Abstract
Idiopathic pulmonary fibrosis (IPF) is a complex lung disease of unknown etiology. Development of IPF is influenced by both genetic and environmental factors. Gene-expression profiling studies have taught us quite a bit about the biology of this fatal disease, but epigenetic marks may be the missing link that connects the environmental exposure in genetically predisposed individuals to transcriptome changes associated with the development of IPF. This review will begin with an introduction to the disease, followed by brief summaries of studies of gene expression in IPF and epigenetic marks associated with exposures relevant to IPF. The majority of the discussion will focus on epigenetic studies conducted so far in IPF, the limitations, challenges and future directions in this field.
Keywords: cigarette smoke exposure, DNA methylation, gene expression, histone modifications, idiopathic pulmonary fibrosis, interstitial pneumonia, miRNAs
Idiopathic pulmonary fibrosis
Pulmonary fibrosis defines a group of fibrosing interstitial lung diseases that can result from environmental exposures (asbestos or silica), connective tissue disease, drug toxicity, or occur as idiopathic pulmonary fibrosis (IPF), in which case the cause is unknown [1]. IPF is a fatal lung disease with a median survival of only 3 years that is characterized by progressive scarring of the pulmonary parenchyma, leading to progressive loss of lung function with dyspnea and hypoxemia, and ultimately respiratory failure and death. The prevalence of IPF is estimated to occur in 14 to 42.7 individuals out of 100,000 [2], with the prevalence and mortality of pulmonary fibrosis increasing [3]. Histologically, IPF is characterized by usual interstitial pneumonia, a fibrosing interstitial pneumonia with a pattern of heterogeneous, subpleural regions of fibrotic and remodeled lung [4]. Development of fibroblastic foci, areas of active fibroproliferation, is a hallmark feature of IPF. Fibroblastic foci consist largely of myofibroblasts, cells that have features of both fibroblasts and smooth muscle cells, are involved in wound healing, and whose differentiation is induced by TGF-β1. Myofibroblasts are the main source of excessive extracellular matrix production in IPF. Another hallmark pathological feature of IPF is microscopic honeycombing, areas of normal lung tissue adjacent to areas of advanced remodeling [5].
The paradigm about disease pathogenesis in IPF has shifted over the years from thoughts that IPF is a result of chronic inflammation to the idea that it results from alveolar epithelial cell injury and subsequent dysregulated repair as well as enhanced epithelial apoptosis. However, it is becoming increasingly clear that the disease process underlying the IPF phenotype is heterogeneous and many different molecular processes may be involved. These include epithelial mesenchymal transition (EMT) [6–8], growth factor regulation [9], apoptosis [10,11], oxidative stress [12], endoplasmic reticulum stress [13,14], cellular senescence associated with aging/telomere shortening [15–17], epithelial stem cell exhaustion [18], intra-alveolar coagulation [19] and potentially impaired mucociliary clearance [20,21].
The disease is likely the result of complex interactions between genetic [20,22,23] and environmental factors such as cigarette smoking and wood or metal dust [23,24], or comorbidities such as gastroesophageal reflux and Type 2 diabetes [25] being risk factors for the development of IPF. Animal models of pulmonary fibrosis recapitulate some but not all pathological features of disease [26]. The bleomycin model of lung injury and subsequent fibrosis in mice has been the most commonly used model to decipher the role of specific genetic factors in the development of the disease.
Gene-expression studies in IPF
Gene-expression profiling studies have demonstrated that transcriptional changes are present in the lung parenchyma of individuals with IPF [27–33]. Gene-expression changes are quite dramatic and involve large numbers of genes, generally around the order of a few thousand differentially expressed genes. In aggregate, these studies have consistently identified similar genes and pathways that are differentially expressed in fibrotic lungs, namely, genes associated with extracellular matrix formation, degradation and signaling, smooth muscle markers, growth factors, and genes encoding immunoglobulins, complements, and chemokines. Some of these studies have successfully identified transcriptional profiles associated with rapid disease progression and acute exacerbations in IPF [28,29,33]. More recent work from our laboratory also identified a subset of IPF subjects with increased expression of cilium genes that is also associated with more extensive microscopic honeycombing, higher expression of the airway mucin gene MUC5B, and better survival in an independent cohort of IPF patients.
Role of environmental exposures in pathophysiology of IPF
Epidemiological studies have shown associations of exposures to inhaled environmental agents and the development of IPF [24,25]. These studies have largely been of case–control design and demonstrate only an association and not causality. Cigarette smoke is the most prevalent exposure that has been linked to the development of IPF. A number of case–control studies, reviewed by Ding et al., have shown a positive association between ever smoking, and specifically former smoking in some studies, and the development of IPF [25]. Smoking history has also been associated with poorer survival with two of these studies demonstrating poorer prognosis for former than current smokers [34–36]. Case– control studies have also established a link between occupational exposure and the development of IPF, including exposure to wood dust, metal dust, silica, textile dust, and possibly agriculture, farming and livestock [24,25]. It is important to note that many individuals with no smoking history or relevant occupation exposure develop IPF, highlighting the fact that genetic predisposition is also a critical factor in the development of this disease.
Modulation of epigenetic marks by environmental exposures
Unlike an individual’s genetic make-up, epigenetic marks can be influenced by exposures, diet and aging. Randy Jirtle’s seminal experiments showed that maternal diet supplemented with methyl donors (folic acid, vitamin B12, choline and betaine) shifts coat color distribution of progeny towards the brown pseudoagouti phenotype, and that this shift in coat color resulted from an increase in DNA methylation in a transposon adjacent to the agouti gene [37,38]. These studies also revealed that mice with yellow coat color are obese and are more prone to develop cancer, suggesting for the first time that changes in DNA methylation caused by diet may be linked to disease development. Other studies have since shown that exposures such as pesticides and fungicides [39] and PM2.5 particles [40] could alter the methylome, and that aging is also associated with changes in DNA methylation and gene expression [41].
Early studies demonstrated the link between exposure to tobacco smoke and lung cancer via methylation of CpG islands associated with cancer genes such as p16 [42]. Several more recent studies have examined the relationship between exposure to cigarette smoke and epigenetic marks in the context of exposure itself and not linked to any disease [43–45]. Cigarette smoke exposure has also been shown to have a significant influence on expression of miRNAs in human bronchial epithelial cells [46], mouse [47] and rat [48] lungs exposed to cigarette smoke. All three studies showed the predominant effect of smoke exposure is downregulation of miRNAs, with substantial overlap between mice and rats and some overlap of rodent miRNA expression changes in the lung with those observed in human airway epithelium. However, the mechanisms linking cigarette smoke to any of these epigenetic changes have not been clearly defined; thus, raising uncertainty about the cause and effect relationship between cigarette smoke and epigenetic marks. Despite some of the similarities in epigenomic profiles of cigarette smoke between human samples and animal models, there is not enough evidence at this point to support the use of animal models of smoke exposure in epigenomic studies of IPF. Finally, in utero exposure to cigarette smoke results in differential methylation [49–51] and downregulation of miRNAs [52] in the placenta, cord blood or peripheral blood of children, suggesting transgenerational effects of smoke exposure.
Role of epigenetic regulation of the immune system
A large body of evidence suggests that epigenetic mechanisms affect the expression of cytokines and binding of transcription factors that control the lineage of Th1, Th2, Treg and Th17 cells [53–58]. Although chronic inflammation may not be as important in disease pathogenesis as it was once assumed, the immune and inflammatory systems are still thought to play a role in the development of IPF. Early studies demonstrated that mononuclear cells were the predominant cell type in interstitial infiltrates from patients with IPF [59] and that CD4 T cells from peripheral blood of patients with IPF had characteristics typical of cell-mediated pathological response [60]. More recent studies have demonstrated global Treg impairment in IPF that strongly correlates with disease severity [61] and an association of CD28 downregulation on circulating CD4 T cells with a poor prognosis in patients with IPF [62]. Therefore epigenetic marks of immune cells may prove to have an important role in the development of IPF.
Epigenetic studies in IPF
Epigenetic mechanisms are likely to be involved in the control of gene expression in IPF, especially given the association of IPF with cigarette smoking and the relationship between cigarette smoke and changes in DNA methylation, histone modifications and miRNAs. Moreover, these epigenetic changes are likely to be important factors in determining transcriptional profiles that directly contribute to pathogenic features of this disease (Figure 1). However, it is important to remember that epidemiological studies that have linked cigarette smoke exposures to disease development have only shown associations and not causality.
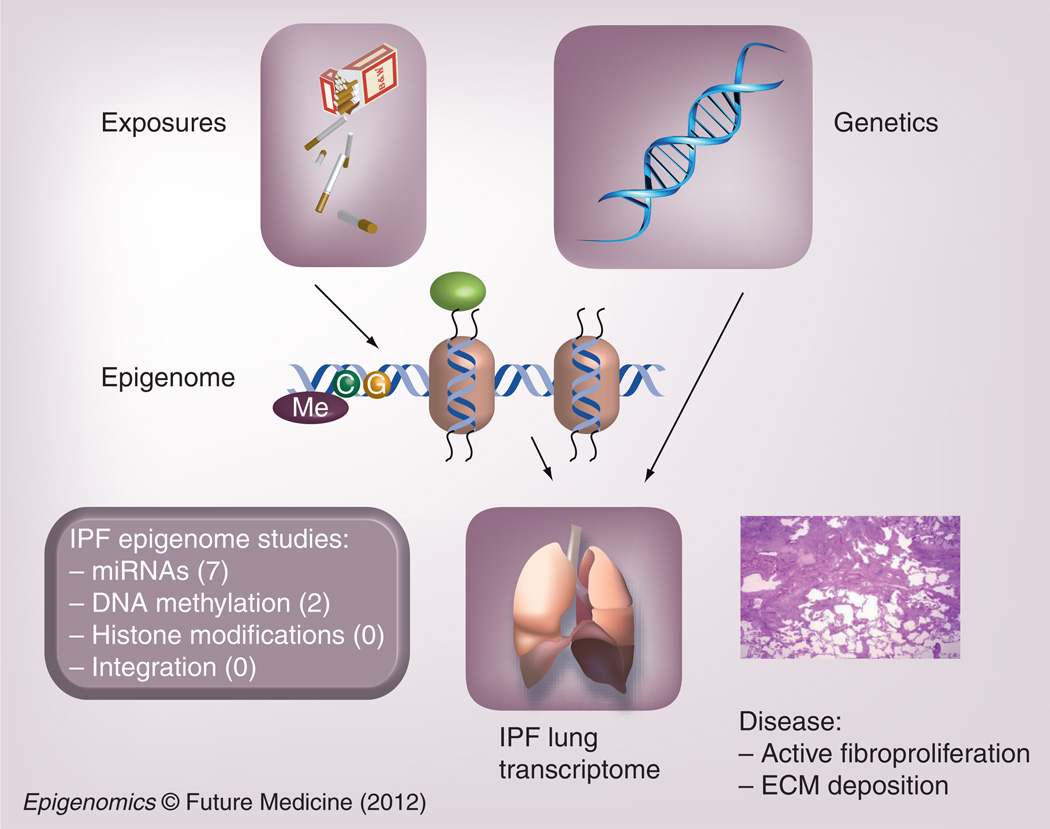
Figure 1. The idiopathic pulmonary fibrosis transcriptome is influenced by both environmental and genetic factors.
The epigenome links environmental exposures to gene-expression changes that lead to disease development. A number of genome-wide miRNA studies in IPF have been published, while DNA methylation and histone modification studies on the genomic scale are just emerging in IPF. ECM: Extracellular matrix; IPF: Idiopathic pulmonary fibrosis; Me: Methyl.
Targeted studies
Several targeted studies have shown that epigenetic modulation regulates expression of genes involved in the pathogenesis of IPF. Defective histone acetylation is responsible for the repression of expression of two antifibrotic genes, COX2 [63] and chemokine IP-10 [64]. Similarly, Thy-1 (CD90) is an important regulator of cell–cell and cell–matrix interactions that is expressed on normal lung fibroblasts but its expression is absent in myofibroblasts within fibroblastic foci in IPF. Thy-1 downregulation in rat lung fibroblasts is controlled by both promoter DNA hypermethylation [65] and histone modifications [66]. Different levels of methylation of three CpG islands in the promoter of the α-smooth muscle actin (α-SMA) in fibroblasts, myofibroblasts, and alveolar epithelial type II cells were shown to correlate with expression of the α-SMA gene in these different cell types [67]. This study also demonstrated that pharmacological- and siRNA-mediated inhibition of DNA methyltransferase activity induced expression of α-SMA in fibroblasts while overexpression of DNA methyltransferase suppressed α-SMA gene expression. Inhibition or overexpression of DNA methyltrasnferase also affected TGF-β1-induced myofibroblast differentiation. A more recent study from the same group showed that MeCP2 binds to the α-SMA gene [68], and that suppression or overexpression of MeCP2 leads to changes α-SMA gene expression in fibroblasts. Furthermore, MeCP2-deficient mice exhibited a significantly decreased alveolar wall thickness, inflammatory cell infiltration, interstitial collagen deposition and myofibroblast differentiation in response to bleomycin. Taken together, these data strongly suggest DNA methylation as an important mechanism that regulates expression of the α-SMA and fibroproliferation.
Epigenomic profiles
Epigenomic studies of DNA methylation profiles in IPF are just emerging. Technologies for collecting epigenomic profiles are either array- or next-generation sequencing-based and have been reviewed elsewhere [69–71], although no study to date in IPF has employed next-generation sequencing. Naftali Kaminski’s laboratory has shown that 625 CpG islands are differentially methylated between IPF (n = 12) and control (n = 10) lungs [72]. Comparison of IPF methylation patterns to lung cancer (n = 10) revealed that IPF lungs display an intermediate methylation profile, partly similar to lung cancer with 402 differentially methylated CpG islands overlapping between IPF and cancer. The Lung Genomics Research Consortium (LGRC [101]) has profiled lung tissue DNA from 100 individuals with IPF and 79 controls using Comprehensive High-Throughput Arrays for Relative Methylation (CHARM) [73,74]. Our results revealed that the majority of differentially methylated regions are hypomethylated in disease, located within CpG island ‘shores’ (areas of lower CpG density near CpG islands) and contained within genes [75]. These findings are different from studies in the lung cancer field that have focused on CpG islands in promoter regions of genes as main methylation sites that control gene expression; we are in the process of correlating genome-wide DNA methylation patterns with global changes in gene expression in IPF lungs. Pathway analysis of genes located within 2 kb from a differentially methylated region identified pathways associated with cancer as well as IPF (e.g., Wnt/β-catenin signaling) [76–78]. To date, no genome-wide histone modification studies have been done in IPF. This is largely due to the fact that it is difficult to obtain fresh cells to perform chromatin immunoprecipitation.
A number of very recent publications studied genomic miRNA profiles in lung tissue from patients with IPF and identified several miRNAs with role in fibroproliferation, EMT, and the TGF-β1 signaling pathway [79–83]. Similar to extensive transcriptional changes in IPF lungs, 10% of miRNAs are differentially expressed in lung tissue from subjects with IPF compared with nondisease controls. Among downregulated miRNAs in IPF are let-7d and miR-29, while miR-155 and miR-21 are upregulated in IPF.
Given the prominent role of TGF-β1 signaling in fibroproliferation, Pandit et al. scanned promoters of differentially expressed miRNAs in IPF lungs for SMAD binding elements and focused on one of the miRNAs whose promoter contains SMAD binding elements, let-7d [79]. They showed that TGF-β1 downregulated let-7d expression, and SMAD3 binding to the let-7d promoter was demonstrated. Inhibition of let-7d in vitro and in vivo by an antagomir for the let-7 family resulted in upregulation of mesenchymal and downregulation of epithelial markers, suggesting a role for the let-7 family of miRNAs in prevention of EMT and profibrotic phenotype.
Two studies utilized the bleomycin model of lung fibrosis to identify miRNAs that play important roles in fibroproliferation. Cushing et al. examined the expression of miRNAs in lungs of bleomycin-treated mice and demonstrated reduced expression of miR-29 in response to bleomycin [81]. Inhibition of miR-29 in human fetal lung fibroblasts led to upregulation of a number of genes associated with fibrotic phenotype including its predicted targets, genes upregulated by TGF-β1 as well as genes independent of TGF-β1, including laminins and integrins. Although the authors did not examine expression of miR-29 in human lung tissue, this miRNA is downregulated in IPF lungs in the dataset from Pandit et al. [79]. These data suggest a prominent role for miR-29 in lung fibrosis by regulation of expression of TGF-β1-inducible or other fibrotic genes. The study by Liu et al. [80] established upregulation of miR-21 in the lungs of mice treated with bleomycin and in the lungs of patients with IPF with expression primarily localized to myofibroblasts. Inhibition of miR-21 expression diminished the severity of bleomycin-induced lung fibrosis in mice, while TGF-β1 enhanced miR-21 expression in primary lung fibroblasts. Overexpression of miR-21 promoted the profibrogenic activity of TGF-β1 in fibroblasts, suggesting a feed-forward loop in which miR-21 amplifies TGF-β1 signaling and fibrosis [84].
In addition to the bleomycin model, cultured human lung fibroblasts have been used to identify novel miRNAs. Pottier et al. examined the expression profiles of miR-155 in human lung fibroblasts stimulated with different cytokines and showed that upregulation of miR-155 correlated with the downregulation of a number of its target genes and that transfection of miR-155 led to fibroblast migration [82]. Among fibroblast-selective targets was keratinocyte growth factor (KGF, FGF-7F); functional in vitro assays experimentally validated that miR-155 can efficiently target KGF 3´-UTR. The authors also demonstrated increased expression of miR-155 in lungs of mice with bleomycin-induced fibrosis, a finding that is also supported by the fact that miR-155 is upregulated in IPF lungs in the study by Pandit et al. [79]. A recent re-analysis of published miRNA data and TargetScan-predicted targets identified a network of dysregulated-miRNAs and their direct targets that belong to pathways associated with IPF (Figure 2) [84].
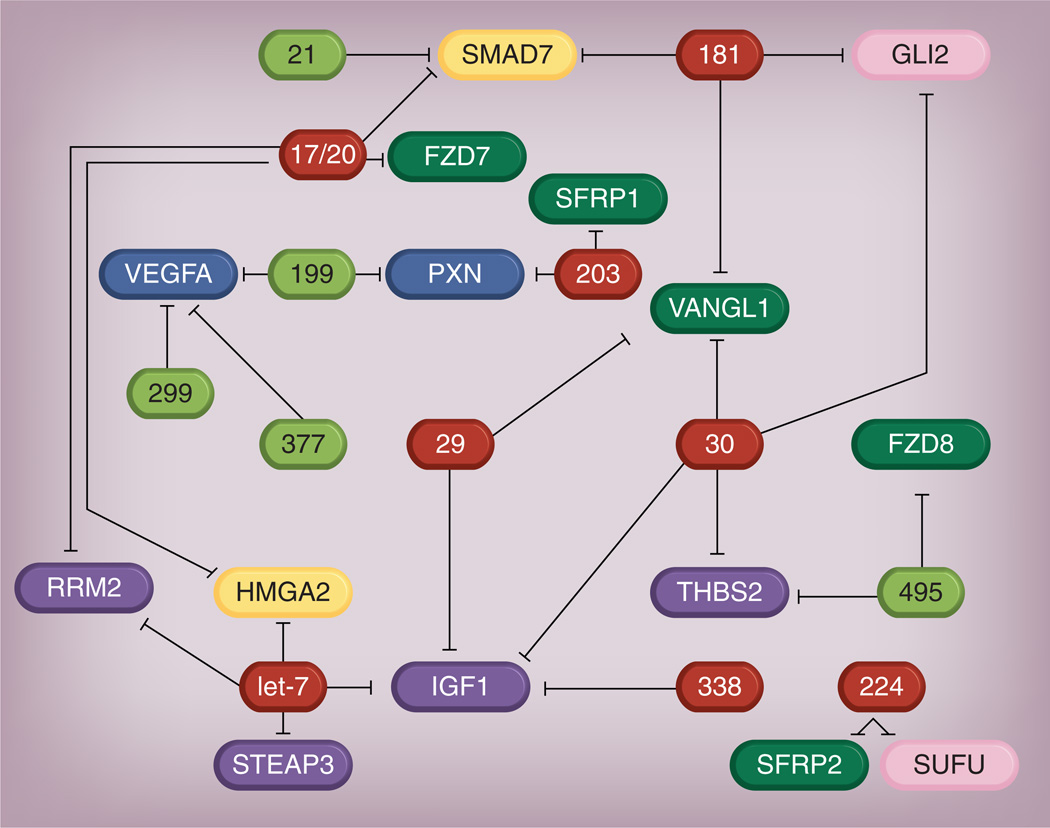
Figure 2. Network of dysregulated miRNAs regulating differentially expressed genes in idiopathic pulmonary fibrosis.
The genes and miRNAs above are differentially expressed in idiopathic pulmonary fibrosis. The black lines represent mRNA–miRNA interactions as predicted by TargetScan. The dark green boxes indicate upregulated miRNAs; the red boxes indicate downregulated miRNAs. The yellow boxes indicate TGF-β1 pathway genes; the purple boxes indicate IGF pathway genes; the blue boxes are VEGF pathway genes; the pink boxes indicate sonic hedgehog pathway genes; the light green boxes are Wnt pathway genes.
Reproduced with permission from [92].
Finally, miRNA expression in IPF lungs has also been correlated with disease severity; five miRNAs (miR-302c, miR-423, miR-210, miR-376c and miR-185) are differentially expressed in lung biopsies of rapidly versus slowly progressing IPF patients [83]. Further studies are needed to provide a functional link between differential expression of these miRNAs and IPF phenotypes.
Conclusion
Evidence for the role of epigenetic regulation of gene expression in the development of IPF is based on studies demonstrating the association of epigenetic marks with exposures such as cigarette smoke and targeted studies of epigenetic marks in specific genes relevant to the profibrotic phenotype. These early studies provide strong support for studies of the IPF epigenome. A few studies to date have examined genome-wide DNA methylation patterns, while genomic studies of miRNA expression patterns have identified specific miRNA and networks of miRNAs that regulate expression of key genes involved in pathogenesis of IPF. Genomic studies of histone modifications in IPF are lacking at the present time.
Future perspective
All epigenomic studies published to date in IPF have used array technologies to assess genome-wide methylation or miRNA patterns. Next-generation sequencing technologies are rapidly becoming more affordable and are likely to be employed in the next wave of studies of epigenetic marks in IPF. They will provide not only better coverage and more accurate data for known epigenetic marks, but they will allow for discovery of novel marks; this is especially true in the area of ncRNAs where many short and long ncRNAs are yet to be identified.
There are several challenges that we will be facing in understanding the role of epigenetic regulation of gene expression in chronic lung diseases such as IPF. First, epigenetic marks are cell-type specific, yet much of the research in this area has been done on the whole-lung tissue because isolation of enough material for specific cell types is often not feasible in human subjects. One approach to address this concern may be to identify epigenetic marks in the whole lung and then attribute them to specific types using immunohistochemistry or confocal microscopy with antibodies to specific epigenetic marks. The disadvantage of this approach is that many important epigenetic marks may be missed in the initial screen because the change in the whole lung may be below detection limits of assays utilized. Another approach is to isolate specific cell types from fresh lung biopsies, which presents feasibility challenges for large-scale studies. This same issue arises in gene-expression data and a recent publication suggested a method to decompose whole-tissue expression into cell-specific components [85]; this approach may prove useful in identifying cell-specific epigenetic marks in complex tissues. The second challenge is the dynamic nature of epigenetic marks; the epigenome needs to be considered in the context of other diseases, exposures, diet and age. Another major challenge with epigenomics will be to integrate the epigenetic mechanisms that affect transcription and translation. For example, evidence for crosstalk between DNA methylation and histone modifications has been rapidly accumulating [86–89]. Similarly, DNA methylation controls expression of miRNAs [90]. Moreover, ncRNAs such as lncRNAs and others are another piece of the epigenetic machinery whose role in regulation of gene expression is just emerging [91], but will need to be considered in future studies. Each of the three epigenetic mechanisms is independently complex but when combined, the complexity of these interactions presents unique experimental and analytic challenges. Epigenetic mechanisms will also need to be considered in the context of genetic variation. Another issue that will need to be addressed is the emerging concept of allele-specific methylation and gene expression [92] where there is an allelic imbalance in expression due to a polymorphism or a DNA methylation mark on one of the DNA strands. Finally, both epigenetic marks themselves and spatial organization of the chromatin (proximity of chromosomal loci) that results from epigenetic modifications appear to be very dynamic [90]. In vivo imaging approaches that are currently under development will enable studies of dynamic chromatin structure [93]. Despite all these challenges, using epigenomic profiling to understand the dynamic biology in the lung, and applying this knowledge to the development of novel diagnostic and therapeutic approaches represent promising approaches for patients with IPF. This may be accomplished by drugs that manipulate DNA methylation state (e.g., 5-aza-2´-deoxycytidine), histone modifications (treatment with histone deacetylase inhibitors) or miRNAs (inhibition or overexpression). Many of these therapeutic approaches are either in use, such as 5-aza-2´-deoxycytidine and histone deacetylase inhibitors in cancer [94] all are being considered for treatment of other diseases [95,96]. Given some of the parallels between IPF and lung cancer, using knowledge from clinical trials of epigenetic-mark-modifying treatments in malignant disease may prove useful in developing therapeutic approaches for IPF.
Executive summary.
Idiopathic pulmonary fibrosis
-
▪
Idiopathic pulmonary fibrosis (IPF) is characterized by progressive scarring of the pulmonary parenchyma that leads to progressive loss of lung function and respiratory failure and death (median survival: 3 years).
-
▪
Pathological hallmarks of IPF are fibroblastic foci and microscopic honeycombing.
-
▪
Many molecular mechanisms have been implicated in the development of IPF.
Gene-expression studies in IPF
-
▪
IPF transcriptome consists of a few thousand differentially expressed genes with consistent changes in expression of genes associated with extracellular matrix formation, degradation and signaling, smooth muscle markers, growth factors and genes encoding immunoglobulins, complements and chemokines.
Role of environmental exposures in pathophysiology of IPF
-
▪
Inhaled environmental agents such as cigarette smoke, wood dust, metal dust, silica, textile dust and possibly agriculture, farming and livestock have been identified as risk factors for development of IPF.
-
▪
However, genetic factors also play a major role in IPF and it is thought that gene–environment interactions are critical in disease development and progression.
Modulation of epigenetic marks by environmental exposures
-
▪
Substantial evidence supports the relationship between exposure to cigarette smoke and epigenetic marks.
Role of epigenetic regulation of the immune system
-
▪
A large body of evidence suggests that epigenetic mechanisms affect the expression of cytokines and binding of transcription factors that control the lineage of Th1, Th2, Treg and Th17 cells.
-
▪
Although chronic inflammation may not be as important in disease pathogenesis as it was once assumed, the immune and inflammatory systems are still thought to play a role in the development of IPF.
Epigenetic studies in IPF
-
▪
Several targeted studies have shown that epigenetic modulation regulates expression of genes involved in pathogenesis of IPF.
-
▪
Epigenomic studies in IPF are just emerging and are limited to a few early DNA methylation studies and a number of recently published miRNA studies.
-
▪
miRNA studies have consistently identified let-7d, miR-29, miR-155 and miR-21 as key regulators of expression of fibrosis genes.
Conclusion
-
▪
Evidence for the role of epigenetic regulation of gene expression in the development of IPF is based on studies demonstrating an association of epigenetic marks with exposures such as cigarette smoke and targeted studies of epigenetic marks in specific genes relevant to the profibrotic phenotype.
-
▪
A few studies of the IPF epigenome have been published to date.
Future perspective
-
▪
Next-generation sequencing approaches are likely to be utilized in the next wave of epigenomic profiles in IPF.
-
▪
Epigenetic marks specific to immune cells may prove to have a role in the development of IPF.
-
▪
Challenges in this field will include determining cell-type specificity of epigenetic marks, accounting for the dynamic nature of epigenetic marks, and integration of epigenetic mechanisms that affect transcription and translation.
-
▪
Epigenetic marks provide targets for the development of novel diagnostic and therapeutic approaches in IPF.
Acknowledgments
This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Financial & competing interests disclosure
The author has no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 2001;345(7):517–525. doi: 10.1056/NEJMra003200. [DOI] [PubMed] [Google Scholar]
- 2.Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2006;174(7):810–816. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 3.Olson AL, Swigris JJ, Lezotte DC, Norris JM, Wilson CG, Brown KK. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am. J. Respir. Crit. Care Med. 2007;176(3):277–284. doi: 10.1164/rccm.200701-044OC. [DOI] [PubMed] [Google Scholar]
- 4.King T, Costabel U, Cordier J-F, et al. American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS) Am. J. Respir. Crit. Care Med. 2000;161(2 Pt 1):646–664. doi: 10.1164/ajrccm.161.2.ats3-00. [DOI] [PubMed] [Google Scholar]
- 5.King TE. In: Interstitial Lung Disease (5th Edition) King TE, Schwarz MI, editors. Shelton, CT, USA: People’s Medical Publishing House; 2010. pp. 895–943. [Google Scholar]
- 6.Willis BC, Liebler JM, Luby-Phelps K, et al. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am. J. Pathol. 2005;166(5):1321–1332. doi: 10.1016/s0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim KK, Kugler MC, Wolters PJ, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc. Natl Acad. Sci. USA. 2006;103(35):13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanjore H, Xu XC, Polosukhin VV, et al. Contribution of epithelial-derived fibroblasts to bleomycin-induced lung fibrosis. Am. J. Respir. Crit. Care Med. 2009;180(7):657–665. doi: 10.1164/rccm.200903-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farkas L, Gauldie J, Voelkel NF, Kolb M. Pulmonary hypertension and idiopathic pulmonary fibrosis: a tale of angiogenesis, apoptosis, and growth factors. Am. J. Respir. Cell Mol. Biol. 2011;45(1):1–15. doi: 10.1165/rcmb.2010-0365TR. [DOI] [PubMed] [Google Scholar]
- 10.Fattman CL. Apoptosis in pulmonary fibrosis: too much or not enough? Antioxid. Redox Signal. 2008;10(2):379–385. doi: 10.1089/ars.2007.1907. [DOI] [PubMed] [Google Scholar]
- 11.Thannickal VJ, Horowitz JC. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc. Am. Thorac. Soc. 2006;3(4):350–356. doi: 10.1513/pats.200601-001TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecker L, Vittal R, Jones T, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat. Med. 2009;15(9):1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson WE, Cheng DS, Degryse AL, et al. Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc. Natl Acad. Sci. USA. 2011;108(26):10562–10567. doi: 10.1073/pnas.1107559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanjore H, Cheng DS, Degryse AL, et al. Alveolar epithelial cells undergo epithelial-to-mesenchymal transition in response to endoplasmic reticulum stress. J. Biol. Chem. 2011;286(35):30972–30980. doi: 10.1074/jbc.M110.181164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsakiri KD, Cronkhite JT, Kuan PJ, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl Acad. Sci. USA. 2007;104(18):7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alder JK, Chen JJ, Lancaster L, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc. Natl Acad. Sci. USA. 2008;105(35):13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cronkhite JT, Xing C, Raghu G, et al. Telomere shortening in familial and sporadic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2008;178(7):729–737. doi: 10.1164/rccm.200804-550OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chilosi M, Doglioni C, Murer B, Poletti V. Epithelial stem cell exhaustion in the pathogenesis of idiopathic pulmonary fibrosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2010;27(1):7–18. [PubMed] [Google Scholar]
- 19.Scotton CJ, Krupiczojc MA, Konigshoff M, et al. Increased local expression of coagulation factor X contributes to the fibrotic response in human and murine lung injury. J. Clin. Invest. 2009;119(9):2550–2563. doi: 10.1172/JCI33288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seibold MA, Wise AL, Speer MC, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N. Engl. J. Med. 2011;364(16):1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boucher RC. Idiopathic pulmonary fibrosis – a sticky business. N. Engl. J. Med. 2011;364(16):1560–1561. doi: 10.1056/NEJMe1014191. [DOI] [PubMed] [Google Scholar]
- 22.Garcia CK. Idiopathic pulmonary fibrosis: update on genetic discoveries. Proc. Am. Thorac. Soc. 2011;8(2):158–162. doi: 10.1513/pats.201008-056MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seibold MA, Schwartz DA. The lung: the natural boundary between nature and nurture. Annu. Rev. Physiol. 2011;73:457–478. doi: 10.1146/annurev-physiol-012110-142212. [DOI] [PubMed] [Google Scholar]
- 24.Taskar VS, Coultas DB. Is idiopathic pulmonary fibrosis an environmental disease? Proc. Am. Thorac. Soc. 2006;3(4):293–298. doi: 10.1513/pats.200512-131TK. [DOI] [PubMed] [Google Scholar]
- 25.Ding Q, Luckhardt T, Hecker L, et al. New insights into the pathogenesis and treatment of idiopathic pulmonary fibrosis. Drugs. 2011;71(8):981–1001. doi: 10.2165/11591490-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chua F, Gauldie J, Laurent GJ. Pulmonary fibrosis: searching for model answers. Am. J. Respir. Cell Mol. Biol. 2005;33(1):9–13. doi: 10.1165/rcmb.2005-0062TR. [DOI] [PubMed] [Google Scholar]
- 27.Kaminski N. Microarray analysis of idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2003;29(Suppl. 3):S32–S36. [PubMed] [Google Scholar]
- 28.Konishi K, Gibson KF, Lindell KO, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2009;180(2):167–175. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selman M, Carrillo G, Estrada A, et al. Accelerated variant of idiopathic pulmonary fibrosis: clinical behavior and gene expression pattern. PLoS One. 2007;2(5):e482. doi: 10.1371/journal.pone.0000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selman M, Pardo A, Barrera L, et al. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am. J. Respir. Crit. Care Med. 2006;173(2):188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo F, Kaminski N, Eugui E, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc. Natl Acad. Sci. USA. 2002;99(9):6292–6297. doi: 10.1073/pnas.092134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang IV, Burch LH, Steele MP, et al. Gene expression profiling distinguishes familial and non-familial forms of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2005;175:45–54. doi: 10.1164/rccm.200601-062OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boon K, Bailey NW, Yang J, et al. Molecular phenotypes distinguish patients with relatively stable from progressive idiopathic pulmonary fibrosis (IPF) PLoS ONE. 2009;4(4):e5134. doi: 10.1371/journal.pone.0005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Sancho Figueroa MC, Carrillo G, Perez-Padilla R, et al. Risk factors for idiopathic pulmonary fibrosis in a Mexican population. A case–control study. Respir. Med. 2010;104(2):305–309. doi: 10.1016/j.rmed.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Antoniou KM, Hansell DM, Rubens MB, et al. Idiopathic pulmonary fibrosis: outcome in relation to smoking status. Am. J. Respir. Crit. Care Med. 2008;177(2):190–194. doi: 10.1164/rccm.200612-1759OC. [DOI] [PubMed] [Google Scholar]
- 36.King TE, Jr, Tooze JA, Schwarz MI, Brown KR, Cherniack RM. Predicting survival in idiopathic pulmonary fibrosis: scoring system and survival model. Am. J. Respir. Crit. Care Med. 2001;164(7):1171–1181. doi: 10.1164/ajrccm.164.7.2003140. [DOI] [PubMed] [Google Scholar]
- 37.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med. 2009;179(7):572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61(8):3419–3424. [PubMed] [Google Scholar]
- 43.Launay JM, Del Pino M, Chironi G, et al. Smoking induces long-lasting effects through a monoamine-oxidase epigenetic regulation. PLoS ONE. 2009;4(11):e7959. doi: 10.1371/journal.pone.0007959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, Killian JK, Yang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29(25):3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips JM, Goodman JI. Inhalation of cigarette smoke induces regions of altered DNA methylation (RAMs) in SENCAR mouse lung. Toxicology. 2009;260(1–3):7–15. doi: 10.1016/j.tox.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Schembri F, Sridhar S, Perdomo C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl Acad. Sci. USA. 2009;106(7):2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23(9):3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23(3):806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit. Care Med. 2009;180(5):462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerrero-Preston R, Goldman LR, Brebi-Mieville P, et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5(6):539–546. doi: 10.4161/epi.5.6.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suter M, Ma J, Harris AS, et al. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6(11):1284–1294. doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5(7):583–589. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuddapah S, Barski A, Zhao K. Epigenomics of T cell activation, differentiation, and memory. Curr. Opin. Immunol. 2010;22(3):341–347. doi: 10.1016/j.coi.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24(4):369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114(18):3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei B, Pei G. microRNAs: critical regulators in Th17 cells and players in diseases. Cell. Mol. Immunol. 2010;7(3):175–181. doi: 10.1038/cmi.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukasa R, Balasubramani A, Lee YK, et al. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32(5):616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 59.Parra ER, Kairalla RA, Ribeiro De Carvalho CR, Eher E, Capelozzi VL. Inflammatory cell phenotyping of the pulmonary interstitium in idiopathic interstitial pneumonia. Respiration. 2007;74(2):159–169. doi: 10.1159/000097133. [DOI] [PubMed] [Google Scholar]
- 60.Feghali-Bostwick CA, Tsai CG, Valentine VG, et al. Cellular and humoral autoreactivity in idiopathic pulmonary fibrosis. J. Immunol. 2007;179(4):2592–2599. doi: 10.4049/jimmunol.179.4.2592. [DOI] [PubMed] [Google Scholar]
- 61.Kotsianidis I, Nakou E, Bouchliou I, et al. Global impairment of CD4+CD25+FOXP3+ regulatory T cells in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2009;179(12):1121–1130. doi: 10.1164/rccm.200812-1936OC. [DOI] [PubMed] [Google Scholar]
- 62.Gilani SR, Vuga LJ, Lindell KO, et al. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PLoS ONE. 2010;5(1):e8959. doi: 10.1371/journal.pone.0008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coward WR, Watts K, Feghali-Bostwick CA, Knox A, Pang L. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol. Cell. Biol. 2009;29(15):4325–4339. doi: 10.1128/MCB.01776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coward WR, Watts K, Feghali-Bostwick CA, Jenkins G, Pang L. Repression of IP-10 by interactions between histone deacetylation and hypermethylation in idiopathic pulmonary fibrosis. Mol. Cell. Biol. 2010;30(12):2874–2886. doi: 10.1128/MCB.01527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders YY, Pardo A, Selman M, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2008;39(5):610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of Thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 2010;45(1):16–23. doi: 10.1165/rcmb.2010-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am. J. Pathol. 2010;177(1):21–28. doi: 10.2353/ajpath.2010.090999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Essential role of MeCP2 in the regulation of myofibroblast differentiation during pulmonary fibrosis. Am. J. Pathol. 2011;178(4):1500–1508. doi: 10.1016/j.ajpath.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 2010;11(7):476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9(3):179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am. J. Respir. Crit. Care Med. 2011;183(10):1295–1301. doi: 10.1164/rccm.201010-1579PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rabinovich E, Yakhini Z, Benos P, et al. Human CpG islands arrays reveal changes in global methylation patterns in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010;181:A2017. [Google Scholar]
- 73.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang I, Hennessy C, Davidson E, et al. Genome-wide DNA methylation patterns in interstitial lung disease (ILD) and chronic obstructive lung disease (COPD) Am. J. Respir. Crit. Care Med. 2011;183:A1049. [Google Scholar]
- 76.Henderson WR, Jr, Chi EY, Ye X, et al. Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc. Natl Acad. Sci. USA. 2010;107(32):14309–14314. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Konigshoff M, Kramer M, Balsara N, et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J. Clin. Invest. 2009;119(4):772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou B, Liu Y, Kahn M, et al. β-catenin/CBP-dependent regulation of TGF-beta-mediated epithelial-mesenchymal transition (EMT) by SMAD3. J. Biol. Chem. 2012 (Epub ahead of print). [Google Scholar]
- 79.Pandit KV, Corcoran D, Yousef H, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010;182(2):220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu G, Friggeri A, Yang Y, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010;207(8):1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cushing L, Kuang PP, Qian J, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2011;45(2):287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pottier N, Maurin T, Chevalier B, et al. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS ONE. 2009;4(8):e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oak SR, Murray L, Herath A, et al. A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLoS ONE. 2011;6(6):e21253. doi: 10.1371/journal.pone.0021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl. Res. 2011;157(4):191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 85.Shen-Orr SS, Tibshirani R, Khatri P, et al. Cell type-specific gene expression differences in complex tissues. Nat. Methods. 2010;7(4):287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 87.Chodavarapu RK, Feng S, Bernatavichute YV, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc. Natl Acad. Sci. USA. 2009;106(52):22187–22192. doi: 10.1073/pnas.0905767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ooi SK, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Milosavljevic A. Emerging patterns of epigenomic variation. Trends Genet. 2011;27(6):242–250. doi: 10.1016/j.tig.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 92.Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum. Mol. Genet. 2010;19(R2):R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sankaranarayanapillai M, Tong WP, Yuan Q, et al. Monitoring histone deacetylase inhibition in vivo: noninvasive magnetic resonance spectroscopy method. Mol. Imaging. 2008;7(2):92–100. [PubMed] [Google Scholar]
- 94.Boumber Y, Issa JP. Epigenetics in cancer: what’s the future? Oncology. 2011;25(3):220–226. 228. [PubMed] [Google Scholar]
- 95.Furdas SD, Kannan S, Sippl W, Jung M. Small molecule inhibitors of histone acetyltransferases as epigenetic tools and drug candidates. Arch. Pharm. (Weinheim) 2012;345(1):7–21. doi: 10.1002/ardp.201100209. [DOI] [PubMed] [Google Scholar]
- 96.Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226(2):365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 101.Lung Genomics Research Consortium. www.lung-genomics.org.