Abstract
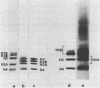
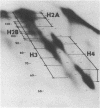
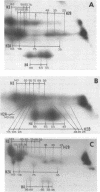
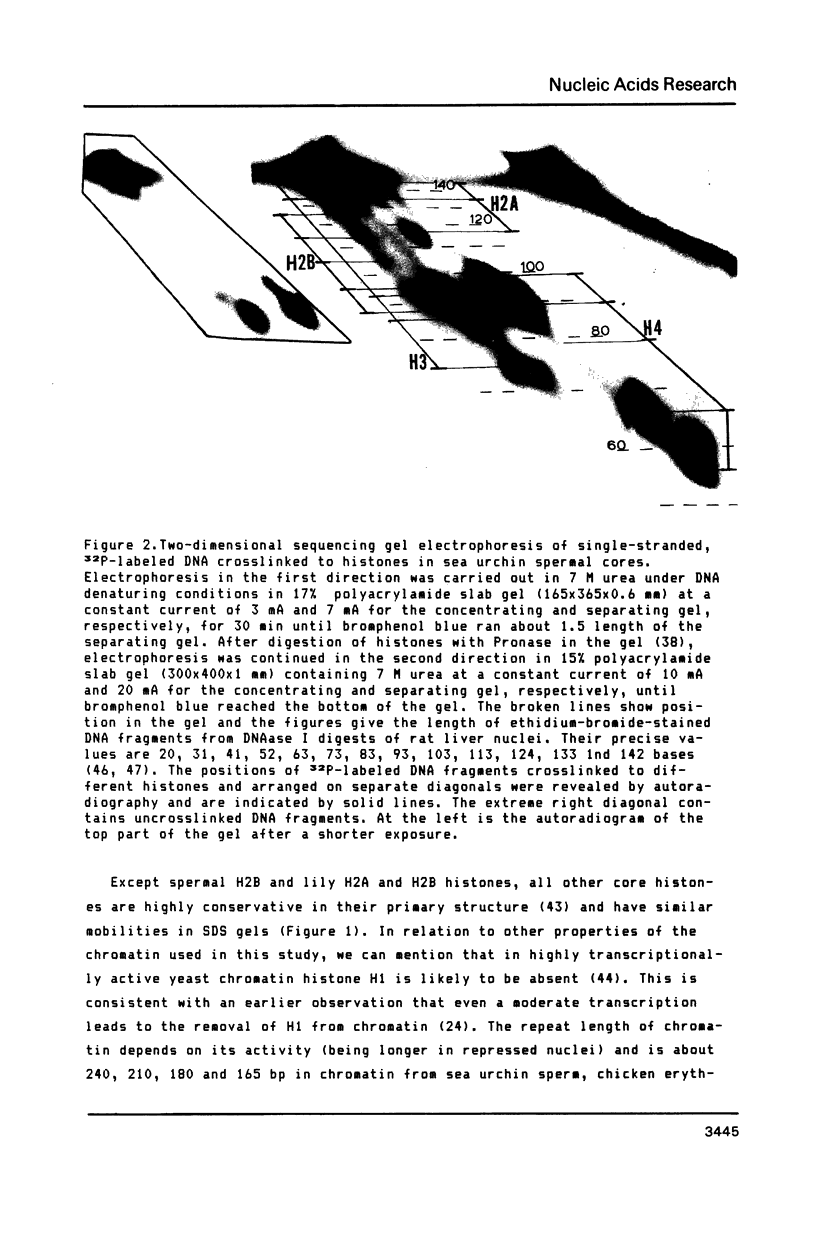
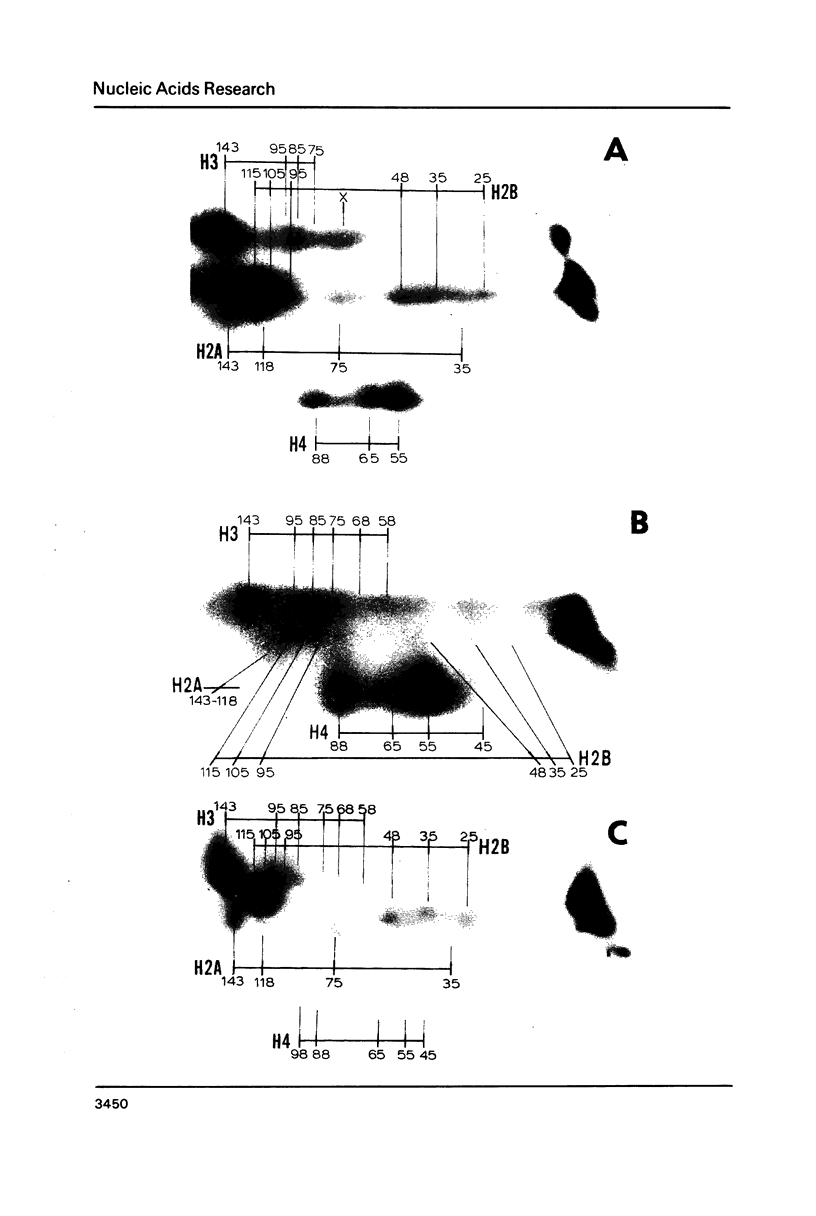
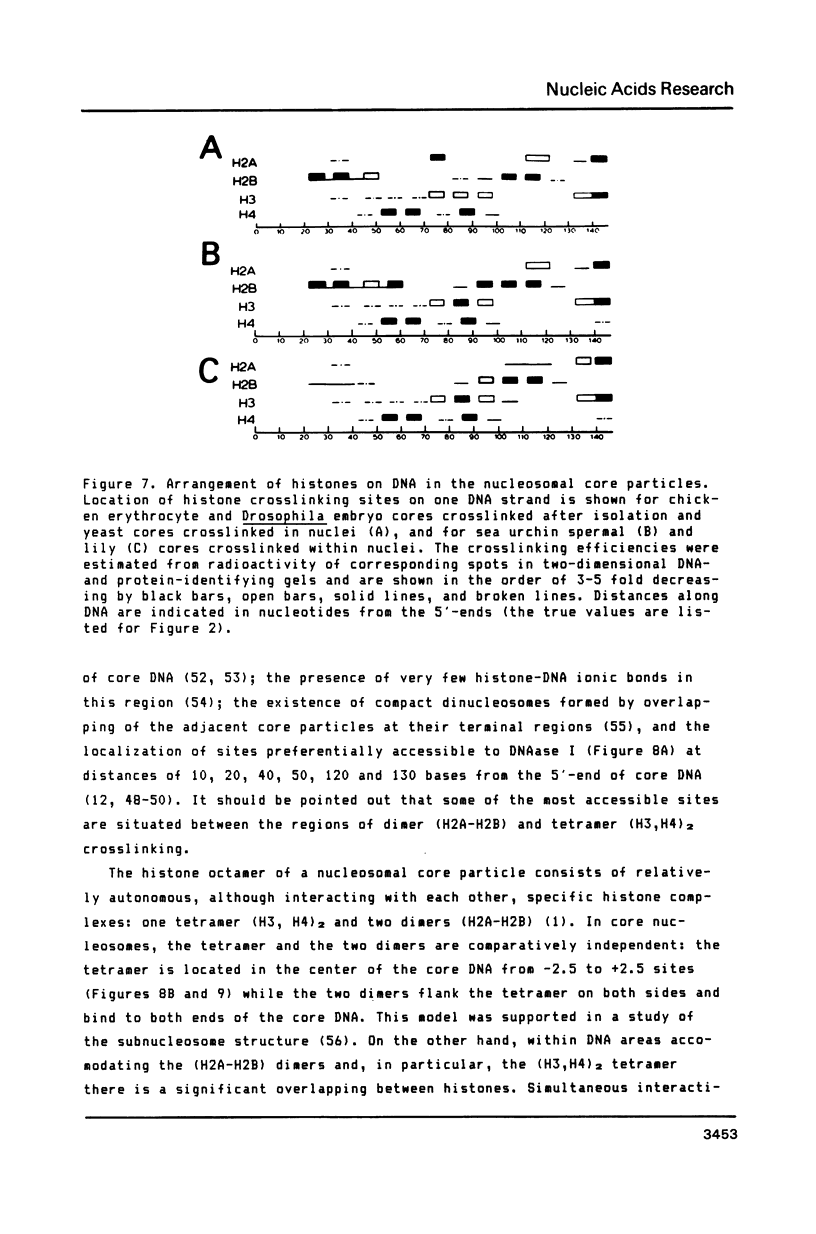
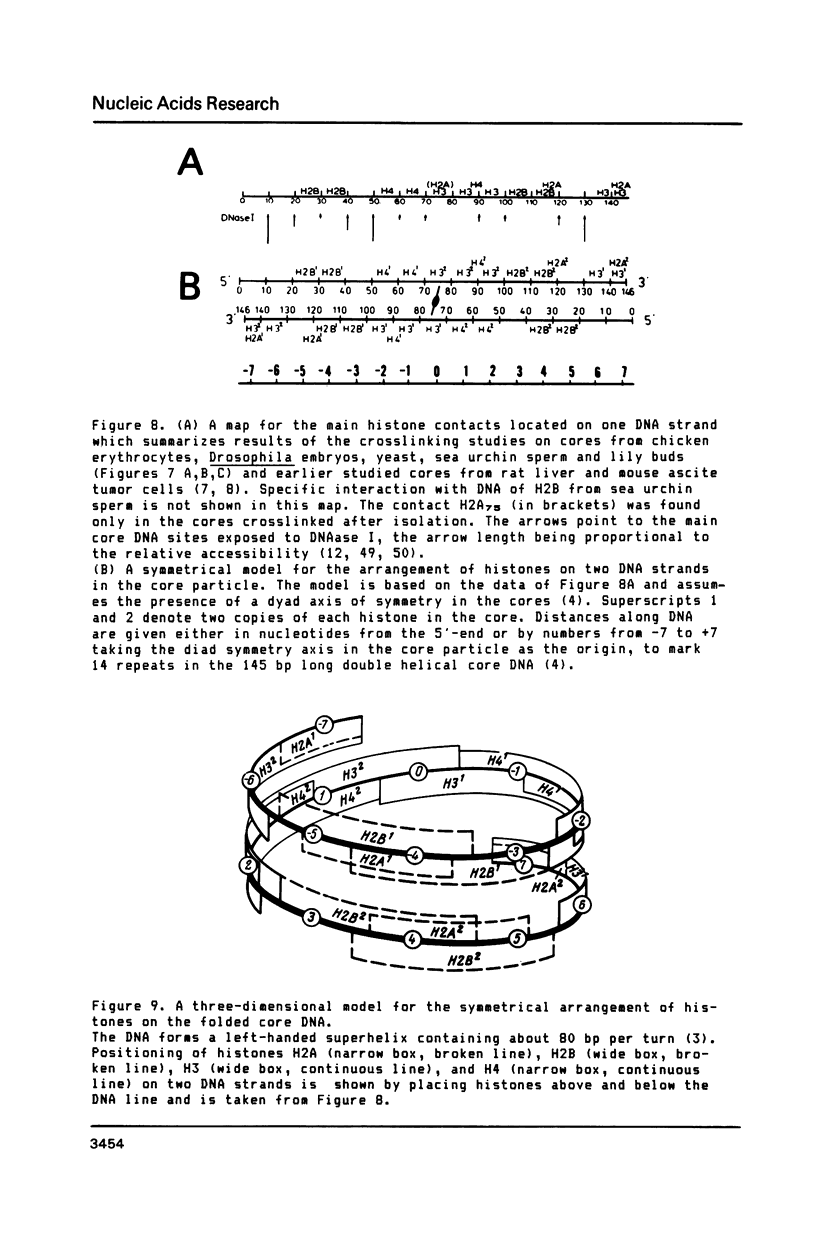
A refined map for the linear arrangement of histones along DNA in nucleosomal core particles has been determined by DNA-protein crosslinking. On one strand of 145-bp core DNA, histones are aligned in the following order: (5') H2B25,35-H455,65-H375,85,95/H488-H2B105,11 5-H2A118-H3135,145/H2A145 (3') (the subscripts give approximate distance in nucleotides of the main histone contacts from the 5'-end). Hence, the histone tetramer (H3,H4)2 and two dimers (H2A-H2B) are arranged on double-stranded core DNA in a symmetrical and rather autonomous way: H2A/H3-(H2A-H2B)-(H3,H4)2-(H2B-H2A)-H3/H2A. The primary organization was found to be very similar in core particles isolated from repressed nuclei of sea urchin sperm and chicken erythrocytes, from active in replication and transcription nuclei of Drosophila embryos and yeast and from somatic cells of lily. These data show that (i) the core structure is highly conserved in evolution and (ii) the overall inactivation of chromatin does not affect the arrangement of histones along DNA and thus does not seem to be regulated on this level of the core structure.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Harborne N., Rau D. C., Gould H. Participation of core histone "tails" in the stabilization of the chromatin solenoid. J Cell Biol. 1982 May;93(2):285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Bakayev V. V., Domansky N. N., Bakayeva T. G. Histone-containing subnucleosomes: possible implications to nucleosome structure. FEBS Lett. 1981 Oct 12;133(1):75–78. doi: 10.1016/0014-5793(81)80474-7. [DOI] [PubMed] [Google Scholar]
- Belyavsky A. V., Bavykin S. G., Goguadze E. G., Mirzabekov A. D. Primary organization of nucleosomes containing all five histones and DNA 175 and 165 base-pairs long. J Mol Biol. 1980 May 25;139(3):519–536. doi: 10.1016/0022-2836(80)90144-8. [DOI] [PubMed] [Google Scholar]
- Bentley G. A., Lewit-Bentley A., Finch J. T., Podjarny A. D., Roth M. Crystal structure of the nucleosome core particle at 16 A resolution. J Mol Biol. 1984 Jun 15;176(1):55–75. doi: 10.1016/0022-2836(84)90382-6. [DOI] [PubMed] [Google Scholar]
- Bolund L. A., Johns E. W. The selective extraction of histone fractions from deoxyribonucleoprotein. Eur J Biochem. 1973 Jun 15;35(3):546–553. doi: 10.1111/j.1432-1033.1973.tb02871.x. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Certa U., Colavito-Shepanski M., Grunstein M. Yeast may not contain histone H1: the only known 'histone H1-like' protein in Saccharomyces cerevisiae is a mitochondrial protein. Nucleic Acids Res. 1984 Nov 12;12(21):7975–7985. doi: 10.1093/nar/12.21.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenchick A., Beabealashvilli R., Mirzabekov A. Topography of interaction of Escherichia coli RNA polymerase subunits with lac UV5 promoter. FEBS Lett. 1981 Jun 1;128(1):46–50. doi: 10.1016/0014-5793(81)81076-9. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Crémisi C., Yaniv M. Sequential assembly of newly synthesized histones on replicating SV40 DNA. Biochem Biophys Res Commun. 1980 Feb 27;92(4):1117–1123. doi: 10.1016/0006-291x(80)90402-7. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Glover J. S., Salter D. N., Shepherd B. P. A study of some factors that influence the iodination of ox insulin. Biochem J. 1967 Apr;103(1):120–128. doi: 10.1042/bj1030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greyling H. J., Schwager S., Sewell B. T., von Holt C. The identity of conformational states of reconstituted and native histone octamers. Eur J Biochem. 1983 Dec 1;137(1-2):221–226. doi: 10.1111/j.1432-1033.1983.tb07818.x. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981 Jan;23(1):121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Karpov V. L., Bavykin S. G., Preobrazhenskaya O. V., Belyavsky A. V., Mirzabekov A. D. Alignment of nucleosomes along DNA and organization of spacer DNA in Drosophila chromatin. Nucleic Acids Res. 1982 Jul 24;10(14):4321–4337. doi: 10.1093/nar/10.14.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Franke W. W. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982 Jul;29(3):799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Labhart P., Koller T. Structure of the active nucleolar chromatin of Xenopus laevis Oocytes. Cell. 1982 Feb;28(2):279–292. doi: 10.1016/0092-8674(82)90346-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee K. P., Baxter H. J., Guillemette J. G., Lawford H. G., Lewis P. N. Structural studies on yeast nucleosomes. Can J Biochem. 1982 Mar;60(3):379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- Levina E. S., Bavykin S. G., Shick V. V., Mirzabekov A. D. The method of crosslinking histones to DNA partly depurinated at neutral pH. Anal Biochem. 1981 Jan 1;110(1):93–101. doi: 10.1016/0003-2697(81)90117-2. [DOI] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982 Feb;28(2):375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B., Dixon G. H. Limited action of micrococcal nuclease on trout testis nuclei generates two mononucleosome subsets enriched in transcribed DNA sequences. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1682–1686. doi: 10.1073/pnas.76.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A. L., Sulimova G. E. Khromatografiia nukleinovykh kislot, belkov i nekotorykh fagov na granulirovannom gidroksiapatite. Biokhimiia. 1975 Jan-Feb;40(1):115–122. [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Another potential artifact in the study of nucleosome phasing by chromatin digestion with micrococcal nuclease. Cell. 1983 Apr;32(4):1205–1215. doi: 10.1016/0092-8674(83)90303-3. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. The number of charge-charge interactions stabilizing the ends of nucleosome DNA. Nucleic Acids Res. 1980 Jun 25;8(12):2751–2769. doi: 10.1093/nar/8.12.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Bavykin S. G., Karpov V. L., Preobrazhenskaya O. V., Ebralidze K. K., Tuneev V. M., Melnikova A. F., Goguadze E. G., Chenchick A. A., Beabealashvili R. S. Structure of nucleosomes, chromatin, and RNA polymerase-promoter complex as revealed by DNA-protein cross-linking. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):503–510. doi: 10.1101/sqb.1983.047.01.060. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Shick V. V., Belyavsky A. V., Bavykin S. G. Primary organization of nucleosome core particle of chromatin: sequence of histone arrangement along DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4184–4188. doi: 10.1073/pnas.75.9.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Rosenblum E. N. Hydroxylapatite chromatography of protein-sodium dodecyl sulfate complexes. A new method for the separation of polypeptide subunits. J Biol Chem. 1972 Aug 25;247(16):5194–5198. [PubMed] [Google Scholar]
- Noll M. DNA folding in the nucleosome. J Mol Biol. 1977 Oct 15;116(1):49–71. doi: 10.1016/0022-2836(77)90118-8. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Seale R. L. In vivo assembly of newly synthesized histones. Biochemistry. 1981 Oct 27;20(22):6432–6437. doi: 10.1021/bi00525a023. [DOI] [PubMed] [Google Scholar]
- Senear A. W., Palmiter R. D. Multiple structural features are responsible for the nuclease sensitivity of the active ovalbumin gene. J Biol Chem. 1981 Feb 10;256(3):1191–1198. [PubMed] [Google Scholar]
- Shick V. V., Belyavsky A. V., Bavykin S. G., Mirzabekov A. D. Primary organization of the nucleosome core particles. Sequential arrangement of histones along DNA. J Mol Biol. 1980 May 25;139(3):491–517. doi: 10.1016/0022-2836(80)90143-6. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Bergman L. W. Structure of sea urchin sperm chromatin core particle. J Biol Chem. 1980 Nov 25;255(22):10702–10709. [PubMed] [Google Scholar]
- Simpson R. T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979 Oct 25;254(20):10123–10127. [PubMed] [Google Scholar]
- Simpson R. T. Modulation of nucleosome structure by histone subtypes in sea urchin embryos. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6803–6807. doi: 10.1073/pnas.78.11.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Spiker S., Key J. L., Wakim B. Identification and fractionation of plant histones. Arch Biochem Biophys. 1976 Oct;176(2):510–518. doi: 10.1016/0003-9861(76)90194-6. [DOI] [PubMed] [Google Scholar]
- Stoeckert C. J., Jr, Beer M., Wiggins J. W., Wierman J. C. Histone positions within the nucleosome using platinum labeling and the scanning transmission electron microscope. J Mol Biol. 1984 Aug 15;177(3):483–505. doi: 10.1016/0022-2836(84)90296-1. [DOI] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Brandt W. F., Von Holt C., Wittmann-Liebold B. The complete amino-acid sequence of histone H2B(3) from sperm of the sea urchin Parechinus angulosus. Eur J Biochem. 1978 Sep 1;89(2):443–452. doi: 10.1111/j.1432-1033.1978.tb12547.x. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Oudet P. Complexes of the arginine-rich histone tetramer (H3)2(H4)2 with negatively supercoiled DNA: electron microscopy and chemical cross-linking. Nucleic Acids Res. 1979 Oct 10;7(3):611–623. doi: 10.1093/nar/7.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. T. Properties of active nucleosomes as revealed by HMG 14 and 17 chromatography. Nucleic Acids Res. 1982 Mar 25;10(6):2017–2042. doi: 10.1093/nar/10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker R. H. New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science. 1969 Jan 10;163(3863):150–160. doi: 10.1126/science.163.3863.150. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Zalenskaya I. A., Pospelov V. A., Zalensky A. O., Vorob'ev V. I. Nucleosomal structure of sea urchin and starfish sperm chromatin. Histone H2B is possibly involved in determining the length of linker DNA. Nucleic Acids Res. 1981 Feb 11;9(3):473–487. doi: 10.1093/nar/9.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayetz V. W., Bavykin S. G., Karpov V. L., Mirzabekov A. D. Stability of the primary organization of nucleosome core particles upon some conformational transitions. Nucleic Acids Res. 1981 Mar 11;9(5):1053–1068. doi: 10.1093/nar/9.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]