Abstract
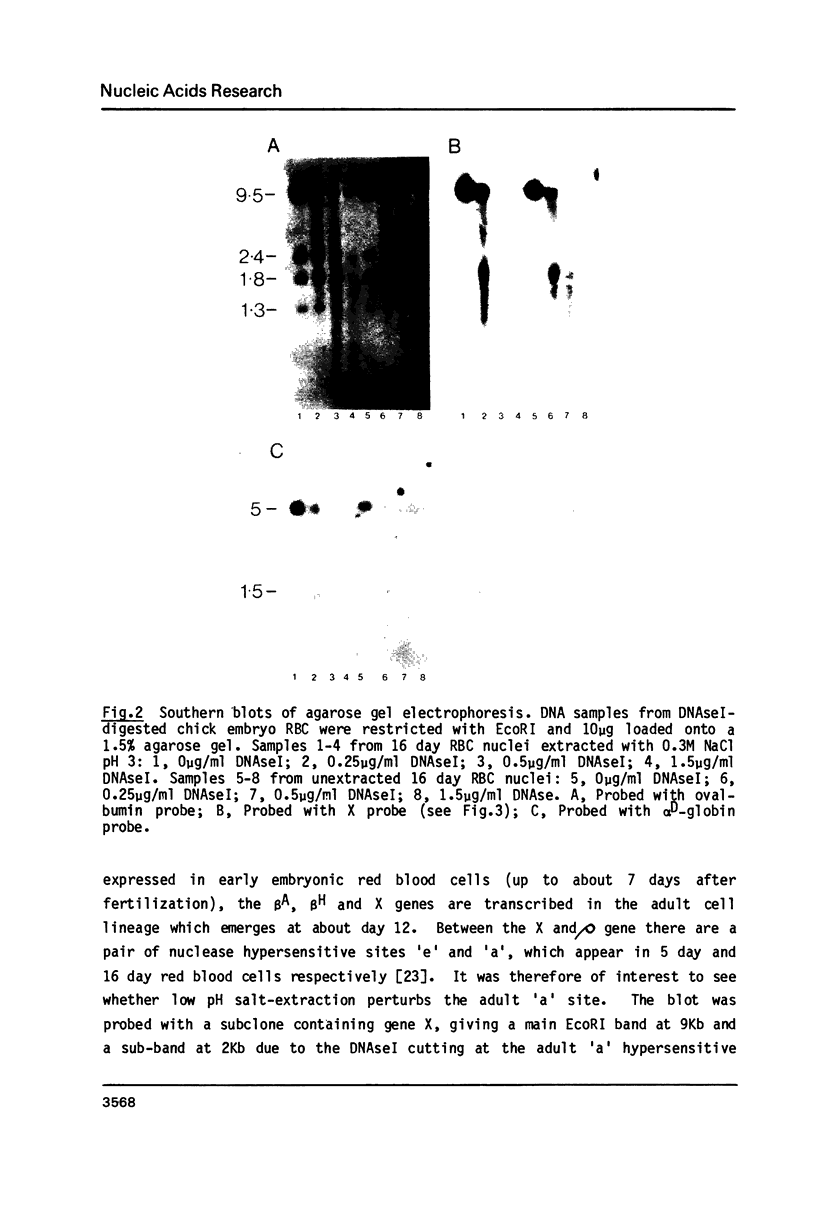
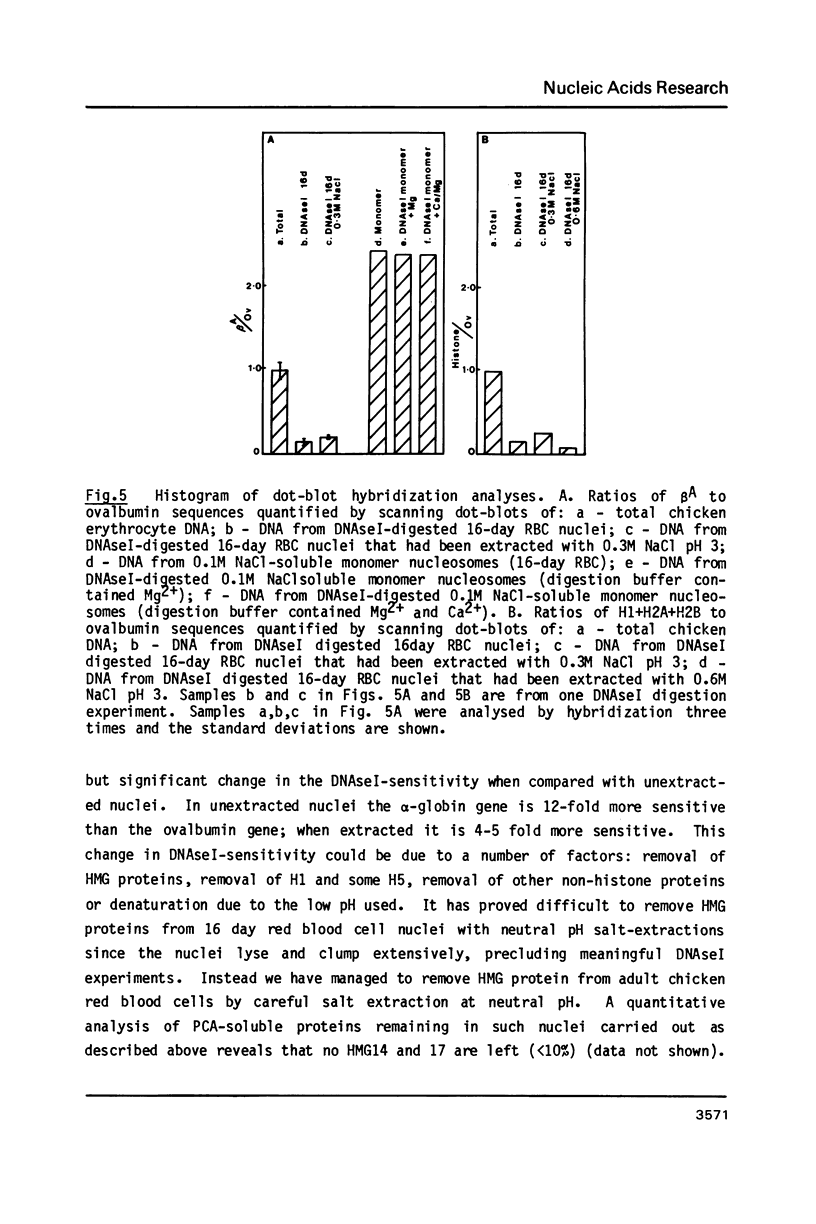
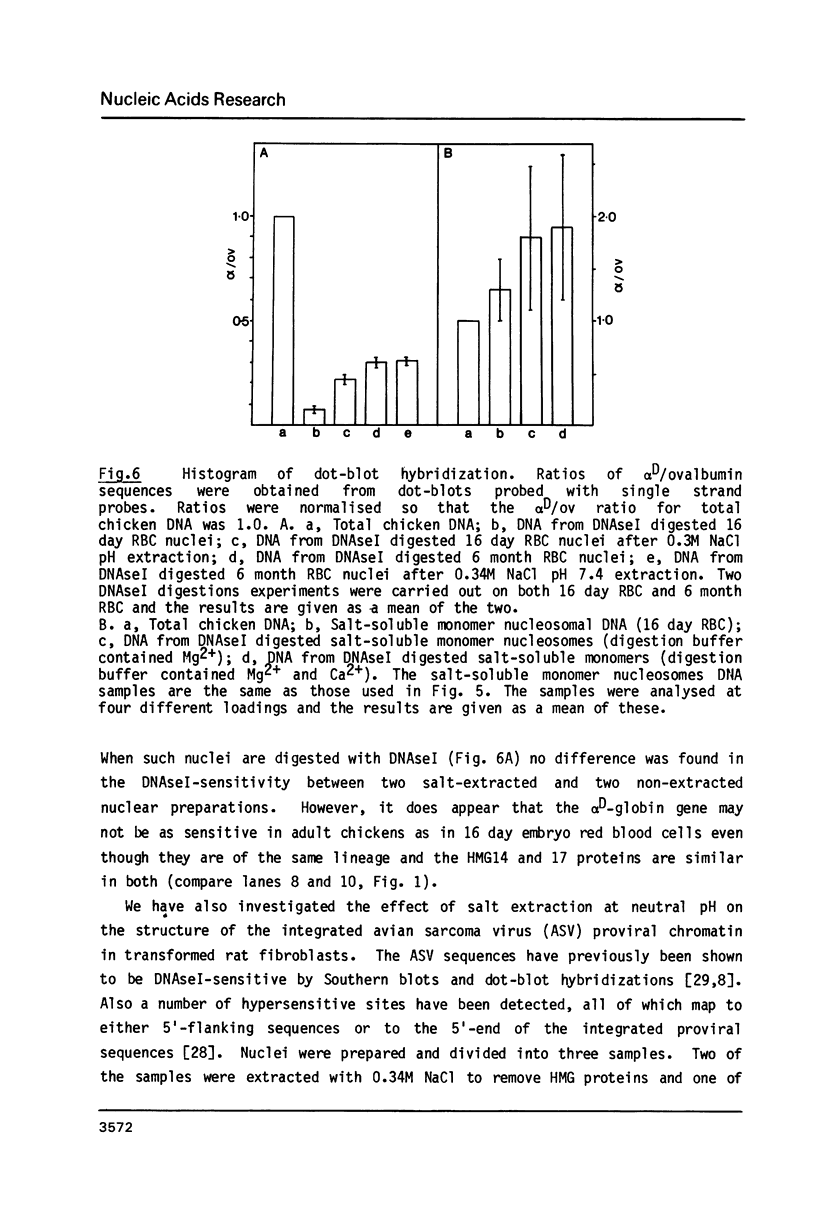
The procedure developed by Lawson and Cole (Biochemistry, 1979, 18 2161-2166) for removing lysine-rich histones from nuclei at low pH also quantitatively extracts proteins HMG14 and 17. The effect of this low pH extraction on the DNAseI-sensitive structures of active genes in avian red blood cells has been investigated. No major perturbation of a developmentally regulated DNAseI hypersensitive site in the beta-globin domain and at the 5' end of the alpha D gene was seen. The overall DNAseI-sensitive conformation of the beta A-globin gene (relative to the ovalbumin gene) is minimally affected by pH3 salt extraction, but there is some loss of sensitivity of the alpha D gene. Removal of HMG proteins at neutral pH had no effect on the sensitivity of active genes in erythroid or fibroblast nuclei. These results, together with those carried out on DNAseI sensitivity and HMG binding to monomer nucleosomes, indicate that there is a major structural feature of active genes responsible for DNAseI-sensitivity which is independent of HMG proteins or nucleosome core particle structure but may be dependent on higher order chromatin structures.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. E. The location of Ultrabithorax transcripts in Drosophila tissue sections. EMBO J. 1983;2(11):2075–2084. doi: 10.1002/j.1460-2075.1983.tb01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J., Levinger L., Varshavsky A. On the chromatin structure of the amplified, transcriptionally active gene for dihydrofolate reductase in mouse cells. J Biol Chem. 1982 May 10;257(9):5274–5282. [PubMed] [Google Scholar]
- Chiswell D. J., Gillespie D. A., Wyke J. A. The changes in proviral chromatin that accompany morphological variation in avian sarcoma virus-infected rat cells. Nucleic Acids Res. 1982 Jul 10;10(13):3967–3980. doi: 10.1093/nar/10.13.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea R., Harvey R., Wells J. R. Vertebrate histone genes: nucleotide sequence of a chicken H2A gene and regulatory flanking sequences. Nucleic Acids Res. 1981 Jul 10;9(13):3119–3128. doi: 10.1093/nar/9.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J. B., Engel J. D. The nucleotide sequence of the adult chicken alpha-globin genes. J Biol Chem. 1983 Apr 10;258(7):4623–4629. [PubMed] [Google Scholar]
- Elgin S. C. Anatomy of hypersensitive sites. Nature. 1984 May 17;309(5965):213–214. doi: 10.1038/309213a0. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Woodhead L., Johns E. W. The presence of high mobility group non-histone chromatin proteins in isolated nucleosomes. FEBS Lett. 1977 Jan 15;73(1):85–88. [PubMed] [Google Scholar]
- Heintz N., Zernik M., Roeder R. G. The structure of the human histone genes: clustered but not tandemly repeated. Cell. 1981 Jun;24(3):661–668. doi: 10.1016/0092-8674(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Humphries P., Cochet M., Krust A., Gerlinger P., Kourilsky P., Chambon P. Molecular cloning of extensive sequences of the in vitro synthesized chicken ovalbumin structural gene. Nucleic Acids Res. 1977 Jul;4(7):2389–2406. doi: 10.1093/nar/4.7.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Mills F. C., Allan J., Gould H. Selective unfolding of erythroid chromatin in the region of the active beta-globin gene. Nature. 1983 Dec 15;306(5944):709–712. doi: 10.1038/306709a0. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lawson G. M., Cole R. D. Selective displacement of histone H1 from whole HeLa nuclei: effect on chromatin structure in situ as probed by micrococcal nuclease. Biochemistry. 1979 May 29;18(11):2160–2166. doi: 10.1021/bi00578a005. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Rau D. C., Felsenfeld G. The high mobility group proteins HMG 14 and 17, do not prevent the formation of chromatin higher order structure. Nucleic Acids Res. 1982 Mar 25;10(6):2007–2016. doi: 10.1093/nar/10.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas R. H., Wright C. A., Cockerill P. N., Wyke J. A., Goodwin G. H. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983 Feb 11;11(3):753–772. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P. A. The essential role of Ca2+ in the activity of bovine pancreatic deoxyribonuclease. J Biol Chem. 1975 Mar 25;250(6):1981–1986. [PubMed] [Google Scholar]
- Reeves R., Chang D. Investigations of the possible functions for glycosylation in the high mobility group proteins. Evidence for a role in nuclear matrix association. J Biol Chem. 1983 Jan 10;258(1):679–687. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R. L., Annunziato A. T., Smith R. D. High mobility group proteins: abundance, turnover, and relationship to transcriptionally active chromatin. Biochemistry. 1983 Oct 11;22(21):5008–5015. doi: 10.1021/bi00290a020. [DOI] [PubMed] [Google Scholar]
- Senear A. W., Palmiter R. D. Multiple structural features are responsible for the nuclease sensitivity of the active ovalbumin gene. J Biol Chem. 1981 Feb 10;256(3):1191–1198. [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Stellwag E. J., Dahlberg A. E. Electrophoretic transfer of DNA, RNA and protein onto diazobenzyloxymethyl (DBM) - paper. Nucleic Acids Res. 1980 Jan 25;8(2):299–317. doi: 10.1093/nar/8.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B., Landes G. M., Pankratz M. J., Martinson H. G. The chicken beta globin gene region. Delineation of transcription units and developmental regulation of interspersed DNA repeats. J Biol Chem. 1982 Sep 25;257(18):11015–11023. [PubMed] [Google Scholar]
- Villeponteaux B., Lasky L., Harary I. Lysine-rich histones and the selective digestion of the globin gene in avian red blood cells. Biochemistry. 1978 Dec 12;17(25):5532–5536. doi: 10.1021/bi00618a031. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. T. Properties of active nucleosomes as revealed by HMG 14 and 17 chromatography. Nucleic Acids Res. 1982 Mar 25;10(6):2017–2042. doi: 10.1093/nar/10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Wood W. I., Felsenfeld G. Chromatin structure of the chicken beta-globin gene region. Sensitivity to DNase I, micrococcal nuclease, and DNase II. J Biol Chem. 1982 Jul 10;257(13):7730–7736. [PubMed] [Google Scholar]