Natural products often possess one or more highly modified deoxysugar moieties, which are generally critical for their biological activities.[1-3] Mithramycin (MTM), an aureolic acid-type anticancer agent, contains five deoxyhexoses, a trisaccharidal chain (d-olivosyl-3-1-d-oliosyl-3-1-d-mycarosyl) and a disaccharidal chain (d-olivosyl-3-1-d-olivosyl) attached at 2- and 6-positions of the aglycon, respectively.[4,5] Among all sugars, d-olivose is the main building component (sugars A, B and C in Scheme 1). In addition to MTM, d-olivose is also an essential component of many biologically active natural products, including all other aureolic acid-family anticancer agents and many angucyclines.[5-8] Recently, increasing effort has been made to alter the deoxysugar moiety of natural products (glycodiversification), as modifications to these important structural motifs can greatly influence their activity and substrate specificity.[3,9-14] This approach requires a thorough understanding of deoxysugar biosynthesis as well as suitable glycosyltransferases (GTs), a group of enzymes able to transfer sugars to a given acceptor substrate.[15-17] However, most deoxysugar biosynthesis and glycosylation events, including those involved in MTM biosynthesis, have not been fully characterized in vitro due to the unavailability of sugar donor substrates and soluble glycosyltransferases.[2,9,18] In the MTM pathway, genes involved in the biosynthesis and attachment of deoxysugars have largely been investigated by in vivo studies.[5,19-23] The presence of five sugar moieties but only four GTs indicated that the MTM pathway does not follow the typical “one GT one glycosylation event” rule. Specifically, a unique GT MtmGIV has been suggested to be responsible for the first and third glycosylation events (attaching sugars C and E, respectively), whereas sugars D, A and B are transferred by MtmGIII, MtmGI and MtmGII, respectively.[22-24] Here, we report an in vitro large-scale preparation of thymidine diphosphate (TDP)-d-olivose based on the investigation of several MTM deoxysugar biosynthetic enzymes.
Scheme 1.
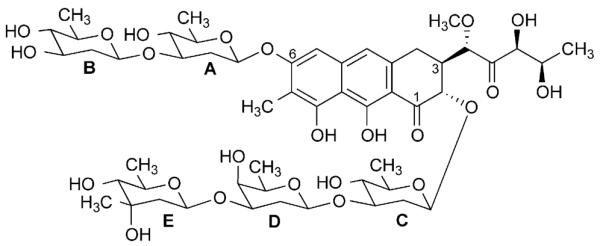
Structure of MTM. The deoxysugar moieties are: d-olivose: A, B and C; d-oliose: D; d-mycarose: E.
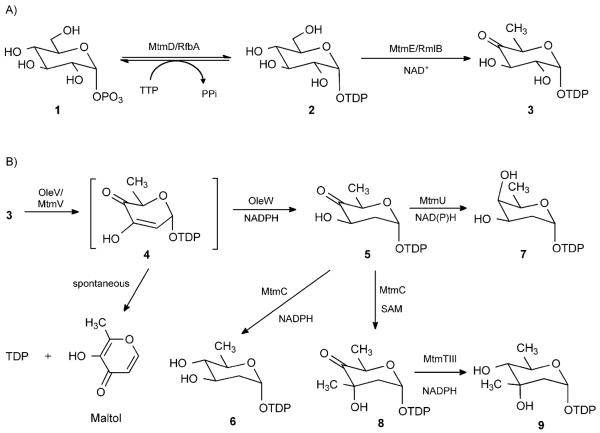
TDP-activated deoxyhexoses represent the majority of sugar donors for natural products. Biosynthesis of TDP-sugars starts from glucose-1-phosphate (1), which is converted to TDP-d-glucose (2) by a thymidylyltransferase, then to TDP-4-keto-6-deoxy-d-glucose (3), the key intermediate for biosynthesis of all TDP-sugars, by a TDP-d-glucose 4,6-dehydratase.[2] In the MTM pathway, these two enzymes were identified as MtmD and MtmE.[9,25] Their functions had been verified in vivo by gene inactivation and incorporation in several deoxysugar biosynthetic cassettes.[21,26] Due to poor expression levels of soluble MtmD and MtmE in Escherichia coli, well-characterized homologues RfbA from E. coli and RmlB from Salmonella typhimurium were used for the in vitro large-scale preparation of intermediate 3 (Scheme 2A).[27]
Scheme 2.
Biosynthetic pathway of MTM deoxysugars. A) Conversion of 1 via 2 to 3 was performed as previously reported.[27] B) Conversion of 3 to 6 was investigated in this study.
TDP-4-keto-2,6-dideoxy-d-glucose (5) was suggested to be a common intermediate for the biosynthesis of all MTM deoxysugars. It is biosynthesized from 3 through the concerted action of a TDP-4-keto-6-deoxy-d-glucose-2,3-dehydratase and a 3-ketoreductase (Scheme 2B). These two enzymes must work together, since the transient product 4 can easily undergo degradation to TDP and maltol.[28,29] Finally, a 4-ketoreductase needs to reduce the 4-keto group of 5 to yield TDP-d-olivose (6). In the MTM gene cluster, mtmV encodes the TDP-4-keto-6-deoxy-d-glucose-2,3-dehydratase;[20] however, genes encoding the 3- and 4-ketoreductases have not yet been identified. It was believed that these functions were encoded by genes elsewhere in the genome of the MTM producer Streptomyces argillaceus. Since repeated efforts failed to obtain soluble MtmV protein in E. coli, its homologue OleV, as well as a 3-ketoreductase OleW involved in l-oleandrose biosynthesis from the oleandomycin pathway[30] were used in this study. OleV and OleW together catalyzed the conversion of 3→5, yielding the desired equatorial 3-OH group.[31] Efficient generation of 5 from 3 was observed by HPLC (Figure 1). Compound 5 was purified and its structure was confirmed by 1H NMR and 1H,1H COSY spectroscopy and MS (see the Supporting Information). To lower the costs of a large-scale preparation of 5, an in situ NADPH regeneration system consisting of glucose-6-phosphate and E. coli-derived glucose-6-phosphate dehydrogenase (G6PD) was utilized.[32]
Figure 1.
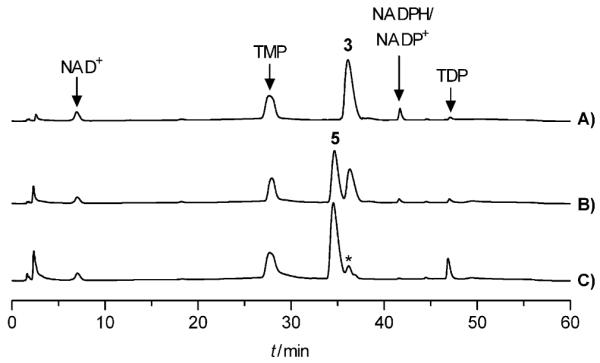
Enzymatic generation of 5 by incubating 3 with OleV and OleW. Aliquots (50 μL) were sampled at different time points and analyzed by HPLC. A) Zero time point; B) 1 h; C) overnight. The peak marked with an asterisk is ADP (product from the TTP regeneration system, see Scheme S1 in the Supporting Information).
The remaining deoxysugar biosynthesis genes identified in the MTM gene cluster are mtmU, mtmC and mtmTIII. Inactivation of the individual genes indicated that MtmU and MtmTIII are both 4-ketoreductases, which reduce the 4-keto group of 5 and 8 to form TDP-d-oliose (7) and TDP-d-mycarose (9), respectively.[19,20] Since the candidate enzyme responsible for 5→6 conversion was missing in the cluster, UrdR was used, which had been suggested by in vivo studies to be a 4-ketoreductase involved in TDP-d-olivose biosynthesis in the urdamycin pathway.[31,33] This enzyme was over-expressed in E. coli and subsequently purified. However, no obvious production of 6 was observed when recombinant UrdR was incubated either with 3, OleV, OleW and NADPH, or with 5 and NADPH. Instead, several new peaks pertaining to a typical thymidine chromophore were observed, indicating a decomposition of starting substrate.
To our surprise, the missing 4-ketoreductase activity for the TDP-d-olivose biosynthesis was identified in the MTM pathway during the parallel work on developing an in vitro preparation system for TDP-d-mycarose (9), for which we explored MtmC. Previous homology searches revealed that MtmC shares high sequence similarity with many S-adenosyl methionine (SAM)-dependent C-methyltransferases involved in deoxysugar biosynthesis, such as SnoG (62.2% of identity) from the nogalamycin pathway, TylCIII (32.4%) from the tylosin pathway and EryBIII (31%) from the erythromycin pathway;[34-37] this suggests that MtmC is a C-methyltransferase and is responsible for the transfer of a methyl group from SAM onto 3-position of 5 to yield 8, the precursor of 9. Gene inactivation results also validated the indispensible role of MtmC in the biosynthesis of d-mycarose.[19,20] The recombinant protein MtmC was heterologously produced in E. coli and purified to near homogeneity. Incubation of 5 with MtmC and SAM resulted in the complete degradation of the sugar substrate and formation of TDP (Figure S2 in the Supporting Information). However, once MtmC was incubated with an enzyme mixture containing OleV, OleW, 3, SAM and NADPH, a new peak at 35.1 min was observed (Figure 2A). The results indicate that MtmC might be able to catalyze two steps in TDP-d-mycarose biosynthesis in a concerted manner: 1) conversion of 5→8 utilizing SAM as methyl donor, and 2) conversion of 8→9 utilizing NADPH. Thereafter, OleV and OleW were excluded from the enzyme mixture. Instead, pure 5 was incubated with MtmC and various combinations of co-enzymes and co-factors, namely SAM+NADPH+NADP+, NADPH+NADP+ and NADPH or NADP+ alone. Interestingly, a new peak was observed under all conditions, except NADP+ alone (Figure 2C–F), regardless of the presence of SAM. These results indicate that SAM is not necessary for the observed conversion, excluding any methylation event, while NADPH is indispensable for the catalysis. The assay was reproduced on a large scale and the product was purified and characterized. The 1H NMR, 1H–1H COSY spectroscopy and MS data surprisingly confirmed the new peak to be TDP-d-olivose (6; see the Supporting Information). The presence of an additional triplet at δ=3.10 clearly proved the existence of an axial 4″-H in 6 and thus clearly differentiated 6 from 5 or 8.
Figure 2.
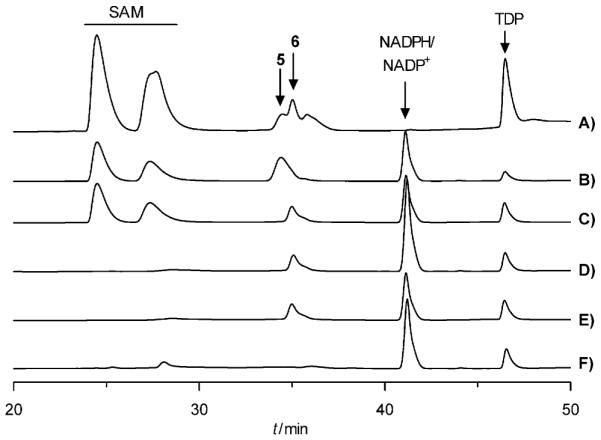
HPLC profiles of MtmC incubated with either A) an enzyme mixture containing OleV, OleW, 3, SAM and NADPH, or pure 5 plus, C) SAM+-NADPH+NADP+, D) NADPH+ NADP+, E) NADPH only, and F) NADP+ only. B) Control containing only sugar substrate 5 and co-enzymes.
Thereafter, large-scale enzymatic preparation of 6 was developed. Purified compound 3 was incubated with enzymes OleV and OleW and the in situ NADPH regeneration system. Complete conversion of 3 to 5 was achieved usually within 2–3 h, as monitored by HPLC. After removal of the proteins by filtration, MtmC as well as G6PD were added to the mixture and incubated overnight. Complete conversion of 5→6 was confirmed by HPLC. Finally, product 6 was purified by using a Bio-Gel P2 column (2 cm × 1000 cm).[18,27]
In conclusion, we have identified the 4-ketoreductase that is responsible for the 4-ketoreduction step of TDP-d-olivose (6) biosynthesis, necessary for the biosynthesis of the d-olivose moieties of MTM, particularly of sugar C.[19] Despite its strong amino acid sequence similarity with only SAM-dependent C-methyltransferases, the isolated enzyme catalyzes the conversion of 5→6 utilizing NADPH as the co-enzyme. This again provides an example that amino acid sequence similarity of an enzyme might not always be sufficient to identify the inherent activity. A large-scale enzymatic preparation of 6 was established. The overall production yield was approximately 70%. This is so far the first report of total enzymatic synthesis of TDP-d-olivose (6), including the first usage of the purified enzyme pair OleV/OleW for the 2-deoxygenation step. Although, a few chemical syntheses of 6 were reported previously,[38,39] the better yield and higher efficiency as well as the environmentally friendly procedure of this enzymatic synthesis make it more appropriate for routine production of TDP-d-olivose. The application of 6 in the in vitro studies of MTM glycosylation steps is currently ongoing in our laboratory.
Experimental Section
All enzymes used in this work were over-expressed in E. coli BL21(DE3), and purified by using immobilized metal affinity chromatography. All enzymatic reactions were carried out at 28°C. For detailed procedures see the Supporting Information.
Supplementary Material
Acknowledgements
This work was supported by the US National Institutes of Health grant CA 091901 to J.R.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201100540.
References
- [1].Thorson JS, Hosted TJ, Jiang JQ, Biggins JB, Ahlert J. Curr. Org. Chem. 2001;5:139. [Google Scholar]
- [2].Thibodeaux CJ, Melancon CE, 3rd, Liu HW. Angew. Chem. 2008;120:9960. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:9814. [Google Scholar]
- [3].La Ferla B, Airoldi C, Zona C, Orsato A, Cardona F, Merlo S, Sironi E, D’Orazio G, Nicotra F. Nat. Prod. Rep. 2011;28:630. doi: 10.1039/c0np00055h. [DOI] [PubMed] [Google Scholar]
- [4].Rohr J, Méndez C, Salas JA. Bioorg. Chem. 1999;27:41. [Google Scholar]
- [5].Lombó F, Menendez N, Salas JA, Mendez C. Appl. Microbiol. Biotechnol. 2006;73:1. doi: 10.1007/s00253-006-0511-6. [DOI] [PubMed] [Google Scholar]
- [6].Krohn K, Rohr J. Topics in Current Chemistry. Vol. 188. Springer; Berlin: 1997. p. 127. [Google Scholar]
- [7].Ostash B, Korynevska A, Stoika R, Fedorenko V. Mini Rev. Med. Chem. 2009;9:1040. doi: 10.2174/138955709788922593. [DOI] [PubMed] [Google Scholar]
- [8].Rohr J, Thiericke R. Nat. Prod. Rep. 1992;9:103. doi: 10.1039/np9920900103. [DOI] [PubMed] [Google Scholar]
- [9].Lombó F, Olano C, Salas JA, Méndez C. Methods Enzymol. 2009;458:277. doi: 10.1016/S0076-6879(09)04811-3. [DOI] [PubMed] [Google Scholar]
- [10].Thibodeaux CJ, Melançon CE, Liu HW. Nature. 2007;446:1008. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- [11].Olano C, Méndez C, Salas JA. Nat. Prod. Rep. 2010;27:571. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- [12].Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat. Biotechnol. 2003;21:1467. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- [13].Hoffmeister D, Wilkinson B, Foster G, Sidebottom PJ, Ichinose K, Bechthold A. Chem. Biol. 2002;9:287. doi: 10.1016/s1074-5521(02)00114-x. [DOI] [PubMed] [Google Scholar]
- [14].Trefzer A, Blanco G, Remsing L, Künzel E, Rix U, Lipata F, Braña AF, Méndez C, Rohr J, Bechthold A, Salas JA. J. Am. Chem. Soc. 2002;124:6056. doi: 10.1021/ja017385l. [DOI] [PubMed] [Google Scholar]
- [15].Härle J, Bechthold A. Methods Enzymol. 2009;458:309. doi: 10.1016/S0076-6879(09)04812-5. [DOI] [PubMed] [Google Scholar]
- [16].Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Glycobiology. 2006;16:29R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- [17].Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu. Rev. Biochem. 2008;77:521. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- [18].White-Phillip J, Thibodeaux CJ, Liu HW. Methods Enzymol. 2009;459:521. doi: 10.1016/S0076-6879(09)04621-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Remsing LL, Garcia-Bernardo J, Gonzalez A, Künzel E, Rix U, Braña AF, Bearden DW, Mendez C, Salas JA, Rohr J. J. Am. Chem. Soc. 2002;124:1606. doi: 10.1021/ja0105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gonzalez A, Remsing LL, Lombo F, Fernandez MJ, Prado L, Braña AF, Künzel E, Rohr J, Méndez C, Salas JA. Mol. Gen. Genet. 2001;264:827. doi: 10.1007/s004380000372. [DOI] [PubMed] [Google Scholar]
- [21].Lombó F, Gibson M, Greenwell L, Braña AF, Rohr J, Salas JA, Méndez C. Chem. Biol. 2004;11:1709. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- [22].Blanco G, Fernandez E, Fernandez MJ, Braña AF, Weissbach U, Künzel E, Rohr J, Méndez C, Salas JA. Mol. Gen. Genet. 2000;262:991. doi: 10.1007/pl00008667. [DOI] [PubMed] [Google Scholar]
- [23].Fernandez E, Weissbach U, Sanchez Reillo C, Braña AF, Méndez C, Rohr J, Salas JA. J. Bacteriol. 1998;180:4929. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nur-e-Alam M, Méndez C, Salas JA, Rohr J. ChemBioChem. 2005;6:632. doi: 10.1002/cbic.200400309. [DOI] [PubMed] [Google Scholar]
- [25].Lombo F, Siems K, Braña AF, Méndez C, Bindseil K, Salas JA. J. Bacteriol. 1997;179:3354. doi: 10.1128/jb.179.10.3354-3357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Perez M, Baig I, Braña AF, Salas JA, Rohr J, Méndez C. ChemBioChem. 2008;9:2295. doi: 10.1002/cbic.200800299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kharel MK, Lian H, Rohr J. Org. Biomol. Chem. 2011;9:1799. doi: 10.1039/c0ob00854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dräger G, Park SH, Floss HG. J. Am. Chem. Soc. 1999;121:2611. [Google Scholar]
- [29].Chen HW, Agnihotri G, Guo ZH, Que NLS, Chen MH, Liu HW. J. Am. Chem. Soc. 1999;121:8124. [Google Scholar]
- [30].Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Braña AF, Méndez C, Salas JA. Antimicrob. Agents Chemother. 2000;44:1266. doi: 10.1128/aac.44.5.1266-1275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rodriguez L, Aguirrezabalaga I, Allende N, Braña AF, Méndez C, Salas JA. Chem. Biol. 2002;9:721. doi: 10.1016/s1074-5521(02)00154-0. [DOI] [PubMed] [Google Scholar]
- [32].Wong CH, Whitesides GM. J. Am. Chem. Soc. 1981;103:4890. [Google Scholar]
- [33].Hoffmeister D, Ichinose K, Domann S, Faust B, Trefzer A, Dräger G, Kirschning A, Fischer C, Künzel E, Bearden D, Rohr J, Bechthold A. Chem. Biol. 2000;7:821. doi: 10.1016/s1074-5521(00)00029-6. [DOI] [PubMed] [Google Scholar]
- [34].Bate N, Butler AR, Smith IP, Cundliffe E. Microbiology. 2000;146:139. doi: 10.1099/00221287-146-1-139. [DOI] [PubMed] [Google Scholar]
- [35].Gaisser S, Bohm GA, Doumith M, Raynal MC, Dhillon N, Cortes J, Leadlay PF. Mol. Gen. Genet. 1998;258:78. doi: 10.1007/s004380050709. [DOI] [PubMed] [Google Scholar]
- [36].Torkkell S, Ylihonko K, Hakala J, Skurnik M, Mäntsälä P. Mol. Gen. Genet. 1997;256:203. doi: 10.1007/s004380050562. [DOI] [PubMed] [Google Scholar]
- [37].Chen H, Zhao Z, Hallis TM, Guo Z, Liu HW. Angew. Chem. 2001;113:627. [Google Scholar]; Angew. Chem. Int. Ed. 2001;40:607. [Google Scholar]
- [38].Amann S, Dräger G, Rupprath C, Kirschning A, Elling L. Carbohydr. Res. 2001;335:23. doi: 10.1016/s0008-6215(01)00195-1. [DOI] [PubMed] [Google Scholar]
- [39].Minami A, Eguchi T. J. Am. Chem. Soc. 2007;129:5102. doi: 10.1021/ja0685250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.