Abstract
MicroRNAs (miRNAs) are small non-coding RNAs of approximately 22 base pairs that regulate the expression of genes by targeting messenger RNA with complementarity with the miRNA base sequence. Regulation of gene expression by miRNAs is crucial in cellular development and differentiation, and recent studies suggest a relationship between human diseases and the breakdown of gene silencing mechanisms induced by miRNA abnormalities. In particular, abnormal miRNA expression has been detected in various types of cancer and the target genes have been identified. These results indicate that miRNAs act in a manner equivalent to oncogenes or tumor suppressors. miRNAs may also serve as diagnostic biomarkers and therapeutic targets. In this review, we introduce the latest findings on miRNAs in human endometrial cancer, a common malignancy, and discuss the potential of miRNAs as biomarkers and targets for molecular therapy.
Keywords: microRNA, endometrial cancer, biomarker, therapeutic drugs
Introduction
In 1993, Lee et al identified a non-coding RNA (ncRNA), lin-42, in C. elegans (1). Subsequently, Fire and Mello found a sequence-specific gene-silencing mechanism, referred to as RNA interference (RNAi), which was induced by double-stranded RNA, and proposed the concept of the regulation of gene expression by ncRNA in 1998 (2). In 2000, let-73 was identified as a short-stranded RNA, termed a microRNA (miRNA), that regulated gene expression, development, cell division and differentiation, and cell homeostasis. Subsequently, miRNAs have been found in various organisms, including viruses and plants, and the number of identified miRNAs continues to rise. A total of 530 human miRNAs were registered (miRecords: http://mirecords.biolead.org/) as of January 2008, and more than 1,000 miRNAs are thought to exist.
miRNA-mediated RNAi is considered to be post-transcriptional regulation, a third means of modulation of gene expression, in addition to transcriptional regulation and post-translational modification. miRNAs generally bind to the 3′ untranslated region (UTR) of a target messenger RNA (mRNA) via base pairing and consequently control gene expression specifically by inhibiting translation and decreasing mRNA stability. In silico analysis has shown that one miRNA controls hundreds of targets, and more than 60% of the approximately 20,000 human genes that code proteins have at least one miRNA target site in the 3′ UTR. However, miRNA target sites are also included in coding regions and in the 5′ UTR, and almost all mRNAs are regulated by miRNAs. This suggests that miRNAs play a key role in the generation of the transcriptome and proteome.
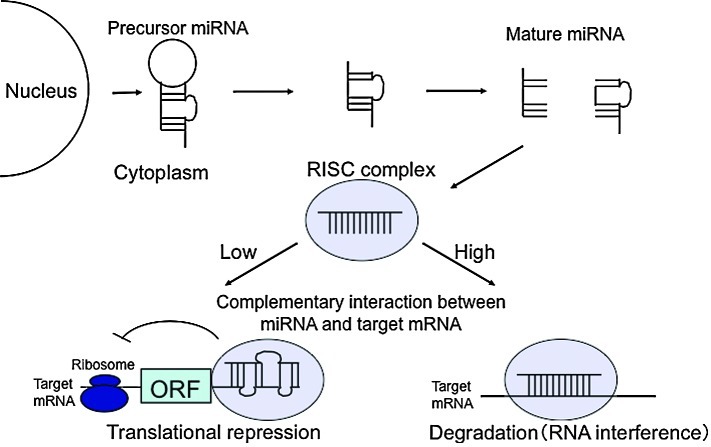
miRNA synthesis starts with the transcription of miRNA genes to create a pri-miRNA, a long (hundreds or thousands of nucleotides) precursor with poly(A) sequences. pri-miRNAs are usually transcribed by RNA polymerase II and then cleaved into one or a number of pre-miRNAs by the microprocessor complex, which contains the nuclear RNase III Drosha-DGCR8 complex with a double-stranded RNA-binding domain. Pre-miRNAs generally have a 70- to 90-bp stem-loop structure. These molecules are transported from the nucleus to the cytoplasm by exportin-5. The pre-miRNA hairpin is cleaved by the Dicer complex, which includes helicase and RNase III domains, and is processed to an approximately 21-bp double-stranded miRNA, which is finally converted to a single-stranded mature miRNA.
The mature miRNA is incorporated into the RNA-induced silencing complex (RISC), a ribonucleoprotein complex, to yield the miRISC, which recognizes target mRNA using the miRNA as a guide and inhibits mRNA translation or cleaves the mRNA. Cleavage occurs when the miRISC has high complementarity with the mRNA, whereas inhibition of mRNA translation occurs when miRISC/mRNA interaction has relatively low complementarity. In animals, almost all miRNAs inhibit translation, whereas cleavage is common in plants. Target mRNA in which translation is inhibited is transported to and degraded in a processing body (P-body), a granule localized in the cytoplasm (Fig. 1).
Figure 1.
miRNA production and mechanism of the gene expression regulation.
miRNA and small interfering RNA (siRNA) guide the RISC to the target mRNA. However, siRNA requires complete complementarity with the target, while miRNA functions with a partial mismatch in base pairing with the target mRNA. Consequently, one miRNA has numerous targets. As described above, different modes of inhibition depend on these differences in complementarity. Translation is inhibited when the base sequence in the 3′ UTR of a target mRNA has incomplete complementarity with the miRNA, but the detailed molecular mechanism has yet to be elucidated.
2. miRNA in carcinogenesis
Relatively early studies on miRNA in cancer revealed the following important findings. First, numerous miRNA genes are located in regions with frequent copy number changes due to amplification and deficit (fragile site) or with chromosomal instability near an integration site of human papillomavirus type 16, suggesting an association of carcinogenesis with genomic abnormalities in the primary structure of miRNA genes. Second, miRNA expression profiles significantly differ among organs and tissues, and miRNA levels in cancer tissues are generally lower than those in normal tissues, suggesting that miRNAs act as tumor suppressors.
It is now known that various miRNAs act in a manner equivalent to oncogenes or tumor suppressor genes in cancer development and progression. An association of miRNAs with cancer stem cells has also gradually become apparent and miRNAs that directly target oncogenes have been identified. miRNAs generally decrease mRNA expression levels, and therefore, miRNAs targeting an oncogene have a tumor suppressor action via control of the cell cycle through reduction of the gene expression. By contrast, certain miRNAs have an oncogene-like function, with a high miRNA expression-inducing oncogenic transformation; i.e., the miRNA acts similar to an oncogene. Recent studies have shown that the expression of tumor suppressor miRNAs is regulated by aberrant DNA methylation, revealing a mechanism in which the miRNA is inactivated by epigenetic abnormalities (methylation) in a similar manner to tumor suppressor (22).
miR-124 is a particular miRNA for which an association with methylation and cancer stem cells has been shown in detail. miR-124 is located in or near a CpG island in the chromosomal regions of 8p23.1, 8q12.3 and 20q13.33. miR-124 expression is inhibited by tumor-specific aberrant DNA methylation in colon cancer HCT116 cells and is reactivated by treatment with 5-aza-2′-deoxycytidine (5-aza-dC). In HCT116 cells, the methyl-CpG binding protein, MeCP2, and methyl-CpG binding domain protein, MBD2, are bound to the promoter of the miR-124 gene, whereas no binding of these proteins occurs in demethylated HCT116 cells following knockout of DNA methyltransferase DNMT1 and DNMT3B genes or treatment with 5-aza-dC. The results suggest that promoter selectivity is due to methylation. The transcriptional product of CDK6 is an oncogene and CDK6 mRNA is a direct target of miR-124. Overexpression of miR-124 or miR-137 has also been shown to induce morphological changes and differentiation into neurons, with an enhanced expression of neuronal differentiation markers in mouse neural stem cells and human glioblastoma (3). The studies suggest that miR-124 is involved in the maintenance of cancer stem cell genotypes, raising the possibility of treatment using the miRNA targeting of cancer stem cells.
The expression of miR-127, located in the CpG island of 14q32.31, is also reactivated by treatment with 5-aza-dC and inhibited by elevated methylation in prostate and colon cancers (4). This miRNA is of interest since it acts as an oncogene in the targeting of BCL6 mRNA (4). However, the most detailed study of an oncogene-type miRNA was performed for miR-17-92 (2). Many miRNAs form clusters in genomes, and miR-17-92 contains the sequence miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92-1, encoded in this order. In B-cell lymphomas, the expression of the cluster is enhanced and induced by the oncogene c-Myc, with studies in mouse models and cell cultures showing subsequent inhibition of apoptosis and enhanced cell division, suggesting that miR-17-92 acts as an oncogene. Of the genes in this cluster, miR-17 and miR-20a target the E2F1 gene, which is related to the apoptosis response, and the down-regulation of E2F1 by miR-17-92 inhibits apoptosis. c-Myc regulates E2F1 transcription and vice versa. Consequently, miR-17-2 forms a feedback loop with c-Myc and E2F1 (5). Expression of miR-17-92 is also regulated by E2F3. This regulation indicates the presence of a comprehensive system for the modulation of gene expression, in which E2F1 activation by c-Myc is inhibited by miR-17-92, resulting in the inhibition of apoptosis. Therefore, this system is crucial for the survival of cancer cells. miR-17-92 also targets various tumor suppressor genes and therefore plays a key role as an oncogene-type miRNA (6).
The miR-15a/16-1 cluster consists of the miRNAs, miR-15a and miR-16-1, and is found in the chromosomal region 13q14. Frequent absence or silencing of this cluster occurs in chronic lymphocytic leukemia. miR-15a and miR-16-1 inhibit BCL2, an oncogene that inhibits apoptosis. Therefore, expression of these miRNAs in leukemia cells induces apoptosis. A recent study showed that miR-15a/16-1 targets the c-Myb and IGSF4 oncogenes, and a microarray analysis showed that the miRNA cluster inhibits a number of cancer-associated genes correlated with cell growth and anti-apoptosis. Another miRNA, let-7, acts as a tumor suppressor by inhibiting the oncogenes K-ras and HMGA2, suggesting that miRNAs also play a key role in the inhibition of oncogenic transformation. As described below, a reduced let-7 expression has been found in patients with lung cancer and the let-7 expression level was correlated with the outcomes in these patients (7).
3. miRNA as a diagnostic biomarker in human cancer
miRNA expression in tissues is regulated by the number of miRNAs present in a cell at a specific period of time. Tissue-specific patterns of miRNA expression have also been shown and analysis of these patterns may produce potential markers for identifying the original source of a cancer, which is crucial in establishing a treatment plan. miRNA analysis may also be useful for distinguishing the primary lesion from metastases.
miRNA expression may serve as a prognostic predictor, since the prognosis of cancer is correlated with the expression levels of numerous miRNAs: has-let-7, has-let-7a-2 and has-miR-155 in lung cancer; has-miR-125b in hepatocellular carcinoma; has-miR-21 in breast, gastric, colon and pancreatic cancers; and has-let-7b and has-miR-205 in head and neck squamous cell carcinoma. These correlations are purely statistical and the fundamental underlying mechanisms have yet to be elucidated. One study showed that methylation of let-7a-3 was related to prognosis in ovarian cancer. Poor outcomes were found in ovarian cancer patients with low levels of mRNA for Dicer and Drosha, which are involved in the maturation of miRNAs. If the miRNA precursor is not processed efficiently, the mature miRNA-induced gene silencing mechanism may not function adequately, resulting in abnormal oncogenic effects. Lung cancer patients with low Dicer expression levels also have a poor prognosis (8).
Numerous studies have been performed with regards to the relationship between miRNA and drug resistance in cancer cells. Meng et al showed that sensitivity to gemcitabine was increased by inhibiting miR-21 and miR-200b, which have a higher expression in bile duct cancer than in benign tumors (9). Yang et al showed that miR-21 introduced into ovarian cancer cells activated AKT through PTEN inhibition and worsened cisplatin resistance (10). The relationships of miRNAs with sensitivity to doxorubicin in breast cancer and sensitivity to endocrine therapy have also been examined. Further studies on multiple drug resistance may facilitate the choice of treatment strategies based on the expression levels of miRNAs.
Since RNA is degraded by RNase in the serum, it was initially thought that miRNAs would be absent. However, a number of miRNAs, including miR-638, were subsequently detected in the serum. The mechanisms by which miRNAs are released into and stabilized in the serum have yet to be determined, but miRNAs appear to be transported selectively based on specific requirements for the molecules in the serum. Lawrie et al found a correlation between serum miR-21 levels and the prognosis of patients with diffuse large B-cell lymphoma (11), and Mitchell et al found high serum miR-141 levels in patients with prostate cancer (12).
Tanaka et al found that the serum of patients with acute leukemia included more miRNAs with low expression levels than patients with high expression levels, in comparison to the serum of healthy volunteers. In particular, the serum miR-92 level was significantly reduced, whereas the miR-92 level in leukemia cells was high (23). To explain these results, Tanaka et al proposed the hypothesis that leukemia cells degrade specific miRNAs or promote the uptake of miR-92 from the blood (23). The correlation between a specific cancer and a serum miRNA level suggests the feasibility for diagnostic and prognostic predictions based on miRNA levels in the blood. This is likely to become a key area of research, since biomarker miRNAs may permit early diagnosis, personalized treatment and prognostic prediction.
4. Applications of miRNA in cancer therapy
Since miRNA expression levels are known to change in cancer cells, inhibition of the overexpression of an oncogene-type miRNA or elevation of the underexpression of a tumor suppressor-type miRNA may have a therapeutic effect. miRNAs are small RNAs of approximately 22 base pairs, thus, antisense oligonucleotides are likely to be effective as base sequence-specific inhibitors. Delivery methods for antisense agents have improved, and these approaches should facilitate the targeting of miRNA, including the use of RNA oligonucleotides with chemical modifications, in order to overcome their lack of in vivo stability.
Stoffel et al (13) synthesized an anti-miRNA oligonucleotide (AMO) (an ‘antagomir’) with 2′-O-methyl, phosphorothioate and cholesterol groups, and showed the efficacy of this molecule in mice. At 24 h after antagomir-16 was injected in the caudal vein, the miR-16 level was reduced in all organs except the brain, indicating that the antagomir did not cross the blood-brain barrier. However, the percutaneous injection of antagomir-16 reduced miR-16 in the brain (13). A study on antagomir-122 targeting liver-specific expression of miR-122 showed that this miRNA is involved in the regulation of cholesterol biosynthesis (14). These results suggest that antagomirs are effective for the treatment of diseases based on the targeting of specific miRNAs.
Locked nucleic acid (LNA) is a 2′-O,4′-C-methylene-linked RNA derivative that is nuclease-resistant and forms highly stable and specific double-stranded helices. Therefore, LNA is used as a probe for hybridization and as a double-dye probe in real-time PCR. LNA is water-soluble and less cytotoxic than other antisense agents, and DNA/LNA mixed oligomers retain the advantage of allowing RNase H activation after hybridization with the target RNA. Use of a DNA/LNA AMO to target miR-21 overexpression in glioblastoma was shown to induce apoptosis, and similar targeting of the overexpression of miR-221 and miR-222 in prostate cancer cells decreased the clonogenicity of the cells (15).
Tumor suppressor miRNAs are used to supplement miRNA underexpression as agonists. In cellular studies, the effects of miRNA have been investigated by transfection with an expression vector, but small RNAs, including AIV10 and siRNA, are preferable for use as miRNA-based therapeutic agents. Inhibition of cell growth has been shown with transfection of sense miR-15/miR-16 oligoRNA in chronic lymphocytic leukemia and transfection of let-7 as dsRNA in lung cancer cells. The design of miR precursors that allows the sense strand of the miRNA to be efficiently incorporated into RISC has been reported using chemically modified dsRNA, and transfection of miR-34a in glioblastoma cells using a pre-miR-34a precursor was shown to increase apoptosis (16).
It remains to be elucidated whether transfected miRNAs have equivalent actions to endogenous miRNA. It is unclear whether exogenous miRNAs have sufficient activity in vivo and there are numerous concerns about adverse reactions and the local distribution of miRNA. Furthermore, although the delivery methods for miRNA (including across the blood-brain barrier) have been greatly improved, only a few options are applicable in practice.
5. miRNA in the endometrium
The endometrium is a tissue that undergoes substantial changes in parallel with the menstrual cycle in humans. This tissue is changed for ovum implantation and embryo development by ovary-releasing sex hormones that regulate gene expression. These changes are histopathologically classified into apoptosis, cell growth, neovascularization, cell differentiation and tissue remodeling. The changes resemble an inflammatory response and changes in the activities of endocrine and paracrine humoral factors, including growth factors, chemokines, cytokines and proteases, as well as alterations in the extracellular matrix are involved. The rapid turnover of the cell cycle is undesirable in the prevention of carcinogenesis. Intrinsic control of gene expression by miRNAs is crucial in this environment and various changes in the expression of miRNAs have been suggested in normal endometrium, endometriotic tissue, metrorrhagia and endometrial cancer.
Pan et al investigated miRNA profiles in the uterine interstitium and granular epithelium and found that 32 miRNAs showed characteristic expression levels (17). In particular, estradiol-17β and progesterone regulated the expression of miR-20a, miR-21, miR-23, miR-26a, miR-18a, miR-181a, miR-206 and miR-142-5p. These miRNAs are involved in the regulation of expression in numerous genes with key actions in the uterine tissue, including transforming growth factor β (TGF-β), TGF-β receptor, estrogen receptor, progesterone receptor and CYP-19A1 (aromatase). A number of inflammation and cell growth-related factors, including nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), are also regulated by miRNAs in local inflammatory microenvironments in the uterine tissue. In this context, inhibition of the expression of tumor necrosis factor α by miR-125b and miR-155 has been analyzed (17).
6. miRNA and endometrial cancer
miRNA-related findings in endometrial cancer have not emerged to the same extent as those in gastrointestinal cancer. However, studies are ongoing in various institutions, including in Japan (Table I). Boren et al found that 13 miRNAs (hsa-let-7i, hsa-miR-221, hsa-miR-30c, hsa-miR-152, hsa-miR-193, hsa-miR-185, hsa-miR-106a, hsa-miR-181a, hsa-miR-210, hsa-miR-423, hsa-miR-103, hsa-miR-107 and hsa-let-7c) and 90 mRNAs derived from human endometrial cancer and normal uterine tissues were involved in endometrial cancer growth, and proposed that the 13 miRNAs recognized 26 (29%) of the 90 mRNAs as targets. A total of 11 of these 26 mRNA genes (KCNMB1, IGFBP-6, ENPP2, TBL1X, CNN1, MYH11, KLF2, TGFB1/1, MYL9, SNCAIP and RAMP1) were shown to form a network and to be involved in cell growth (18).
Table I.
Endometrial cancer-associated miRNAs.
Myatt et al found that the expression of the tumor suppressor gene FOXO1 in endometrial cancer cells was down-regulated in comparison to that in the normal endometrium (19). Immunohistochemical studies showed that the level of FOXO1 was highest in the normal endometrium, lower in endometrial hyperplasia and further reduced in an endometrial tumor. However, the FOXO1 level is relatively high in HEC-1B endometrial cancer cells and lower in Ishikawa endometrial cancer cells. In HEC-1B cells, the levels of miR-9, miR-27, miR-96, miR-153, miR-182, miR-183 and miR-186 were high, whereas little expression of miR-29a, miR-128, miR-152 and miR-486 was found. Therefore, this pattern of miRNA expression appears to produce only a limited inhibition of FOXO1 expression. In Ishikawa cells, the levels of miR-27, miR-96, miR-128, miR-153, miR-182, miR-183 and miR-186 were higher than the respective levels in HEC-1B cells, which may reflect the lower FOXO1 level in Ishikawa cells compared to HEC-1B cells. Furthermore, in Ishikawa cells, inhibition of the expression of miR-9, miR-27, miR-96, miR-153, miR-183 and miR-186 induced FOXO1-dependent G1 cell cycle arrest and cell death with a gradual decrease of RNAi with FOXO1 expression. The results suggest that miRNA-dependent FOXO1 down-regulation plays a key role in the survival of endometrial tumor cells.
Myatt et al concluded that the abnormal expression of miRNAs affect the regulation of apoptosis-associated genes and plays a specific role in the development of endometrial cancer (19). This study is notable as it showed that miRNAs in a gene network developed in silico inhibited tumor suppressor genes and induced tumorigenesis. The results also showed that the tumor size is reduced by inhibiting these miRNAs, which suggests the potential of targeting of miRNAs as a therapeutic strategy in patients with endometrial cancer.
Hiroki et al conducted a comparative study of carcinoma and normal endometrial tissues derived from 21 patients with serous endometrial adenocarcinoma between January 2001 and December 2006 and analyzed the relationship of miRNA levels with clinical outcomes, including clinicopathological features and survival rate (20). Out of 120 miRNAs that were analyzed, the expression levels of 54, including miR-101, miR-10b*, miR-152 and miR-29b, were down-regulated, and those of the remaining 66, including miR-200a, miR-200b and miR-205, were up-regulated. miR-205 was also investigated as a predictor for head and neck squamous cell carcinoma and significant correlations were found between inhibition of the expression of miR-10b*, miR-29b and miR-455-5p and vascular invasion (P=0.048, P=0.013 and P=0.032, respectively). A low expression of miR-101, miR-10b*, miR-139-5p, miR-152, miR-29b and miR-455-5p was a significant prognostic factor for poor overall survival (P<0.05) and a low expression of miR-152, miR-29b and miR-455-5p was a significant prognostic factor for poor disease-free survival (P<0.05). Results of the multivariate analysis revealed that a low expression of miR-152 was an independent risk factor for poor overall survival (P=0.021), and a low expression of miR-101 and miR-152 was an independent risk factor for poor disease-free survival (P=0.016 and P=0.010, respectively). In vitro studies showed that transfection with miR-101 and miR-152 precursors inhibited cell growth in serous endometrial adenocarcinoma (P<0.0001 and P<0.01, respectively), and a strong positive correlation was found between miR-101 down-regulation and the immune response to cyclooxygenase-2 (COX-2) (P=0.035). Hiroki et al concluded that these data showed that insufficient regulation of miRNA expression was an effective indicator for predicting a poor prognosis in patients with serous endometrial adenocarcinoma (20). This study is crucial since it indicates an actual relationship between prognosis and the miRNA levels in endometrial cancer.
Chung et al (21) investigated miRNA expression and clinicopathological features of endometrial cancer in 30 specimens collected from patients with endometrioid endometrial adenocarcinoma in Hong Kong. The results were compared to those for 22 normal tissue specimens. Carcinoma was detected in 25 specimens of stages I and II, and 5 of stage III. High levels of miR-95, miR-103, miR-106a, miR-151, miR-155, miR-182, miR-183, miR-194, miR-200a, miR-200c, miR-203, miR-205, miR-210 and miR-223 were found. A total of 30 miRNAs correlated with features associated with prognosis, including disease period, invasion into muscle layers, recurrence and invasion of lymph nodes. A total of 68 genes were identified as potential targets of the 30 miRNAs. Chung et al (21) noted that the expression level of miR-205 in cancer tissue was extremely high, at 27.2-fold that in normal tissues, also noted by Hiroki et al (20). The miR-205 level showed a correlation with invasion into muscle layers and recurrence. Chung et al showed that the transfection of sequences inhibiting miR-205 into the endometrial cancer cell line RL95-2 inhibited the expression of miR-205 by 64.9% and increased the expression of the tumor suppressor gene JPH4 by 14.2% (21). JPH4 shows abnormalities in endometrioid endometrial adenocarcinoma and these in vitro and in vivo results suggest that it is a target of miR-205. The results of Chung et al are significant since they show the relationship between miRNA levels and clinicopathological features in endometrioid adenocarcinoma, which accounts for 80–90% of endometrial cancer.
7. Conclusion
This review evaluated recent findings on miRNA expression and activities and on the relationship between miRNA and endometrial cancer. Techniques for diagnosis and treatment based on miRNA have yet to be established and the majority of results remain at the in vitro level. However, the utility of miRNAs as prognostic predictors has been shown. The use of miRNAs for the treatment of malignant tumors involves delivery of a single miRNA that may modify the expression of approximately 100 genes. This type of treatment is significantly different from conventional gene therapy, in which only one gene is targeted, despite a malignant tumor being a phenotype of cumulative abnormalities. This phenomenon is a limitation of conventional gene therapy that may be overcome by treatment with miRNAs. However, inhibition of the expression of genes unrelated to carcinogenesis, but with an mRNA sequence similar to the target gene, may present a difficulty. As for conventional antitumor agents, a delivery system with selectivity for local lesions is required for the targeting of miRNA drugs to focus on the tumor and avoid effects on other tissues that may cause serious adverse reactions.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;91:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Coller HA, Forman JJ, Hegesse-Miller A. Myc’ed messages: myc induces transcription of E2Fl while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 10.Yang N, Kaur S, Volinia S, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 16.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 17.Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26:479–493. doi: 10.1055/s-0028-1096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boren T, Xiong Y, Hakam A, Wenham R, Apte S, Wei Z, Kamath S, Chen DT, Dressman H, Lancaster JM. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110:206–215. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 19.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2009;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, Sasano H, Yaegashi N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung TK, Cheung TH, Huen NY, et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osaki M, Takeshita F, Ochiya T. MicroRNAs as biomarker and therapeutic drugs in human cancer. Biomarkers. 2008;13:658–670. doi: 10.1080/13547500802646572. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu W, Lin Z, Zhang Z, Liang X. Expression profile of mammalian microRNAs in endometrial carcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]