficient delivery of nucleic acids as compared with processes associated with the endolysosomal pathway.24 For example, efficient siRNA has also been speculated to use uptake methods which fall outside of traditional caveolae- and clathrin-mediated endocytosis.25 All told, decoupling membrane lytic potential from pH may provide a useful enhancement to the intracellular delivery of siRNA with CPPs.
We recently described the synthesis of cationic helical polypeptide derivatives of polyglutamic acid with unprecedented helical stability under a variety of ionic, pH, and temperature conditions.26 Through slight modification of the synthetic procedure, we were able to generate a small library of these cationic helical polypeptides differing only in the hydrophobic/hydrophilic balance of the groups appended to the helical polypeptide backbone (Figure 1a).27 Library screening revealed that one particular polymer—so-called PVBLG267-8—possessed the ability to act as an effective delivery agent for plasmid DNA in a variety of cell lines (Figure 1b). A mechanistic examination using helical PVBLG267-8 and its random coil analogue revealed that DNA transfection efficiency was strongly linked to the formation of an α-helix by PVBLG267-8. Helical PVBLG267-8 was able to act as a membrane disruptive agent while the random coil version was not. Unlike CPPs such as penetratin and oligoarginine, the stable helix of PVBLG267-8 allowed it to retain its membrane disruptive properties regardless of pH environment. As escape from endocytic vesicles is believed to be one of the limiting factors of siRNA delivery, we hoped that decoupling membrane activity from pH would allow analogues of PVBLG267-8 to function as an effective siRNA-delivery agent.28
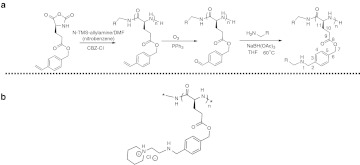
Figure 1.
Reaction scheme for PVBLGn-X synthesis. (a) Scheme for the generation of PVBLGn-X polymers. (b) Chemical structure of PVBLGn-8.
Results
Design and synthesis of PVBLGn-8 polymer
The ring-opening polymerization of γ-(4-vinylbenzyl)-l-glutamate N-carboxyanhydride (VB-Glu-NCA) was used to form poly(γ-(4-vinylbenzyl)-L-glutamate) (PVBLG). With its vinyl group, PVBLG served as a reactive template that, through subsequent ozonation, hydroamination and reduction, allowed the generation of a small library of cationic polypeptides (PVBLGn-X, where n is the degree of polymerization and X refers to the grafted amine). Due to its glutamate residues, PVBLG has a propensity to adopt an α-helical secondary structure in water. Typically, cationic polypeptides such as poly-L-lysine are unable to adopt helical conformations at physiological pH due to side chain charge disruption. However, we recently reported that the helical structure of cationic PVBLG polypeptides can be stabilized by maintaining a minimum separation distance of 11 σ-bonds between the polypeptide backbone and side chain charge (Figure 1a).26 The PVBLGn-X polymers synthesized for this study have their amine charge exactly 11 σ-bonds from the backbone, resulting in a helix structure that is stable over a broad range of pH values and salt concentrations for a wide variety of side chains and polypeptide molecular weights (Figure 2a). 26,27
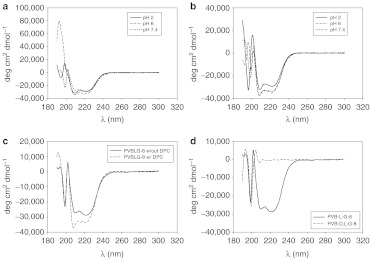
Figure 2.
PVBLGn-8 adopts a pH-independent helical conformation. (a) Circular dichroism spectra of PVBLG267-8 in water at pH 2, 6, or 7.4 buffer. (b) Circular dichroism spectra of PVBLG267-8 in water at pH 2, 6, or 7.4 along with lipid dodecylphosphcholine (DPC) membranes. (c) Circular dichroism spectra of PVBLG267-8 in water at pH 7.4 with and without lipid DPC membranes. (d) Circular dichroism spectra of helical PVBLG267-8 and random coil PVBDLG150-8 in water.
In a previous study, we used the reaction scheme described above to generate over 30 unique cationic helical polypeptides which differed only in the structure of the pendant amines.27 It was hoped that the inclusion of a variety of amines with unique hydrophobic/hydrophilic balances would yield a material which had the appropriate DNA binding strength (hydrophilic characteristic) and membrane disruption strength (hydrophobic characteristic) to yield efficient DNA delivery. Library screening yielded one particular PVBLG derivative—so-called PVBLGn-8 with an aminoethyl piperidine side chain—that outperformed 25-kDa polyethylenimine in cells generally amenable to transfection (i.e., COS-7, HEK293) and outperformed lipofectamine 2000 in traditionally hard-to-transfect H9 human embryonic stem cells.27 Using the same synthetic protocol as before, PVBLGn-8 was synthesized at a variety of molecular weights (n = 100, 150, and 267). Circular dichroism (CD) confirmed that PVBLG267-8 possessed a helical structure at pH 2, 6, and 7.4 both in the presence and absence of lipid membranes composed of dodecylphosphocholine (DPC) (Figure 2a,b). In fact, further characterization revealed that the helix is actually stabilized—indicated by a decrease in the measured ellipticity—by interaction with DPC lipid membranes (Figure 2c). Previous characterization studies dealing with PVBLGn-X indicate that the trend of stable helices is to be expected for all of the degrees of polymerization examined in this study.26,27 In addition to helical materials, an analogue of PVBLGn-8 with disrupted conformation was synthesized using DL-VB-Glu-NCA monomers. CD confirmed that the racemic configuration of amino acids (1:1 ratio) prevented the formation of a helical secondary structure in the resulting PVBDLG150-8 polymer (Figure 2d).
Interaction of PVBLGn-8 with siRNA
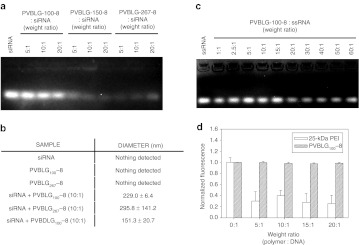
The interaction of PVBLGn-8 with siRNA was evaluated for complex size, stability, and protection against nucleolytic degradation. An agarose gel retardation assay revealed that PVBLG100-8 was largely unable to form complexes that were stable against the electrophoretic force applied during electrophoresis even at polymer:siRNA weight ratios as high as 20:1 (Figure 3a). The higher molecular weight polymers PVBLG150-8 and PVBLG267-8, however, formed complexes which were better able to remain intact under the applied voltage, as evidenced by the decreased intensity of the bands. With more amines per molecule, PVBLG150-8 and PVBLG267-8 have an increased binding strength that seemingly is able to resist complex disruption during electrophoresis, thus explaining their reduced fluorescence as compared to PVBLG100-8. The inability of PVBLG100-8 to form stable complexes was surprising as the very same polymer was able to prevent the migration of plasmid DNA on an agarose gel at ratios lower than those tested here (data not shown). To explore if complexes—albeit weakly bound—were indeed being formed between PVBLG100-8 and siRNA, dynamic light scattering (DLS) was used to probe for the formation of measurable particles when the polymer and siRNA were mixed. The results of Figure 3b indicate that complexes are indeed being formed with PVBLG100-8 as well as PVBLG267-8 and siRNA. Neither siRNA, PVBLG100-8, or PVBLG267-8 produced detectable particles when measured independently. However, when mixed at a 10:1 polymer:siRNA weight ratio, complexes of ~230 and 296 nm were detected. Complexes formed between PVBDLG100-8 and siRNA were substantially smaller with a measured diameter of ~151 nm.
Figure 3.
PVBLGn-8 forms complexes with short interfering RNA (siRNA). (a) Agarose gel electrophoresis of PVBLGn-8:siRNA complexes formed at a variety of PVBLGn-8:siRNA weight ratios (listed above lanes). (b) Dynamic light scattering data of aqueous solutions of free siRNA, free PVBLGn-8, or siRNA mixed with PVBLGn-8 or PVBDLG100-8. (c) Agarose gel electrophoresis of PVBLG100-8:ssRNA (single-stranded RNA) complexes formed at a variety of PVBLG100-8:ssRNA weight ratios (listed above lanes). (d) Normalized fluorescence of ethidium bromide mixed with polymer:siRNA complexes formed with 25-kDa polyethylenimine (PEI) or PVBLG100-8 at a variety of weight ratios.
To explore the possibility that the helical secondary structure of the polymer as well as the short persistence length of siRNA contribute to the weak binding, an agarose gel retardation assay was performed using PVBLG100-8 and flexible single-stranded RNA (ssRNA).29 Somewhat surprisingly, siRNA retardation was not observed for polymer:single-stranded RNA weight ratios as high as 50:1 (Figure 3c). While our DLS data still suggests that particles are formed between PVBLG100-8 and siRNA, it would seem that the materials are inherently weak binding regardless of secondary structure.
An in situ ethidium bromide exclusion assay performed using PVBLG100-8 also did not reveal any siRNA condensation with PVBLG100-8:siRNA weight ratios as high as 20:1 (Figure 3d). By comparison, the addition of 25-kDa branched polyethylenimine showed a 70% drop in ethidium bromide fluorescence at polymer:siRNA weight ratios as low as 5:1. Since DLS data indicates that siRNA and PVBLG100-8 do in fact form measurable complexes, it is likely that siRNA is at least partially exposed in the complexes, thereby allowing free ethidium bromide to interact with the siRNA.
Protection against nucleolytic degradation
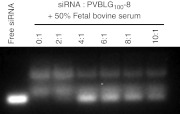
Since siRNA has only weak interaction with PVBLG100-8, we were concerned with the ability of PVBLG100-8 to adequately protect siRNA from nucleolytic degradation. Thus, complexes were formed between TAMRA-labeled siRNA and PVBLG100-8 at a variety of polymer:siRNA weight ratios between 0:1 and 10:1. The complexes were then incubated for 30 minutes at 37 °C in the presence of 50% fetal bovine serum—a source of digestive RNase enzymes—and run on an agarose gel. Since PVBLG100-8 was unable to retard the migration of siRNA under agarose gel electrophoresis (Figure 3a), we were able to visualize the siRNA without performing any additional steps to de-complex the siRNA and polymer. The results of Figure 4 reveal that PVBLG100-8 is unable to completely prevent the nucleolytic degradation of siRNA. Nonetheless, it does appear to retard siRNA degradation. This is evidenced by the stronger intact siRNA band seen at polymer:siRNA weight ratios of 4:1 and higher. We are unable to discern if the siRNA:polymer complex is disrupted by the presence of excess anionic serum proteins prior to partial degradation or if the siRNA is partially digested while it is still bound to PVBLG100-8.
Figure 4.
PVBLGn-8 provides protection against nucleolytic degradation. Agarose gel electrophoresis of PVBLG100-8:siRNA-TAMRA complexes formed at a variety of PVBLG100-8:siRNA-TAMRA weight ratios following a 30-minute incubation with fetal bovine serum (50% final concentration). siRNA, short interfering RNA.
siRNA delivery with helical PVBLGn-8
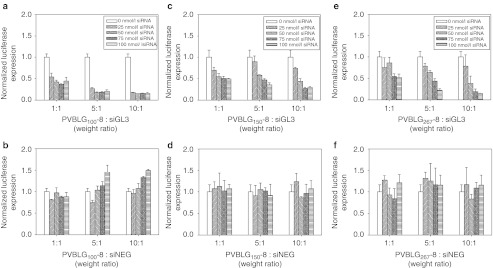
Since even the lowest molecular weight PVBLGn-8 polypeptide appeared to form complexes with siRNA that were somewhat resistant to nucleolytic degradation, we investigated the ability of PVBLGn-8 to mediate luciferase knockdown in HeLa cells stably producing the GL3 luciferase gene (HeLa-Luc). PVBLGn-8 polymers with degrees of polymerization of 100, 150, and 267 were used to form complexes with siRNA specific to the GL3 gene (siGL3) at polymer:siRNA weight ratios of 1:1, 5:1, and 10:1. These complexes were then incubated with HeLa-Luc cells at siRNA concentrations of 25, 50, 75, and 100 nmol/l. As a control, identical experiments were conducted using siRNA that did not interfere with the expression of any known gene products (siNEG). The results can be seen in Figure 5. PVBLGn-8 with degrees of polymerization of 100, 150 and 267 were all capable of mediating successful siGL3 delivery to HeLa-Luc cells. Experiments with siNEG verified that the observed luciferase knockdown was specific and not due to material toxicity. Although PVBLG100-8, PVBLG150-8 and PVBLG267-8 were capable of achieving similar luciferase knockdown levels of ~70–80%, PVBLG100-8 did so at lower siRNA concentrations—82% knockdown with 25 nmol/l siGL3 at a 10:1 polymer:siRNA weight ratio as compared to 25 and 20% with PVBLG150-8 and PVBLG267-8 at identical concentrations and weight ratios. The dominant performance of PVBLG100-8 was surprising as PVBLG100-8 appeared to form the weakest complexes with siRNA as measured by agarose gel electrophoresis (Figure 3a). The data also indirectly confirms that, although unable to condense siRNA on an agarose gel, PVBLG100-8 is capable of effectively protecting siRNA against nucleolytic degradation in cell culture experiments.
Figure 5.
In vitro transfection of HeLa-Luc cells with PVBLGn-8. In vitro transfection of HeLa-luc cells with (a,c,e) luciferase siGL3 or negative control (b,d,f) siNEG at a variety of polymer:siRNA weight ratios and siRNA concentrations. siRNA, short interfering RNA.
Membrane permeation and toxicity of PVBLGn-8
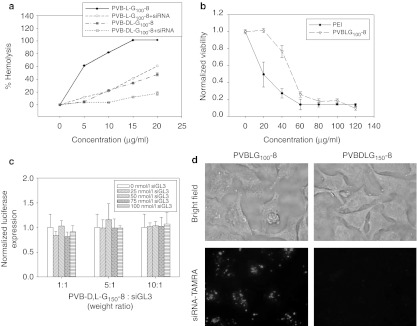
Since the PVBLGn-8 polymers were designed to have an α-helical architecture similar to that found in peptides capable of disrupting membranes, we examined the ability of PVBLG100-8 to cause pore formation in cell membranes. The leakage of hemoglobin—measured as the absorbance of 530 nm light—from murine erythrocytes incubated with various concentrations of free and complexed PVBLG100-8 was used to evaluate the membrane disruptive ability of the polymer. Similar experiments also were performed with PVBDLG100-8, the analogue of helical PVBLG-8 that has disrupted conformation. In all cases, the absorbance readings were normalized to the absorbance of cells which were completely lysed through incubation with the surfactant Triton X-100. The results of the erythrocyte leakage assay can be seen in Figure 6a. Whereas both PVBLG100-8 and PVBDLG100-8 result in membrane permeation, the helical polypeptide does so more efficiently. A concentration of 10 µg/ml of the helical polypeptide results in 80% hemoglobin leakage while only 20% of the hemoglobin is released using the same concentration of the random coil polymer. The partial membrane permeation observed with the random coil polypeptide could be due, in part, to the general toxicity of polycationic materials as well as the formation of small helical domains within the polypeptide. Complexation with siRNA generally reduces the membrane disrupting potential of both PVBLG100-8 and PVBDLG100-8. In the case of PVBLG100-8, complexation reduces hemoglobin leakage by 40–80% (Figure 6b). This may be due to charge neutralization of the PVBLG100-8 by the siRNA which would act to prevent electrostatic attraction to the cell membrane. Despite its hemolytic potential, it should be noted that PVBLG100-8 shows less toxicity than the commonly used polycationic material 25-kDa polyethylenimine (Figure 6b).
Figure 6.
Helical PVBLGn-8 disrupts membranes more efficiently than random coil PVBDLGn-8. (a) Hemoglobin leakage from murine erythrocytes incubated with free or complexed helical PVBLG100-8 or random coil PVBDLG100-8 at a variety of concentrations. (b) In vitro toxicity of PVBLG100-8 and 25-kDa polyethylenimine (PEI) in HeLa-Luc cells as determined by MTT. (c) In vitro transfection of HeLa-Luc cells with luciferase siGL3 using random coil PVBDLG150-8 at a variety of polymer:siRNA weight ratios and short interfering RNA (siRNA) concentrations. (c) Fluorescence microscopy of HeLa-Luc cells incubated with complexes formed with TAMRA-labeled siRNA and either helical PVBLG100-8 or random coil PVBDLG150-8. In both cases, the final siRNA concentration was 100 nmol/l and the polymer:siRNA weight ratio used was 10:1.
siRNA delivery by PVBDLGn-8 with disrupted conformation
To determine whether a helical structure—and thus an enhanced ability to permeabilize cell membranes—is necessary for effective siRNA delivery with PVBLGn-8, transfections were performed using PVBDLG150-8, a polypeptide that has identical chemical structure as PVBLGn-8 but does not adopt helical conformation. When PVBDLG150-8 was mixed with siRNA specific to the luciferase gene and administered to HeLa-Luc cells, no luciferase knockdown was detected 24 hours post-transfection (Figure 6c). This suggests that vector helicity is an essential characteristic for effective siRNA delivery with PVBLGn-8. To determine whether the difference in performance between the helical and random coil polymer was due to intracellular performance or cell entry, uptake of complexes formed between TAMRA-labeled siRNA and either PVBLG100-8 or PVBDLG150-8 was measured by fluorescence microscopy. The results of Figure 6d reveal that complexes formed between PVBDLG150-8 and siRNA-TAMRA do not enter HeLa-Luc cells. Thus, it would seem that PVBLG100-8, with its helix-associated membrane permeation properties, is facilitating direct membrane transduction of siRNA. Part of the absence of internalization with PVBDLG150-8 complexes may be due to the inability of PVBDLG150-8 to form stable complexes with siRNA (data not shown). Although PVBLG100-8 is also unable to form very stable complexes, it is able to translate extracellularly displaced polymer into membrane disruption. In the case of PVBDLG150-8, the displaced polymer has greatly reduced membrane lytic potential and is unable to create pores for siRNA migration into the cell cytosol.
Exploration of direct membrane transduction of siRNA/PVBLG100-8 complexes
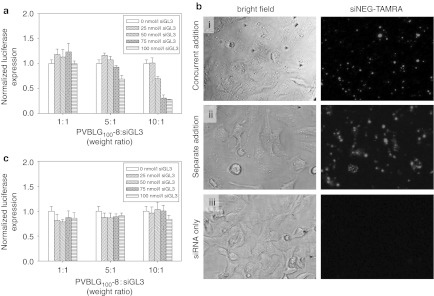
Because PVBLG100-8 appeared to facilitate direct membrane transduction of siRNA, we examined siRNA uptake under various scenarios. We considered the possibility of polymer/siRNA complexes forming in situ prior to cell uptake as well as the possibility of free polymer permeabilizing the cell to allow siRNA diffusion into the cytosol. To explore in situ complexation, free PVBLG100-8 and free siGL3 were added simultaneously to HeLa-Luc cells at appropriate concentrations to yield 1:1, 5:1, and 10:1 polymer:siRNA (weight ratio) complexes at siRNA concentrations from 0 nmol/l to 100 nmol/l siRNA. The results in Figure 7a reveal that complexation before addition to the cell media is not necessary to deliver functional siRNA. It should be noted, however, that a higher siRNA dose (100 nmol/l siGL3) was necessary to achieve the same 80% luciferase knockdown as when the polymer and siRNA were mixed before adding to cells. An analogous experiment performed using TAMRA-labeled siNEG added concurrently with suitable PVBLG100-8 to achieve at 10:1 weight ratio confirmed uptake in HeLa-Luc cells (Figure 7b, concurrent addition). By comparison, the addition of free fluorescent siRNA to the cell media at 100 nmol/l did not result in any observable uptake (Figure 7b, siRNA only). As PVBLG100-8 was demonstrated to destabilize membranes, we also investigated the possibility that the polymer was causing pore formation in cell membranes through which free and uncomplexed siRNA was able to diffuse into the cell cytosol. To examine this, free PVBLG100-8 was added to the cell media at concentrations corresponding to the formation of complexes with polymer:siRNA weight ratios of 1:1, 5:1, and 10:1. After incubation at 37 °C for 1 hour, the cell media was aspirated and the cells were washed with phosphate-buffered saline (PBS). Free siRNA was then added at concentrations between 0 nmol/l and 100 nmol/l. After 24 hours, the cells were lysed and analyzed for luciferase knockdown. Unfortunately, no luciferase knockdown was detected (Figure 7c). However, when an analogous experiment was performed using TAMRA-labeled siNEG, it was clear that siRNA was able to diffuse into the permeabilized cells (Figure 7b, Separate Addition). Thus, it would seem that PVBLG100-8 can facilitate cell entry of siRNA by porating membranes. However, because siRNA which had diffused into the cell did not mediate any gene knockdown, it also seems that PVBLG100-8 performs an additional function in the cytosol, possibly including protecting the siRNA from degradation within the cell.
Figure 7.
Free short interfering RNA (siRNA) diffuses into HeLa-Luc cells permeabilized with PVBLGn-8. (a) In vitro transfection of HeLa-Luc cells with in situ formed complexes of PVBLG100-8 and luciferase siGL3 at a variety of polymer:siRNA weight ratios and siRNA concentrations. (b) Fluorescence microscopy of HeLa-Luc incubated with complexes formed with TAMRA-labeled siRNA and PVBLG100-8. Free polymer and free siRNA were added sequentially in the pane i. Free polymer was added, incubated and removed prior to addition of free siRNA in pane ii. Free siRNA was incubated without any poymer in pane iii. (c) In vitro transfection of HeLa-Luc cells with free luciferase siGL3 following incubation and removal of free PVBLG100-8 at a variety of effective polymer:siRNA weight ratios and siRNA concentrations.
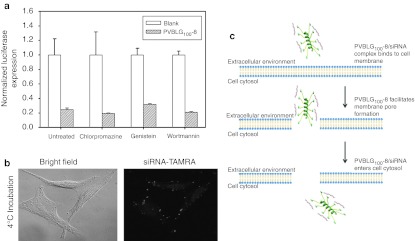
To better understand the cellular uptake mechanism of PVBLG100-8:siRNA particles, transfections were performed on cells which had been pretreated with small-molecule drugs capable of inhibiting various endocytic pathways. HeLa-Luc cells were incubated with 5 µg/ml of chlorpromazine, 50 µg/ml of genistein, or 100 nmol/l wortmannin for 30 minutes prior to PVBLG100-8:siRNA nanoparticle addition to inhibit clathrin-mediated, caveolae-mediated or macropinocytic uptake, respectively.30 After 4 hours, the cells were washed and the growth media was replaced. The cells were assayed for luciferase expression 24 hours post-transfection. Effective drug concentrations were based on previous studies and confirmed by the inhibition of uptake of folate-labeled beads (caveolae-mediated endocytosis) and free transferrin (clathrin-mediated endocytosis) as measured by flow cytometry (data not shown).24 The results of Figure 8a show that none of the inhibitor drugs have any effect on the performance of PVBLG100-8:siRNA complexes. Fluorescence microscopy also was used to examine the uptake of complexes formed between PVBLG100-8 and TAMRA-labeled siRNA when the transfection was conducted at 4 °C. Under low temperature conditions, active forms of endocytosis—including caveolae- and clathrin-mediated endocytosis as well as macropinocytosis—are inhibited. The results of Figure 8b clearly show that TAMRA-labeled siRNA is internalized even at low temperatures. Combined with the drug inhibition data, this suggests that siRNA delivery with PVBLG100-8 is independent of all three common types of endocytosis and thus supports the previous observation that PVBLG100-8 penetrates cell membranes to allow the diffusion of either free or complexed siRNA into the cell cytosol.
Figure 8.
Modes of PVBLG100-8:siRNA complex uptake. (a) In vitro transfection of HeLa-Luc cells with luciferase siGL3 using helical PVBLG100-8 in the presence or absence of endocytic inhibitor drugs. PVBLG100-8:siRNA complexes were formed at a 10:1 PVBLG100-8:siRNA weight ratio. Complexes between lipofetamine 2000 (LFA) and siRNA were formed at a 5:1 LFA:siRNA weight ratio. The final short interfering RNA (siRNA) concentration was 100 nmol/l in all cases. Combined the data indicates that PVBLG100-8:siRNA complexes do not use any of the traditional forms of cell uptake. (b) Fluorescence microscopy of HeLa-Luc cells transfected at 4 °C with complexes formed with TAMRA-labeled siRNA and helical PVBLG100-8 to demonstrate uptake even in the absence of active endocytosis. (c) In a proposed model for PVBLG100-8:siRNA complex entry into cells, small siRNA molecules are first weakly bound to the larger and cationic PVBLG100-8 which in turn is electrostatically attracted to anionic cell membranes. PVBLG100-8 then facilitates cell membrane disruption, allowing PVBLG100-8:siRNA complexes to diffuse into the cell cytosol.
Discussion
Shortly after its discovery and characterization, siRNA was considered for use as a therapeutic tool to treat a variety of diseases.4,5 Like other nucleic acid-based therapies, efficient delivery has been the greatest challenge to the clinical application of the technology. Although still challenging, successful siRNA delivery can be considered to pose fewer barriers than its DNA counterpart. Most notably, siRNA has a dramatically reduced size compared to plasmid DNA (~104 Da for siRNA versus ~106 Da for plasmid DNA). siRNA also needs only to be delivered to the cell cytosol while plasmid DNA must reach the nucleus of the cell, meaning a second membrane (i.e., the nuclear membrane) must be negotiated to realize successful DNA delivery. The added barriers to DNA delivery have proven difficult to overcome, suggesting that siRNA may be a more immediate nucleic acid-based therapy. Indeed, siRNA has already recently been shown to have benefits in clinical trials for cancer treatment.10,31
Whether applied in vivo or in vitro, successful siRNA delivery requires that cell and endosomal membranes be traversed. This aspect of delivery—particularly crossing endosomal membranes—has been considered to be one of the most challenging barriers preventing efficient siRNA delivery.28 With this in mind, the carrier vehicle described here was designed with two basic criteria in mind: (i) the ability to complex with anionic siRNA and (ii) the ability to efficiently and consistently penetrate biological membranes. Association between siRNA and its delivery vehicle can be accomplished by incorporating cationic entities into the vector. In regards to the second criterion, one such class of materials that has been demonstrated to effectively cross membranes is CPPs. A structural characteristic shared by a broad range of CPPs is the ability to form a helical secondary structure. For CPPs like KALA and GALA, the helix formed is facially amphiphilic.32,33 When acting to disrupt cell membranes, the hydrophilic face is believed to induce binding with negatively charged membranes while the hydrophobic face is believed to intermix with the lipid bilayer to cause pore formation. For many CPPs the conformational shift from random coil to helix is driven by a drop in pH.32,33 Considering that many common forms of cell uptake result in the acidification of endocytic vesicles, reliance on acidification to drive helix formation seems appropriate.24,34 However, it has recently been demonstrated that pathways which avoid acidification, namely caveolae-mediated uptake, yield efficient delivery of nucleic acids.24 Furthermore, it has been speculated that functional siRNA delivery may proceed by an internalization pathway that is distinct from traditional clathrin- and caveolae-mediated endocytosis.25 Thus, to permit cytosolic delivery regardless of uptake pathway, we sought to develop helical structures whose pore forming ability (i.e., helical structure) was decoupled from the surrounding pH environment.
We recently described the synthesis of cationic helical polypeptide derivatives of polyglutamic acid with unprecedented helical stability under a variety of ionic, pH and temperature conditions.26 By modifying the synthetic procedure, we were able to devise a parallel reaction scheme which allowed us to generate a small library of cationic helical polypeptides differing only in the hydrophobic/hydrophilic balance of the groups appended to the helical polypeptide backbone (Figure 1a). By varying the hydrophobic/hydrophilic balance in the pendant groups, we hoped to strike a balance—much like the amphiphilic balance in KALA and GALA—that would impart membrane disruption properties to the polypeptide. Library screening revealed that one particular constituent—PVBLG267-8—possessed the appropriate hydrophobic/hydrophilic balance in the side group to allow the molecule to act as an effective cell-penetrating material.27 With its ability to disrupt membranes, we demonstrated that PVBLG267-8 also was able to function as an effective delivery agent for plasmid DNA in a variety of cell lines. Due to the similar requirements of gene and siRNA delivery, we reasoned that polymers based on PVBLG267-8 could also function as effective siRNA-delivery vehicles.
PVBLGn-8 has a secondary and tertiary amine on its pendant group and thus possesses a net cationic charge. We previously demonstrated that PVBLGn-8 is capable of binding and condensing plasmid DNA.27 The affinity of PVBLGn-8 and DNA is due to their mutual electrostatic attraction. As siRNA possesses a negatively charged backbone akin to DNA, we anticipated that PVBLGn-8 would similarly bind and condense RNA. Surprisingly, our agarose gel shift assay showed incomplete siRNA condensation regardless of polymer molecular weight (Figure 3a). Condensation generally increased with molecular weight, but even PVBLG267-8—which possessed the highest cationic charge per molecule—failed to completely retard siRNA migration. Nonetheless, DLS revealed that complexes were indeed formed between even low molecular weight PVBLG100-8 and siRNA, although the binding was sufficiently weak to allow them to be disrupted under the application of an electrophoretic force. Despite its weak siRNA binding, even PVBLG100-8 was able provide some protection against nucleolytic degradation of siRNA (Figure 4).
Luciferase knockdown experiments revealed that PVBLGn-8 of all molecular weights tested was able to mediate up to 80% gene silencing in HeLa-Luc cells (Figure 5). Control experiments demonstrated the knockdown to be gene-specific and not due to associated material toxicity. Despite being the weakest siRNA binder, PVBLG100-8 surprisingly was more efficient at mediating gene knockdown than its larger molecular weight counterparts. In fact, trends indicated that as the ability to bind and condense siRNA increased, the efficiency of gene silencing dropped. Flow cytometry and fluorescent microscopy revealed uptake levels of complexes formed with siRNA and either PVBLG100-8 or PVBLG267-8 to be similar (data now shown). Thus, it would seem that reduced binding strength is beneficial for siRNA delivered using PVBLGn-8 materials.
With its helical secondary structure, PVBLGn-8 was designed to mimic the behavior of CPPs. As hoped, helical PVBLG100-8 possessed the ability to efficiently destabilize membranes while its helix-disrupted analogue, PVBDLG100-8, did not (Figure 6a). Helical structure was also shown to be essential for successful siRNA mediated knockdown (Figure 6c), implying that membrane permeation is an essential component of PVBLGn-8 mediated siRNA delivery. We expected this membrane permeation to aid endosomal escape, as it was revealed to do for plasmid DNA delivery.27 However, when examining uptake of siRNA complexes of helical PVBLG100-8 versus helix-disrupted PVBDLG150-8, we were surprised to see no siRNA uptake using the helix-disrupted polymer (Figure 6d). This suggested that membrane permeation was facilitating the passive diffusional entry of siRNA complexes into HeLa-Luc cells and not endosomal escape as we had expected. To examine whether siRNA could indeed diffuse across cell membranes that were permeabilized by PVBLGn-8, we separately incubated HeLa-Luc cells with free PVBLG100-8 and then, following polymer removal by washing with PBS, added free siRNA. While free siRNA is unable to enter untreated cells, it was able to enter into HeLa-Luc cells which were pretreated with PVBLG100-8 (Figure 7b). This suggests that free siRNA is able to diffuse across the cell membrane into the cell cytosol via membrane pores formed by PVBLG100-8. Despite successful cell uptake, free siRNA which had diffused across the cell membrane was unable to mediate knockdown (Figure 7c). However, when free siRNA and free PVBLG100-8 were simultaneously administered to cell media without any premixing, both siRNA uptake as well as specific gene silencing were observed (Figure 7a,b). The observation that coincubation with PVBLG100-8 is necessary for knockdown but not uptake suggests that PVBLG100-8 plays an additional role within the cell, including the possible protection of internalized siRNA from intracellular nucleolytic degradation.
To more thoroughly examine the possibility that PVBLG100-8 creates pores in cell membranes to permit the free diffusion of siRNA into the cell cytosol, we examined the gene knockdown performance of PVBLG100-8:siRNA complexes in cells which were treated with inhibitor drugs to prevent either clathrin, caveolae, or macropinocytic uptake. According to Figure 8a, none of the three drugs had a detrimental effect on the performance of PVBLG100-8 to mediate luciferase knockdown in HeLa-Luc cells. Similarly, uptake of TAMRA-labeled siRNA was confirmed even in cells which were transfected under low temperature conditions as a means to prevent active endocytosis (Figure 8b). Combined, the data further supports our notion that siRNA uptake with PVBLGn-8 occurs via diffusion across pores in the cell membrane caused by the helical polymer. This diffusional mechanism also may explain the decreased knockdown observed with the higher molecular weight PVBLG150-8 and PVBLG267-8 polymers. It is possible that the larger complexes formed between siRNA and PVBLG150-8 or PVBLG267-8 are too large to efficiently diffuse through the porated cell membrane while complexes of the shorter PVBLG100-8 and siRNA move through the cell pore more readily (Figure 8c).
In conclusion, we have presented a synthetic polypeptide modeled after natural CPPs for siRNA delivery. Unlike many other cell-penetrating materials, the helical structure—and by association membrane disruptive capabilities of the synthetic polypeptide—is not sensitive to local pH environment. This lack of sensitivity allows the polymer to disrupt cell membranes at the near-neutral pH of the extracellular environment. Our results suggest that, unlike most other materials for siRNA delivery, PVBLG100-8 operates by causing pore formation in the cell membrane through which siRNA is able to diffuse. This nonspecific and direct form of entry into the cell cytosol may prove useful when trying to deliver siRNA to cells which have proven to be difficult to transfect by more traditional materials.
Materials and Methods
Instrumentation. NMR spectra were recorded on a Varian UI400 MHz, a UI500NB MHz or a VXR-500 MHz spectrometer. Tandem gel permeation chromatography experiments were performed on a system equipped with an isocratic pump (Model 1100; Agilent Technology, Santa Clara, CA), a DAWN HELEOS 18-angle laser light scattering detector (also known as multi-angle laser light scattering detector; Wyatt Technology, Santa Barbara, CA) and an Optilab rEX refractive index detector (Wyatt Technology). The detection wavelength of HELEOS was set at 658 nm. Separations were performed using serially connected size exclusion columns (100 Å, 500 Å, 103 Å, and 104 Å Phenogel columns, 5 µm, 300 × 7.8 mm; Phenomenex, Torrance, CA) at 60 °C using DMF containing 0.1 mol/l LiBr as the mobile phase. The multiangle laser light scattering detector is calibrated using pure toluene with no need for external polymer standards and can be used for the determination of the absolute molecular weights. The molecular weights of all polymers were determined based on the dn/dc value of each sample calculated offline by using the internal calibration system processed by the ASTRA V software (version 5.1.7.3; Wyatt Technology). CD measurements were carried out on a JASCO J-700 or a JASCO 720 CD Spectrometer. Ozone was produced by an OZV-8S ozone generator manufactured by Ozone Solutions (Hull, IA). Lyophilization was performed on a FreeZone lyophilizer (Labconco, Kansas City, MO).
Synthesis of γ-(4-vinylbenzyl)-L-glutamate NCA (VB-Glu-NCA). γ-(4-Vinylbenzyl)-L-glutamate (VB-Glu) was synthesized through a modified procedure. VB (2.45 g, 10 mmol) was dried under vacuum for 2 hours. This solid was suspended in anhydrous tetrahydrofuran (30 ml). Phosgene (20% in toluene, 7 ml) was added dropwise over 5 minutes under nitrogen atmosphere. The suspension was stirred at 50 °C for 2–3 hours and the solvent was removed under reduced pressure. The residue was dissolved in anhydrous tetrahydrofuran in a glove box and centrifuged to remove the unreacted amino acid. The resulting supernatant was decanted and the solvent was removed under vacuum. The residue (containing a mixture of NCA was dissolved in tetrahydrofuran (10 ml) followed by addition of anhydrous ether (100 ml). The solution was cooled to –30 °C in a glove box and a dark oily residue was removed. The clear solution containing NCA was combined and concentrated. VB-Glu-NCA (3.4 mmol, 1.0 g, 34%) was obtained as a white crystal through recrystallization three times using tetrahydrofuran/hexane. 1H NMR (CDCl3, 500 MHz): δ7.40 (d, 2H, J = 8.0 Hz, ArH), 7.30 (d, 2H, J = 8 Hz, ArH), 6.73–6.67 (m, 2H, NH and C6H4CH=CH2), 5.76 (d, 1H, J = 17.5 Hz, C6H4CH = CH2), 5.27 (d, 1H, J = 11 Hz, C6H4CH=CH2), 5.11 (s, 2H, ArCH2), 4.38 (t, 1H, J = 6.0 Hz, CHCH2CH2COOCH2), 2.58 (t, 2H, CH2CH2COOCH2), 2.26 (m, 1H, CH2CH2COOCH2), 2.12 (m, 1H, CH2CH2COOCH2). 13C NMR (CDCl3, 500 MHz): δ172.4, 169.3, 151.9, 137.8, 136.1, 134.6, 128.6, 126.4, 114.6, 66.8, 56.9, 29.8, 26.8. ESI MS analysis (with NaCl) Calcd: m/z 289.2 (M); found: m/z 312.3 (M+Na). Anal. Calcd. For C15H15NO5: 62.29%C, 5.21%H, 4.84%N; found: 62.06%C, 5.12%H, 4.83%N.
General procedure for the polymerization of VB-Glu-NCA. In a glove box, VB-Glu-NCA (29 mg, 0.1 mmol) was dissolved in a mixture of DMF (1 ml) and nitrobenzene (50 µl) (nitrobenzene was used to inhibit potential radical reactions on the vinyl group). The VB-Glu-NCA solution was added to a DMF solution of N-TMS allylamine (20 µl, 0.1 mmol/ml) and stirred for 15 hours at room temperature. An aliquot of the polymerization solution was diluted to 10 mg PVBLG/ml using DMF (containing 0.1 mol/l LiBr) and analyzed by gel permeation chromatography. The Mn and MWD were assessed by gel permeation chromatography (Mn = 26,200 g/mol; MWD = 1.03). The remaining PVBLG was precipitated with ether (15 ml). The obtained PVBLG was sonicated for 5 minutes in ether and centrifuged to remove the solvent. After the sonication/centrifugation procedure was repeated twice more, PVBLG was collected and dried under vacuum. 1H NMR (TFA-d, 500 MHz): δ7.53 (d, 2H, J = 7.0 Hz, ArH), 7.39 (d, 2H, J = 7.0 Hz, ArH), 6.84 (dd, 1H, J1 = 11.0 Hz, J2 = 18.0 Hz C6H4CH=CH2), 5.91 (d, 1H, J = 18.0 Hz, C6H4CH=CH2), 5.43 (d, 1H, J = 11.0 Hz, C6H4CH=CH2), 5.26 (m, 2H, ArCH2), 4.80 (m, 1H, CHCH2CH2COOCH2), 2.68 (m, 2H, CHCH2CH2COO), 2.30 (m, 1H, CHCH2CH2COO), 2.12 (m, 1H, CHCH2CH2COO) (Supplementary Figure S1).
Synthesis of PVBLG-8. Poly(γ-(4-aldehydebenzyl-L-glutamate) (20 mg), 2-aminoethyl piperidine (3–5 molar equivalents relative to the Glu repeating unit) and the reducing agent NaBH (5–10 molar equivalents) were mixed in tetrahydrofuran (3 ml). The reaction mixtures were stirred at 50–60 °C for 24–72 hours. The mixture was then poured into 3 mol/l HCl (3 ml), followed by dialysis against water and lyophilization. Grafting efficiencies were analyzed by 1H NMR.
General procedure for the analysis of polymer by CD. CD experiments were performed on a JASCO J-700 or on a J-720 CD spectrometer. Polymer samples were prepared at concentrations of 0.05 mg/ml in DI water at pH 3 unless otherwise specified. The solution was placed in a quartz cell with light path of 0.2 cm. The mean residue molar ellipticity of each polymer was calculated based on the measured apparent ellipticity, the molar concentration of polymer, and the molecular weight of the repeating unit. Experiments were carried out at room temperature unless otherwise specified. For the experiments using DPC, liposomes were prepared by dissolving DPC in trifluoroethanol and then drying under a nitrogen stream. DPC liposomes were formed when rehydrated at 1 mg/ml. For the pH-dependency studies, the pH of the polymer solution was tuned by the addition of a concentrated HCl or NaOH solution.
Cells and siRNA. HeLa-Luc cells which stably produced the GL3 luciferase gene were a kind gift from the lab of Dr Daniel Pack at the University of Illinois, Urbana-Champaign. The cells were cultured according to standard ATCC protocols at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM). The growth medium was supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. siRNA specific to the GL3 luciferase gene (siGL3) was obtained from Dharmacon (Lafayette, CO). Negative control siRNA (siNEG) and tetramethylrhodamine-labeled negative control siRNA (siNEG-TAMRA or siRNA-TAMRA) was obtained from Qiagen (Valencia, CA).
Agarose gel retardation studies. A solution of siRNA or single-stranded RNA was mixed with a solution of PVBLGn-8 polypeptide at various concentrations in 150 mmol/l NaCl, 20 mmol/l HEPES at pH 7.2 to achieve the desired polymer:siRNA weight ratio. Complexes were then incubated at room temperature for 15 minutes, after which loading dye was added and the solution was run on a 1% agarose gel (70 V, 70 minutes). The resulting gel was stained with ethidium bromide and visualized on a Gel Doc imaging system (Biorad, Hercules, CA).
In situ ethidium bromide exclusion. A solution of siRNA was prepared in 150 mmol/l NaCl, 20 mmol/l HEPES at pH 7.2. Appropriate amounts of each polymer dissolved in double distilled water were added to the siRNA solution to achieve the desired weight ratio. Polyplexes were then incubated in wells of an opaque 96-well microplate for 15 minutes, after which ethidium bromide was added to each aliquot of polyplex solution. The polyplexes and ethidium bromide were then incubated at room temperature for another 10 minutes before exciting the polyplexes at 510 nm and reading the fluorescence at 595 nm using a PerkinElmer multilabel counter (Waltham, MA). The fluorescence values were normalized to wells containing only siRNA and ethidium bromide. Experiments were performed in triplicate.
Complex formation and transfection. HeLa-Luc cells were cultured in DMEM supplemented with 10% horse serum and 1% penicillin–streptomycin according to standard ATCC protocols and plated in 96-well plates at 1 × 104 cells/well 24 hours before transfection. Polymer:siRNA complexes were prepared at room temperature by dissolving siRNA in 150 mmol/l NaCl, 20 mmol/l HEPES and adding an equal volume of PVBLGn-8 in 150 mmol/l NaCl, 20 mmol/l HEPES to achieve the desired weight ratio. After incubation at room temperature for ~15 minutes, the complexes were diluted in serum-free DMEM to achieve the desired final siRNA concentrations. Immediately before siRNA transfection, the cells were washed with PBS and the solution of polymer:siRNA complexes was added. The HeLa-Luc cells and complexes were incubated at 37 °C for 4 hours, after which the cells were washed with PBS and the growth media was replaced. For low temperature studies, the cells and media were prechilled at 4 °C and the cells were incubated for 2 hours at 4 °C prior to sample fixation and visualization. For studies involving drug treatment (genistein at 50 µg/ml, chlorpromazine at 5 µg/ml, wortmannin at 100 nmol/l), complexes were diluted in serum-free DMEM containing the appropriate drug. Luciferase expression was quantified 24 hours post-transfection using the Promega Bright-Glo luciferase assay system (Madison, WI). Luciferase activity was measured in relative light units using a PerkinElmer multilabel counter with luminescence capabilities (Waltham, MA). To verify specific knockdown, identical experiments were conducted for both GL3 luciferase siRNA (siGL3) and well as negative control siRNA (siNEG)
DLS. Free siRNA, free PVBLGn-8 or PVBLGn-8:siRNA complexes were formed in 150 mmol/l NaCl, 20 mmol/l HEPES and incubated for 10 minutes before adding to doubled distilled water. The diluted materials were subjected to size measurement on a Brookhaven Instruments Corporation 90 Plus Particle Size Analyzer (Holtsville, NY). Five sets of measurements were performed for each sample.
Erythrocyte disruption assay. Red blood cells were collected from freshly killed C57BL6 mice and stored on ice. Following clot removal by decantation, the blood was diluted in PBS and mixed with various concentrations of free or complexed helical and random coil polymer. Following incubation at 37 °C for 1 hour, the intact red blood cells were pelleted via centrifugation and the supernatant was collected. The leakage of hemoglobin—measured as the supernatant absorbance of 530 nm light—was used to evaluate the membrane disruptive ability of the polymers (PerkinElmer). The background-subtracted absorbance readings were normalized to the absorbance readings of cells which had been incubated in the presence of the surfactant Triton X-100 to achieve complete lysis.
Toxicity. The cytotoxicity of PVBLG100-8 was characterized using the MTT cell viability assay (Sigma-Aldrich, St Louis, MO). Cells (HeLa-Luc) were seeded in 96-wells plates at 1 × 104 cells/well and grown overnight at 37 °C, 5% CO2 in medium containing 10% fetal bovine serum and 1% penicillin–streptomycin. Approximately 24 hours after seeding the medium was replaced with serum-free DMEM and the polymer was added to the cells at final concentrations between 0 and 120 µg/ml. After four hours of incubation, the medium was replaced with serum-containing medium and grown for another 20 hours, after which reconstituted 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT, 10 µl) was added. The plates were then incubated for another 4 hours and MTT solubilization solution (100 µl; Sigma-Aldrich) was added and the absorbance at 570 nm was read using a PerkinElmer plate reader (Waltham, MA). The background absorbance of cells killed with ethanol was subtracted from the viable cell absorbance and normalized to cells grown in DMEM. Each experiment was repeated four times at each concentration.
Nucleolytic digestion assay. Free siRNA-TAMRA or siRNA-TAMRA: PVBLGn-8 complexes were formed at various weight ratios in 150 mmol/l NaCl, 20 mmol/l HEPES. Following incubation at room temperature for 10 minutes, the material was diluted with equal parts fetal bovine serum. Following incubation at 37 °C for 1 hour, loading dye was added to the mixtures and they were run on a 1% agarose gel (70 V, 70 minutes). siRNA-TAMRA was visualized on a Gel Doc imaging system (Biorad).
Fluorescence microscopy. HeLa-Luc cells were cultured in DMEM supplemented with 10% horse serum and 1% penicillin–streptomycin according to standard ATCC protocols and plated in 6-well plates containing coverslips at 20 × 104 cells/well 24 hours prior to transfection. siRNA-TAMRA complexes were formed as described above. After incubation at room temperature for ~15 minutes, the complexes were diluted in serum-free DMEM to achieve the desired final siRNA-TAMRA concentrations. The diluted complexes were then added to the wells of HeLa-Luc cells which had been washed with PBS. After 2 hours, the cells were washed with PBS and formaldehyde (3.7%, 1–2 ml) was added to each well. Following a 10-minute incubation, the cells were rinsed with PBS and mounted on glass slides. For experiments involving pre-treatment with polymer, free polymer was incubated with cells for 1 hour before washing with PBS and adding free siRNA-TAMRA. Cells were visualized with a Zeiss Axiovert 40 CFL fluorescence microscope equipped with a 20× objective (Thornwood, NY).
SUPPLEMENTARY MATERIAL Figure S1. 1H NMR of PVBLG20 in CDC13/TFA-d (v/v: 5/1).
Acknowledgments
J.C. acknowledges support from the NIH (1R21EB013379 and Director's New Innovator Award 1DP2OD007246) and NSF (CHE-0809420). N.P.G. is currently an Institute for Genomic Biology Fellow at University of Illinois at Urbana-Champaign. H.L. is currently a Jake Wetchler Foundation Fellow for Pediatric Innovation of the Damon Runyon Cancer Research Foundation.
Supplementary Material
1H NMR of PVBLG20 in CDC13/TFA-d (v/v: 5/1).
REFERENCES
- Buckley RH. Gene therapy for SCID–a complication after remarkable progress. Lancet. 2002;360:1185–1186. doi: 10.1016/S0140-6736(02)11290-6. [DOI] [PubMed] [Google Scholar]
- Hollon T. Researchers and regulators reflect on first gene therapy death. Nat Med. 2000;6:6. doi: 10.1038/71545. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM., and, Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126:231–235. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar S., and, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z., and, Corey DR. Challenges for RNAi in vivo. Trends Biotechnol. 2004;22:390–394. doi: 10.1016/j.tibtech.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N.et al. (2008A combinatorial library of lipid-like materials for delivery of RNAi therapeutics Nat Biotechnol 26561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY., and, McMillan NA. Lipidic systems for in vivo siRNA delivery. AAPS J. 2009;11:639–652. doi: 10.1208/s12248-009-9140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM.et al. (2005Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors Nat Biotechnol 23709–717. [DOI] [PubMed] [Google Scholar]
- Crombez L, Aldrian-Herrada G, Konate K, Nguyen QN, McMaster GK, Brasseur R.et al. (2009A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells Mol Ther 1795–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E.et al. (2006Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras Nat Biotechnol 241005–1015. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J., and, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M.et al. (2004Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs Nature 432173–178. [DOI] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL.et al. (2007Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes Proc Natl Acad Sci USA 10412982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svintradze DV., and, Mrevlishvili GM. Fiber molecular model of atelocollagen-small interfering RNA (siRNA) complex. Int J Biol Macromol. 2005;37:283–286. doi: 10.1016/j.ijbiomac.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Dominska M., and, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123 Pt 8:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- Davidson TJ, Harel S, Arboleda VA, Prunell GF, Shelanski ML, Greene LA.et al. (2004Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation J Neurosci 2410040–10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratovska A., and, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Christensen LV, Jo S, Yockman JW, Jeong JH, Kim YH.et al. (2006Cholesteryl oligoarginine delivering vascular endothelial growth factor siRNA effectively inhibits tumor growth in colon adenocarcinoma Mol Ther 14343–350. [DOI] [PubMed] [Google Scholar]
- Björklund J, Biverståhl H, Gräslund A, Mäler L., and, Brzezinski P. Real-time transmembrane translocation of penetratin driven by light-generated proton pumping. Biophys J. 2006;91:L29–L31. doi: 10.1529/biophysj.106.083881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocky TB, Menon AK., and, Gellman SH. Cytoplasmic and nuclear delivery of a TAT-derived peptide and a beta-peptide after endocytic uptake into HeLa cells. J Biol Chem. 2003;278:50188–50194. doi: 10.1074/jbc.M308719200. [DOI] [PubMed] [Google Scholar]
- Gabrielson NP., and, Pack DW. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J Control Release. 2009;136:54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Lu JJ, Langer R., and, Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Wang J, Bai Y, Lang JW, Liu S, Lin Y.et al. (2011Ionic polypeptides with unusual helical stability Nat Commun 2206. [DOI] [PubMed] [Google Scholar]
- Gabrielson NP, Lu H, Yin L, Li D, Wang F., and, Cheng J. Reactive and bioactive cationic a-helical polypeptide template for nonviral gene delivery. Angew Chem Int Ed Engl. 2012;51:1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg P, El-Andaloussi S, Sütlü T, Johansson H., and, Langel U. Delivery of short interfering RNA using endosomolytic cell-penetrating peptides. FASEB J. 2007;21:2664–2671. doi: 10.1096/fj.06-6502com. [DOI] [PubMed] [Google Scholar]
- Kebbekus P, Draper DE., and, Hagerman P. Persistence length of RNA. Biochemistry. 1995;34:4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- van der Aa MA, Huth US, Häfele SY, Schubert R, Oosting RS, Mastrobattista E.et al. (2007Cellular uptake of cationic polymer-DNA complexes via caveolae plays a pivotal role in gene transfection in COS-7 cells Pharm Res 241590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA.et al. (2010Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles Nature 4641067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nicol F., and, Szoka FC., Jr GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- Wyman TB, Nicol F, Zelphati O, Scaria PV, Plank C., and, Szoka FC., Jr Design, synthesis, and characterization of a cationic peptide that binds to nucleic acids and permeabilizes bilayers. Biochemistry. 1997;36:3008–3017. doi: 10.1021/bi9618474. [DOI] [PubMed] [Google Scholar]
- Forrest ML., and, Pack DW. On the kinetics of polyplex endocytic trafficking: implications for gene delivery vector design. Mol Ther. 2002;6:57–66. doi: 10.1006/mthe.2002.0631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H NMR of PVBLG20 in CDC13/TFA-d (v/v: 5/1).