Abstract
N-glycans were released from the SARS coronavirus (SARS-CoV) spike glycoprotein produced in Vero E6 cells and their structures were determined by a combination of matrix-assisted laser desorption/ionization (MALDI) mass spectrometry, negative ion electrospray collision-induced dissociation time-of-flight mass spectrometry and normal-phase high-performance liquid chromatography with exoglycosidase digestion. Major glycans were high-mannose (Man5–9GlcNAc2), hybrid and bi-, tri- and tetra-antennary complex with and without bisecting GlcNAc and core fucose. Complex glycans with fewer than the full complement of galactose residues were present and sialylation was negligible. Treatment with the glucosidase inhibitor N-butyl-deoxynojirimycin (NB-DNJ) inhibited N-glycan processing as evidenced by the appearance of glycans of composition Glc3Man7–9GlcNAc2. However, some complex glycans remained suggesting the presence of an α-endomannosidase. Our data in tissue culture indicate that inhibition of N-glycan processing may be considered as a therapeutic strategy against SARS CoV infections.
Abbreviations: 2AB, 2-Aminobenzamide; ACE2, angiotensin converting enzyme 2; CoV, coronavirus; CPE, cytopathic effects; DHB, 2,5-dihydroxybenzoic acid; FBS, fetal bovine serum; GU, glucose unit; HPLC, high-performance liquid chromatography; HRP, horseradish peroxidase; IgG, immunoglobulin; MALDI, matrix-assisted laser desorption/ionization; MOI, multiplicity of infection; MS, mass spectrometry; NB-DNJ, N-butyl-deoxynojirimycin; NP, normal phase; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PNGase F, peptide N-glycosidase F; PTFE, polytetrafluoroethylene; PVDF, polyvinylidene fluoride; Q, -quadrupole; S, spike protein; SARS, severe acute respiratory syndrome; SDS, sodium dodecyl sulphate; TOF, time-of-flight
Keywords: SARS, Spike glycoprotein, Glycosylation, N-Linked, Mass Spectrometry, Negative ion fragmentation
Introduction
The aetiological agent of the severe acute respiratory syndrome (SARS) is a coronavirus (SARS-CoV) which was responsible for causing a worldwide pandemic of lower respiratory tract infection between November 2002 and July 2003 (Drosten et al., 2003, Ksiazek et al., 2003, Marra et al., 2003, Rota et al., 2003). During this time, there were approximately 8100 probable SARS cases and approximately 800 recorded fatalities (http://www.wpro.who.int/sars/). Rapid identification of individuals presenting with the disease as well as appropriate nursing techniques and quarantine procedures allowed the outbreak to be controlled by July of 2003. There have since been smaller outbreaks of the disease, but these have been contained with minimal secondary transmission. In the clinic, SARS-CoV infection presents as atypical pneumonia, with patients reporting symptoms of high fever (>38 °C), dry cough, shortness of breath (dyspnea) and/or more severe breathing difficulties, which may occasionally require mechanical ventilation. The viral reservoirs are presumed to be bats (Wang and Eaton, 2007, Wang et al., 2006) and the palm civet cat has been implicated as a carrier of the virus, though there may also be other asymptomatic animal carriers (Shi and Hu, 2008).
Of the several proteins found in the SARS coronavirus (Tan et al., 2006, Ying et al., 2004), the large SARS-CoV spike glycoprotein (S) (Tripet et al., 2004) located on the virion surface is involved in both target cell attachment and fusion processes (Yang et al., 2004). As with several coronaviruses, the spike glycoprotein can be proteolytically cleaved into two non-covalently associated glycoproteins: the virion-associated S1 (666 amino acid residues) containing the receptor attachment site and the transmembrane-anchored S2 (583 residues) responsible for fusion activity (Bergeron et al., 2005, Follis et al., 2006, Li et al., 2005).
The SARS-CoV S glycoprotein is a 1255-amino acid precursor polypeptide as deduced from the published genome sequence and has 23 potential N-linked glycosylation sites (Eickmann et al., 2003, Marra et al., 2003, Rota et al., 2003, Xiao et al., 2003). Previous studies have demonstrated that several of these sites are occupied and possess both complex-type and high-mannose type N-glycans (Krokhin et al., 2003, Song et al., 2004, Ying et al., 2004). However, full glycan analysis has not yet been performed.
SARS-CoV uses the angiotensin converting enzyme 2 (ACE2) as the primary receptor (Chen and Subbarao, 2007, Li et al., 2003, Moore et al., 2004, Wang et al., 2004) suggesting that oligomannose-containing N-glycans on the SARS-CoV S glycoprotein surface may be involved in mediating virus attachment to cells expressing this cell surface lectin. The C-type lectins DC-SIGN and L-SIGN (DC/L-SIGN) can enhance viral entry (Marzi et al., 2004, Yang et al., 2004). Site-directed mutagenesis analyses have identified seven glycosylation sites on the S protein critical for DC/L-SIGN-mediated virus entry. They include asparagine residues 109, 118, 119, 158, 227, 589, and 699, which are distinct from residues of the ACE2-binding domain (amino acids 318–510) (Han et al., 2007). Recently, L-SIGN (DC-SIGNR) has been shown to act as an entry receptor for SARS-CoV infection (Jeffers et al., 2004), and its expression in lung alveolar and liver cells correlates with the disease pathogenesis observed in these organs in many SARS patients.
Given that oligomannose N-glycans may be involved in viral attachment to cells it is of interest to study the effects of modifications to the N-glycan profile with the object of possibly identifying therapeutic strategies. This report describes the first complete N-glycosylation analysis of SARS-CoV S glycoprotein produced in Vero E6 cells, a cell line derived from African green monkeys, and demonstrates the inhibitory effect of the α-glucosidase inhibitor N-butyl-deoxynojirimycin (NB-DNJ) on virus replication in this cell line.
Results and discussion
Characterization of SARS-CoV spike protein
Viral proteins from purified viral particles were separated on 9% SDS-PAGE gels and stained with Coomassie blue (Fig. 1 ). The proteins S, M and N were detected but the gel was not dense enough to detect the smaller (7 kDa) E protein. Western blotting using the SARS-CoV S glycoprotein antibody (Imgenex) confirmed the identity and molecular weight (180 kDa) of the S protein which was consistent with earlier reports (Krokhin et al., 2003, Song et al., 2004, Xiao et al., 2003) and indicated the presence of the fully modified glycoprotein. There was no apparent difference in molecular weight between the glycoproteins produced with and without NB-DNJ.
Fig. 1.
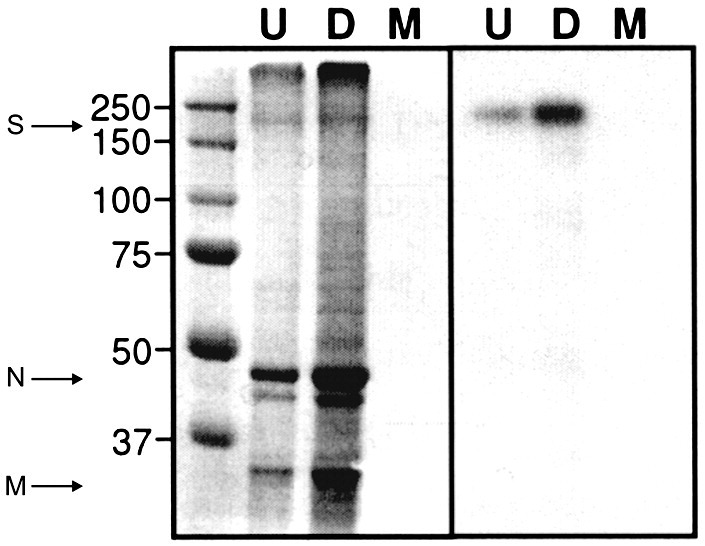
SDS-PAGE and western blotting analysis of purified SARS-CoV (strain Tor2) preparation. Left panel: 9% SDS-PAGE analysis of SARS-CoV virion preparation. S = SARS-CoV spike glycoprotein, N = SARS-CoV nucleoprotein, M = SARS-CoV matrix protein. Molecular weight markers are shown on the left. Right panel: Western blotting analysis of SARS-Co V virion preparation using an anti-SARS-CoV S antibody (Imgenex). U, untreated preparation; D, drug(NB-DNJ) treated preparation; M, mock infected preparation.
Identification of N-glycans
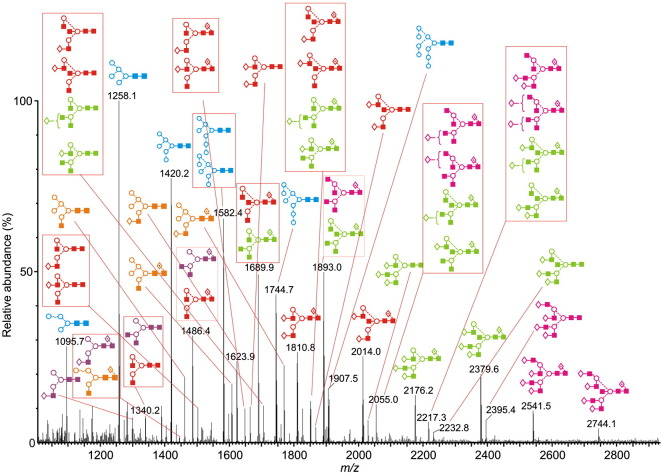
The spike glycoprotein-containing band was cut from the gel for N-glycan analysis by in-gel PNGase F digestion followed by HPLC (as 2AB derivatives), exoglycosidase digestion, MALDI-TOF and nanoelectrospray MS/MS as described below. The HPLC trace of the SARS-CoV glycans obtained without NB-DNJ treatment is shown in Fig. 2a. Traces obtained before and after digestion with Arthrobacter ureafaciens sialidase (Fig. 2b) were virtually identical showing negligible incorporation of sialic acid in these glycans. Previous work (Feldmann et al., 1994) has also shown a lack of sialylated N-glycans in Marburg virus grown in Vero E6 cells. The positive ion MALDI (Fig. 3 ) and negative ion nanoelectrospray spectra were similar to each other and to the HPLC trace although more glycans were resolved by mass spectrometry. Table 1 lists the identified glycans and their percentage of the total glycan pool as measured by MALDI-TOF MS and Table 2 lists the percentage of various compound glycan types. Because a few of the glycans with different structures had the same mass and were, thus, not separated by mass spectrometry, the figures should be taken as approximate. Nevertheless, they reflect the composition of the mixture reasonably accurately.
Fig. 2.
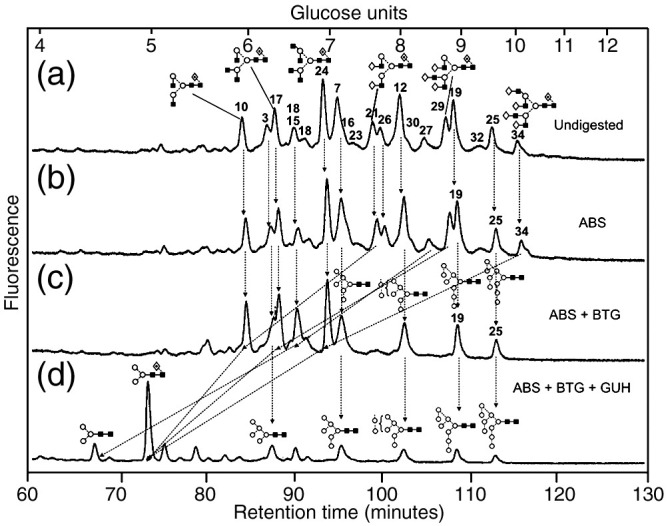
(a) NP-HPLC analysis of SARS-CoV S N-glycans. (b) HPLC analysis of SARS-CoV N-glycans following digestion with Arthrobacter ureafaciens sialidase (ABS, EC 3.2.1.18), (c) HPLC analysis following digestion with a mixture of ABS and bovine testis β-galactosidase (BTG, EC 3.2.1.23) and (d) HPLC analysis following incubation with a mixture of ABS, BTG and Streptococcus pneumoniae hexosaminidase (GUH, EC 3.2.1.30). The broken lines connecting the peaks in the various traces show movement of the peaks following digestion. Symbols used for the structural formulae: ■ = GlcNAc, ○ = mannose, ◊ = galactose,  = fucose. The angle of the lines connecting the symbols shows the linkage with full and broken lines specifying β- and α-linkages, respectively. Further details are given in the paper by Harvey et al. (2009).
= fucose. The angle of the lines connecting the symbols shows the linkage with full and broken lines specifying β- and α-linkages, respectively. Further details are given in the paper by Harvey et al. (2009).
Fig. 3.
Positive ion MALDI-TOF mass spectrum ([M+Na]+ions) of the N-linked glycans obtained from the SARS-CoV spike protein. Peaks are identified in Table 1 and the key to the structural symbols is in the legend to Fig. 2 with the addition of □ = glucose. Glycan types are coloured: blue = high mannose, orange = hybrid, purple = monoantennary, red = biantennary, green = triantennary, pink = tetra-antennary.
Table 1.
N-linked glycans with the masse of their [M+Na]+ and [M+H2PO4]− ions.
| No.a | GUb |
m/z |
Composition |
Structuree | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MALDI ([M+Na]+)c |
Electrospray ([M+H2PO4]−)d |
Hex | HexNAc | dHex | ||||||
| Found | Calc. | Found | Calc. | |||||||
| 1 | — | 1095.7 | 1096.0 | — | 1169.3 | 4 | 2 | 0 |
 f f
|
|
| 2 | — | 1136.9 | 1137.0 | — | 1210.4 | 3 | 3 | 0 |
 f f
|
|
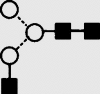
| 3 | 6.17 | 1258.1 | 1258.1 | 1331.4 | 1331.4 | 5 | 2 | 0 | 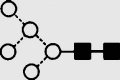 |
|
| 4 | — | 1283.8 | 1283.2 | 1356.6 | 1356.4 | 3 | 3 | 1 | 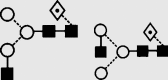 |
|
| 5 | — | 1299.4 | 1299.2 | — | 1372.4 | 4 | 3 | 0 |
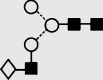 f f
|
|
| 6 | — | 1339.9 | 1340.2 | 1413.5 | 1413.5 | 3 | 4 | 0 | 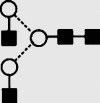 |
|
| 7 | 7.05 | 1420.2 | 1420.2 | 1493.5 | 1493.5 | 6 | 2 | 0 | 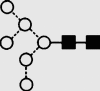 |
|
| 8 | 6.40 | 1444.7 | 1445.3 | 1518.5 | 1518.5 | 4 | 3 | 1 | 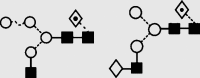 |
|
| 9 | — | 1461.4 | 1461.3 | 1534.5 | 1534.5 | 5 | 3 | 0 | 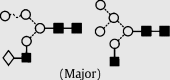 |
|
| 10 | 5.89 | 1486.4 | 1486.4 | 1559.5 | 1559.5 | 3 | 4 | 1 | 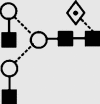 |
|
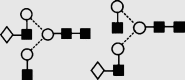
| 11 | — | 1502.0 | 1502.4 | 1575.5 | 1575.5 | 4 | 4 | 0 |
 f f
|
|
| 12 | 7.94 | 1582.4 | 1582.4 | 1655.5 | 1655.5 | 7 | 2 | 0 | 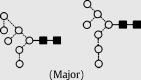 |
|
| 13 | — | 1607.7 | 1607.4 | 1680.5 | 1680.5 | 5 | 3 | 1 | 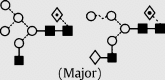 |
|
| 14 | — | 1623.9 | 1623.4 | 1696.5 | 1696.5 | 6 | 3 | 0 | 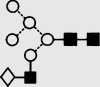 |
|
| 15 | 6.63 6.81 |
1648.3 | 1648.5 | 1721.6 | 1721.6 | 4 | 4 | 1 | 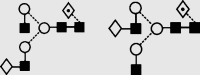 |
|
| 16 | 7.13 | 1664.4 | 1664.5 | 1737.6 | 1737.6 | 5 | 4 | 0 | 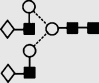 |
|
| 17 | 6.17 6.27 |
1689.9 | 1689.6 | 1762.6 | 1762.6 | 3 | 5 | 1 | 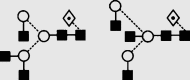 |
|
| 18 | 6.50 | 1705.3 | 1705.5 | 1778.5 | 1778.6 | 4 | 5 | 0 | 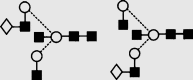 |
|
| 19 | 8.80 | 1744.7 | 1744.5 | 1817.6 | 1817.6 | 8 | 2 | 0 | 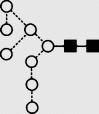 |
|
| 20 | — | 1769.9 | 1769.6 | 1842.6 | 1842.6 | 6 | 3 | 1 | 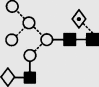 |
|
| 21 | 7.54 | 1810.8 | 1810.6 | 1883.6 | 1883.6 | 5 | 4 | 1 | 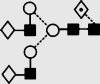 |
|
| 22 | — | 1851.9 | 1851.7 | 1924.6 | 1924.6 | 4 | 5 | 1 | 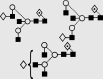 |
|
| 23 | 7.24 | 1867.7 | 1867.7 | 1940.6 | 1940.6 | 5 | 5 | 0 | 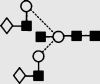 |
|
| 24 | 6.86 | 1893.0 | 1892.7 | 1965.6 | 1965.7 | 3 | 6 | 1 | 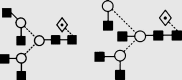 |
|
| 25 | 9.48 | 1907.5 | 1906.7 | 1979.6 | 1979.6 | 9 | 2 | 0 | 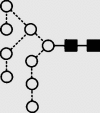 |
|
| 26 | 7.65 | 2014.0 | 2013.8 | 2086.7 | 2086.7 | 5 | 5 | 1 | 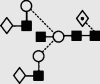 |
|
| 27 | 8.32 | 2029.8 | 2029.8 | 2102.7 | 2102.7 | 6 | 5 | 0 | 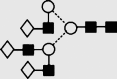 |
|
| 28 | 8.02 | 2055.0 | 2054.9 | 2127.7 | 2127.7 | 4 | 6 | 1 | 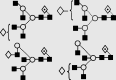 |
|
| 29 | 8.67 | 2176.2 | 2176.0 | 2248.7 | 2248.8 | 6 | 5 | 1 | 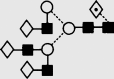 |
|
| 30 | — | 2217.3 | 2217.0 | 2289.7 | 2289.8 | 5 | 6 | 1 | 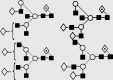 |
|
| 31 | — | 2232.8 | 2233.0 | — | 2305.8 | 6 | 6 | 0 | 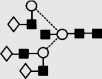 |
|
| 32 | 9.26 | 2379.6 | 2379.2 | 2451.8 | 2451.8 | 6 | 6 | 1 | 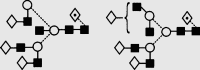 |
|
| 33 | — | 2395.4 | 2395.2 | — | 2467.8 | 7 | 6 | 0 | 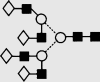 |
|
| 34 | 9.95 | 2541.5 | 2541.3 | 2613.8 | 2613.9 | 7 | 6 | 1 | 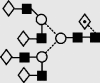 |
|
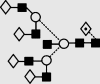
| 35 | — | 2744.1 | 2744.5 | — | 2817.0 | 7 | 7 | 1 |
 f f
|
|
| 36g | 10.27 | — | — | — | — | 10 | 2 | 0 |
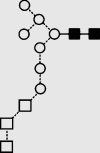 g g
|
|
| 37g | 11.16 | — | — | — | — | 11 | 2 | 0 |
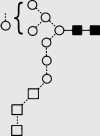 g g
|
|
| 38g | 11.80 | — | — | — | — | 12 | 2 | 0 |
 g g
|
|
Peak numbers as in the MALDI-TOF spectrum (Fig. 3).
Glucose unit.
Average mass.
Monoisotopic mass.
Symbols used for the structural formulae: ■ = GlcNAc, ○ = mannose, ◊ = galactose,  = fucose, □ = glucose. The angle of the lines connecting the symbols shows the linkage with full and broken lines specifying β- and α-linkages, respectively.
= fucose, □ = glucose. The angle of the lines connecting the symbols shows the linkage with full and broken lines specifying β- and α-linkages, respectively.
In the presence of NB-DNJ only.
Not confirmed by fragmentation.
Table 2.
Percentages of various groups of N-glycan in SARS spike glycoprotein.
| Glycan type | % Total | |
|---|---|---|
| High-mannose | — | 30 |
| Hybrid | — | 28 |
| Complex | Total | 42 |
| Biantennary | 19 | |
| Triantennary | 12 | |
| Tetraantennary | 11 | |
| Truncated hybrid and complex (no galactose) | — | 21 |
The glycans were identified by exoglycosidase digestion with monitoring by HPLC and by negative ion MS/MS of their [M+H2PO4]− ions (Harvey, 2005a) using electrospray sample introduction (Harvey, 2005b,Harvey, 2005c, Harvey et al., 2008b). The HPLC traces obtained after ABS and bovine testis β-galactosidase (BTG) treatment (Fig. 2c) confirmed attachment of galactose at the non-reducing termini of the glycan antenna. Further incubation with hexosaminidase (Fig. 2d) reduced the profile to Man3GlcNAc2, Man3GlcNAc2Fuc1 and a series of unaffected high-mannose glycans. A few compounds were present at concentrations that were too weak to permit acquisition of informative MS/MS spectra. However, their structures could be deduced from the structures of related compounds, e.g., complex glycans with different numbers of galactose residues. Such compounds are identified in Table 1. Ion nomenclature in the following discussion follows that introduced by Domon and Costello (1988) as modified by Harvey et al. (2004) by the use of the subscript “R” to specify cleavages adjacent to the reducing-terminal GlcNAc residue and “R-1” to specify cleavages adjacent to the penultimate GlcNAc in order to avoid confusion by the subscript number changing as a function of antenna length.
High-mannose glycans
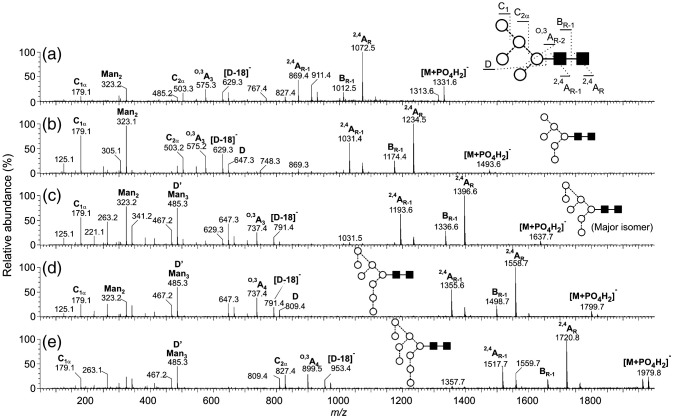
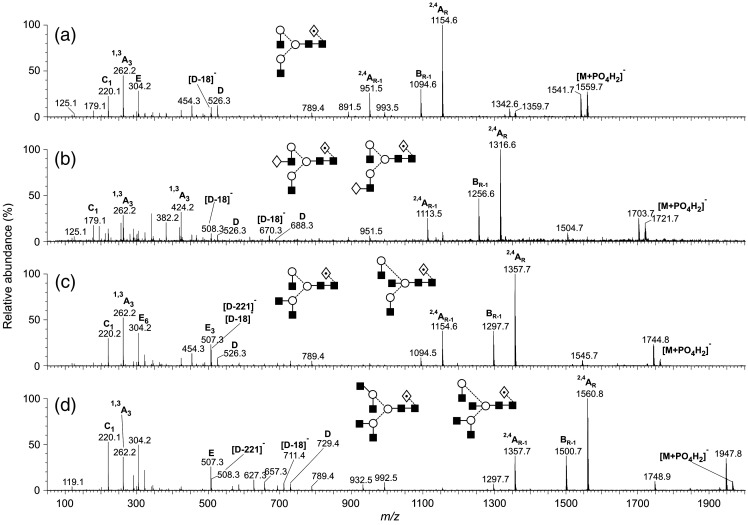
High-mannose glycans containing from four to nine mannose residues were present in the glycan mixture (Fig. 3). Incubation with Aspergillus saitoi a-mannosidase (data not shown) reduced the larger compounds to Man5GlcNAc2 (compound 3, Table 1) confirming the α-mannose linkages. The negative ion MS/MS spectra of the high-mannose glycans Man5–9GlcNAc2 (phosphate adducts) are shown in Fig. 4 . The cross-ring 2,4AR and 2,4AR-1 fragments together with the BR-1 ion at the high-mass end of the spectra define the 1→4-linked chitobiose core (Harvey, 2005b, Harvey et al., 2008b) and the single C1 fragment at m/z 179 confirmed hexose (mannose) as the only non-reducing end residue. The ions towards the centre of the spectrum were mainly derived from the 6-antenna and allowed the distribution of the mannose residues between the two antennae to be determined. Predominant among these ions were the D-ion (formal loss of the 3-antenna from the CR-1 ion), the [D-18]− ion and the O,3AR-2 cross-ring cleavage fragment (core mannose residue) (Harvey, 2005b). Thus, Man5GlcNAc2 (3) produced a spectrum with these ions at m/z 647, 629 and 575, respectively. Further confirmation of the substitution pattern was provided by the C2 ion at m/z 503 (three mannose residues). In the spectrum of Man6GlcNAc2 (7, Fig. 4b) the D, [D-18]− and O,3AR-2 ions were in the same position as in the spectrum of Man5GlcNAc2 showing that the sixth mannose was attached to the 3-antenna. These three diagnostic ions all shifted by 162 mass units to higher mass in the spectrum of Man7GlcNAc2 (12, Fig. 4c) showing that the seventh mannose was attached to the 6-antenna and the high abundance of the ion at m/z 485 was consistent with its location on the 6-branch of this antenna. This ion, termed D′, is thought to be equivalent to ion D as the result of the same substitution pattern of the branched mannose of the 6-antenna to that at the mannose of the core. This same pattern of D, [D-18]−, O,3AR-2 and D′ ions was present in the spectrum of Man8GlcNAc2 (19, Fig. 4d) confirming attachment of the 8th mannose to the 3-antenna and indicating that this compound had the classical d1,d3-substitution pattern. Finally, in the spectrum of Man9GlcNAc2 (25, Fig. 4e), the three 6-antenna-derived diagnostic ions shifted upwards by another 162 mass units to m/z 971, 953 and 899 as the result of the extra mannose residue in the 6-antenna. The presence of a second set of the D, [D-18]−, O,3AR-2 ions at m/z 647, 629 and 575 in the spectrum of Man7GlcNAc2 (12) indicated the presence of a second isomer with three mannose residues in the 3-antenna. A fragmentation spectrum was not obtained for Man4GlcNAc2 (1).
Fig. 4.
Negative ion MS/MS spectra of the [M+H2PO4]− ions from the high-mannose glycans Man5–9GlcNAc2. Symbols for the structural diagrams are defined in the legend to Fig. 2. Ions are labelled according to the scheme introduced by Domon and Costello (1988).
Hybrid glycans
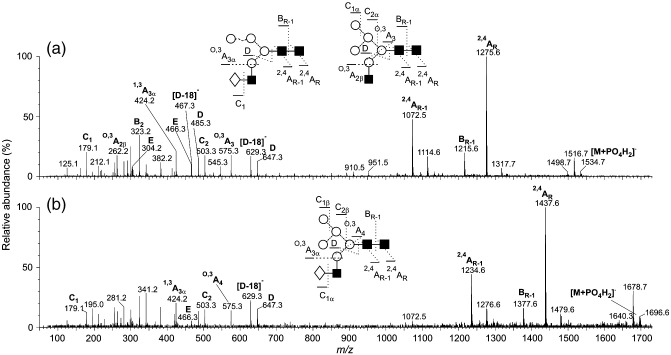
Hybrid glycans (numbers 9, 13, 14 and 20, Table 1) were detected in the mass spectra but were not apparent in the HPLC trace of the total glycans. However, they became visible by HPLC following incubation with Jack bean α-mannosidase which removed all of the mannose residues from the 6-antenna to leave the chitobiose core and 3-antenna which consisted of GlcNAc and Gal-GlcNAc (data not shown). Fig. 5 shows the negative ion MS/MS spectra of the hybrid glycans 9 (Fig. 5a) and 14 (Fig. 5b). The pattern of mannose substitution on the 6-antenna was determined by the masses of the D, [D-18]−, O,3AR-2, D′ and [D′-18]− ions in the negative ion MS/MS spectra as described above. The spectrum of the glycan corresponding to Hex5GlcNAc3 (9, Fig. 5a) showed the presence of two compounds as evidenced by the appearance of two sets of D and [D-18]− ions (m/z 647/629 and 485/467) indicating three and two mannose residues respectively in the 6-antenna. The linkage of the mannose residue in the compounds with two mannose residues in the 6-antenna was not determined. Location of the fucose at position 6 of the terminal GlcNAc residue, when present, was determined by the masses of the abundant 2,4AR, 2,4AR-1 and BR-1 ions in the MS/MS spectra (Harvey, 2005c, Harvey et al., 2008b) all of which were formed by loss of the fucose residue as the result of its attachment to the eliminated neutral fragment. Structures are shown in Table 1. No antenna-substituted fucose was detected in these or in the spectra of the complex glycans. The composition of the 3-antenna was specified by the mass of an E-type cross-ring fragment ion (substituents plus 101 mass units from the mannose residue (Harvey, 2005c, Harvey et al., 2008b)). These ions were of low abundance but were present at m/z 304 and 466 in the spectrum shown in Fig. 5a and at m/z 466 in Fig. 5b confirming the presence of GlcNAc and Gal-GlcNAc antennae respectively. The 3-antenna composition was further defined by an O,3A cross-ring cleavage ion formed by fragmentation of the mannose residue and which appeared at 59 mass units above the mass of the substituents linked to the mannose (m/z 262 for a GlcNAc-containing antenna and m/z 424 for Gal-GlcNAc, see Fig. 5).
Fig. 5.
Negative ion MS/MS spectra of the [M+H2PO4]− ions from the hybrid glycans 9 and 14. Symbols for the structural diagrams are defined in the legend to Fig. 2.
Complex glycans
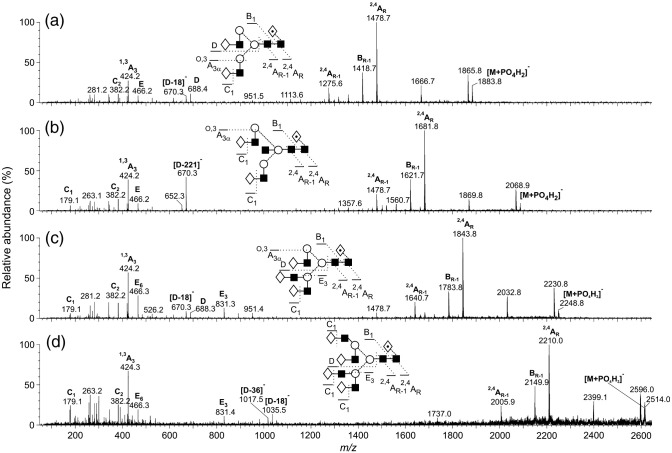
Representative spectra are shown in Fig. 6 . Bi-, tri- and tetra-antennary glycans were found, with and without core fucose and with and without a bisecting GlcNAc residue. Exoglycosidase digestion with ABS, BTG and GUH (Fig. 2) showed that the antennae consisted of galactose and GlcNAc only. Their negative ion MS/MS spectra were identical to those from reference standards. The branching structure of the triantennary glycan (29, Fig. 6c) was revealed by the mass of the E3 ion (m/z 831, the subscript “3” indicates its origin from the 3-antenna) (Harvey, 2005c, Harvey et al., 2008a). This ion was also present in the spectrum of the tetra-antennary glycan (34, Fig. 6d) but, whereas the D and [D-18]− ions from the 6-antenna in the spectrum of the triantennary glycan were at the same m/z values (688 and 670 respectively) as in the spectrum of the biantennary glycan (Fig. 6a), they moved to m/z 1053 and 1035 in the spectrum of the tetra-antennary glycan reflecting the presence of the additional Gal-GlcNAc moiety in the 6-antenna. An additional [D-36]− ion at m/z 1017 was also characteristic of this branched structure on the 6-antenna. The fucose substitution pattern on the core GlcNAc residue was defined by the masses of the 2,4AR, 2,4AR-1 and BR-1 ions as described above. The occurrence of a bisecting GlcNAc residue was determined by the presence of an abundant [D-221]− ion (e.g., m/z 670 in Fig. 6b), equivalent in mass to the [M-18]− ion from the corresponding compound that did not contain a bisecting GlcNAc residue, as described earlier (Harvey et al., 2008b).
Fig. 6.
Negative ion MS/MS spectra of the [M+H2PO4]− ions from (a) the fucosylated biantennary glycan 21, (b) the bisected, fucosylated biantennary glycan 26, (c) the fucosylated triantennary glycan 29 and (d) the fucosylated tetra-antennary glycan 34. Symbols for the structural diagrams are defined in the legend to Fig. 2.
These basic complex structures were accompanied by relatively large amounts of analogues with fewer numbers of galactose residues (Table 1), confirmed by incubations with bovine testis β-galactosidase. Four representative spectra are shown in Fig. 7 . All illustrated compounds contain a fucose residue attached to the core GlcNAc as shown by the masses of the 2,4AR, 2,4AR-1 and BR-1 ions as above. The spectrum of the biantennary glycan lacking both galactose residues (10, Fig. 7a) contained D and [D-18]− ions at m/z 526 and 508, respectively, consistent with the absence of galactose in the 6-antenna. The C1 ion was at m/z 220 showing GlcNAc at the non-reducing terminus. Addition of one galactose residue (Fig. 7b) produced a mixed spectrum of glycans with the galactose attached to either antenna (15) as shown by the two pairs of D and [D-18]− ions at m/z 526/508 (major isomer) and at m/z 688/670. Two C1 ions at m/z 179 and 220 confirmed both hexose (galactose) and GlcNAc, respectively, at the non-reducing terminus. The 1,3A3 ion at m/z 424 was consistent with the presence of a Gal-GlcNAc-containing antenna.
Fig. 7.
Negative ion MS/MS spectra of the [M+H2PO4]− ions from (a) the fucosylated biantennary glycan lacking galactose (10), (b) a mixture of the fucosylated biantennary glycan with one galactose residue on either antenna (15), (c) a mixture of the fucosylated triantennary glycan and the bisected biantennary glycan lacking galactose (17) and (d) a mixture of the fucosylated tetra-antennary glycan and the bisected triantennary glycan lacking galactose (24). Symbols for the structural diagrams are defined in the legend to Fig. 2.
Most of the other spectra from the glycans containing low galactose were of mixtures. They were interpreted by postulation of the expected ions, based on the structures of the identified fully galactosylated compounds and finding the relevant diagnostic ions in the spectra. Thus, the spectrum of the compounds having the composition Man3GlcNAc5Fuc1 (17, Fig. 7c) would be expected to contain contributions from the bisected biantennary glycan and the triantennary glycan, both lacking galactose. The spectrum contained a prominent ion at m/z 507 at the expected mass of the E ion from the triantennary glycan lacking galactose and D and [D-18]− ions at m/z 526/508 from the 6-antenna. The enhanced relative abundance of the ion at m/z 508 suggested the presence of a small amount of the bisected biantennary glycan, this ion corresponding to [D-221]−. The two compounds (24) producing the spectrum in Fig. 7d were the degalactosylated tetra-antennary glycan and the bisected triantennary compound. The former compound produced the characteristic D, [D-18]− and [D-36]− triplet at m/z 729, 711 and 693 from the 6-antenna and the E ion at m/z 507. The other, minor compound was characterized by the [D-221]− ion at m/z 508. The ion corresponding to the antenna mass plus 59 mass units (C2H3O2 from the attached mannose) also defined the antenna composition (the specific cross-ring cleavage producing these A-type ions varied with the linkage position of the antenna).
Effect of NB-DNJ treatment
Effect of NB-DNJ against SARS-CoV infection
Following infection with SARS-CoV (strain Tor2, MOI = 10-4) and incubation with the iminosugar inhibitor NB-DNJ, the integrity of the Vero E6 cell monolayer was monitored daily by light microscopy. At NB-DNJ concentrations of 100 μg/mL and above, the cell monolayer remained intact and showed no signs of virally-induced cytopathic effects (CPE). However, no significant toxicity was seen at this drug concentration as shown by the XTT assay (data not shown).
Effect of NB-DNJ treatment on N-glycan profiles
Fig. 8b shows the NP-HPLC profile of the N-glycans from the spike protein that was obtained in the presence of NB-DNJ at 100 μg/mL, the concentration that protected the cells against CPE. The glycans of higher molecular weight than were observed in the sample without NB-DNJ were identified by HPLC and exoglycosidase digestion (A. ureafaciens sialidase, bovine testis β-galactosidase S. pneumonia N-acetyl-hexosaminidase, Jack bean α-mannosidase and bovine kidney α-fucosidase) as Glc3Man7GlcNAc2, Glc3Man8GlcNAc2 and Glc3Man9GlcNAc2 showing that NB-DNJ at the concentration used (100 μg/mL) was able to partially inhibit glycan processing by blocking the first stage, namely removal of the terminal glucose residues from the 3-antenna. α-Mannosidase was, however, still able to remove the outer mannose residues from the 6-antenna to give the observed compounds. Although processing of the 3-antenna was blocked, complex glycans were still found in the profile. The most likely reason would be the presence of an endo-mannosidase that was able to split off Glc3Man from the 3-antenna. Such an enzyme has been reported from rats (Lubas and Spiro, 1987, Lubas and Spiro, 1988, Moore and Spiro, 1990) but there appears to be no information on its possible occurrence in the African green monkey. However, the high abundance of the peak labelled as 29 in Fig. 8b is accounted for by the isomer of Man8GlcNAc2 that has lost a mannose residue from the 3-antenna (same GU value as the fucosylated triantennary glycan) supporting this proposal.
Fig. 8.
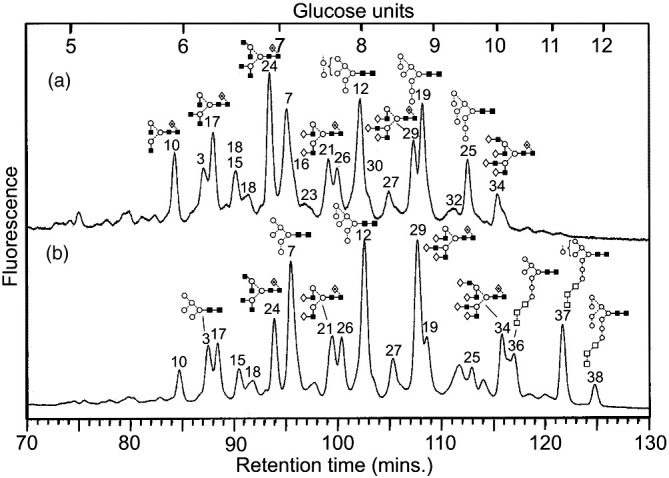
(a) NP-HPLC analysis of SARS-CoV N-glycans obtained without (a) and with (b) 100 μg/mL NB-DNJ. Peaks are identified in Table 1. The three glycans with residual glucose residues are highlighted.
Although the postulated endomannosidase activity permits extensive N-glycan processing with the production of complex glycans and a nearly normal profile in the presence of NB-DNJ, the drug prevents formation of the mono-glucosylated N-glycans that are necessary for binding to the chaperones calnexin and calreticulin that are essential for correct protein folding. Viral surface glycoproteins are dependent on such an interaction for correct folding (Vigerust and Shepherd, 2007). Thus, α-glucosidase inhibition will prevent this interaction and potentially lead to regional or global glycoprotein misfolding. Therefore, the observation that the SARS-CoV S glycoprotein can undergo extensive N-glycan processing to complex structures in the presence of NB-DNJ does not necessarily correlate with proper glycoprotein folding. Unfortunately, the necessary conformationally sensitive anti-SARS-CoV S antibodies for testing this conclusion were not available at the time this study was performed. Improper folding induced by NB-DNJ could explain the protective effect of the drug towards viral infection but the ability to retain complex N-glycan processing could contribute to the drug's low cytotoxicity.
Conclusions
This study is the first to describe fully the range of N-glycans present on the SARS-CoV spike glycoprotein. They consisted mainly of high-mannose (Man5–9GlcNAc2, about 30% of the total glycans), hybrid (28%) and bi-, tri- and tetra-antennary complex glycans (42%) with and without both bisecting GlcNAc and core fucose. Complex glycans with fewer than the full complement of galactose residues were present in relatively high abundance with 49% of the complex glycans carrying no galactose. Sialylation was negligible. No evidence was found for glycans with N-acetyllactosamine extensions or with fucose attached to an antenna. The relatively high abundance of complex glycans lacking one or more of the normal galactose residues might indicate the involvement of the lectin, LSECtin in viral binding, as recently found for Ebola virus (Powlesland et al., 2008).
Treatment with the glucosidase inhibitor NB-DNJ inhibited N-glycan processing as evidenced by the appearance in the profiles of glycans of composition Glc3Man7–9GlcNAc2. However, some complex glycans still appeared in the glycan profile, suggesting the presence of an α-endomannosidase that was able to hydrolyse the Glc3-Man3 chain of the 3-antenna allowing attachment of GlcNAc in the first step towards complex glycan biosynthesis. Protection of cells against the cytopathic effects of the virus, possibly as the result of the inhibition of glycan processing, suggests a possible use of glucosidase inhibitors to treat SARS-CoV infections.
Materials and methods
Materials
Methanol was obtained from BDH Ltd. (Poole, UK). Ammonium phosphate and 2,5-dihydroxybenzoic acid (DHB) were from Aldrich Chemical Co. Ltd. (Poole); water was distilled before use in a quartz apparatus. NB-DNJ was a gift from Monsanto Searle (St Louis, USA).
Preparation of SARS-CoV
All work with infectious SARS-CoV was performed in the high biocontainment laboratory of the National Microbiology Laboratory (NML) of the Public Health Agency of Canada (PHAC). Vero E6 cells (ATCC CRL-1586, kidney epithelial cells derived from African green monkeys (Cercopithecus aethiops)) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and subsequently infected with the Tor2 strain of the SARS-CoV (Marra et al., 2003) at an MOI of 10−4 with and without the presence of different concentrations (0, 100, 500 and 1000 mg/mL) of NB-DNJ. After 1 h of virus adsorption, cells were washed 3 times and DMEM supplemented with 2% FBS (with and without NB-DNJ) was added. Forty-eight hours post-infection, the cell supernatant was harvested and cells were cleared by low-speed centrifugation. Virus particles were pelleted through a 20% sucrose cushion at 20,000 rpm for 2 h, washed with PBS and resuspended and inactivated in 1 × sample loading buffer with 1.5% SDS. Infectivity was further inactivated by heating the specimens at 100 °C for 15 min prior to removal from high biocontainment.
Assay for cellular toxicity
Vero E6 cells were cultured in 96-well plates in DMEM supplemented with 10% FBS. At 80% confluency the medium was removed and replaced with DMEM/2% FBS containing various concentrations (10–1000 μg/mL) of NB-DNJ, 5 wells per each concentration. Drug cytotoxicity was assessed on days 1, 2 and 3 post-treatment using the XTT metabolic assay (Roche, Laval, Canada) according to the manufacturer's directions.
SDS-PAGE analysis of SARS-CoV preparation
The SARS-CoV preparation was analysed by 9% polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining. The stained protein band corresponding to the SARS-CoV S glycoprotein was cut from the gel for enzymatic digestion with peptide N-glycosidase F (PNGase F, Roche Diagnostics Ltd., Basel, Switzerland) and subsequent N-glycan analysis (see below).
Western blotting analysis of the SARS-CoV S glycoprotein
To confirm that the band selected for N-glycan analysis was that corresponding to the SARS-CoV S glycoprotein, an anti-SARS-CoV S antibody was used in a western blotting protocol. Briefly, the SARS-CoV preparation was subjected to SDS-PAGE on a 9% gel and the proteins were transferred to PVDF membranes using the semi-dry transfer technique. The membranes were blocked in 5% milk solution in 1× phosphate-buffered saline (PBS) overnight then incubated with a mouse anti-SARS IgG1 (clone 16F1071, Imgenex, CA, USA) diluted 1:1000 in 3% milk solution at room temperature for at least 1 h. The membranes were washed three times in PBS–0.05% Tween-20 before 1 h incubation at room temperature with a horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (Sigma Chemicals) diluted 1:1000 in 3% milk solution. The membranes were washed three times in PBS–0.05% Tween-20, then once with PBS. The SARS-CoV glycoprotein band was detected by developing the membranes using the ECL Western blotting detection kit (Amersham Biosciences, Little Chalfont, UK) according to the manufacturers instructions and followed by exposure to X-ray film (CL-X Posure film, Pierce).
N-glycan release using peptide N-glycosidase F
The procedure essentially followed that described by Küster et al. (1997). The glycoproteins were separated by SDS-PAGE with a BioRad Mini-Protean II apparatus with 80 × 80 × 0.75 mm plates and a 9% resolving gel. Developed gels were stained with Coomassie Blue R-250. The SARS-CoV S glycoprotein gel bands were excised from the gel and cut into pieces about 1 mm2, washed with 300 μL 20 mM NaHCO3 (BDH Limited, Poole, UK) and soaked in the buffer for 30 min. This procedure was repeated twice; on the third wash, 45 mM dithiothreitol (20 μL) was added and incubated for 30 min at 60 °C to reduce disulfide bonds. Iodoacetamide (100 mM; 20 μL) was added and the solution was incubated for 30 min in the dark. Then a 1:1 (v/v) solution (300 μL) of aqueous acetonitrile:20 mM NaHCO3, pH 7.0, was added and incubated for 60 min followed by vacuum drying. PNGase F (EC 3.2.2.18, 30 mL, 100 U/mL; Roche Diagnostics Ltd.) and NaHCO3 buffer (enough to cover the gel, 70–100 μL) was added to the dried gel pieces which were incubated at 37 °C for 12–16 h (overnight). The supernatant was retained and the gel was washed twice with water (200 μL), and then with acetonitrile (200 μL, Riedel de Häen Laboratory, Seelze, Germany); the washes were collected and desalted by incubation with 50 μL H+-activated AG-50, then centrifuged. The supernatant was filtered through 0.45 μm Millipore LH hydrophilic PTFE syringe-tip filter then vacuum dried in preparation for direct mass spectrometric analysis or 2-aminobenzamide (2AB) labelling followed by HPLC analysis. Samples for mass spectrometric analysis were further cleaned by drop dialysis with a Nafion 117 membrane (Börnsen et al., 1995).
Glycan derivatisation with 2-aminobenzamide
Glycans were labelled with 2AB by reductive amination (Bigge et al., 1995) using a 2AB labelling kit from Glyko (now part of Prozyme at Hayward, CA, USA). The 2AB labelling solution was prepared according to the manufacturer's instructions and stored at −20 °C. Five microliters of this solution was added to the dried sugars and incubated at 65 °C for 2 h. Residual 2AB reagent was removed by ascending paper chromatography on 3MM chromatography paper (Millipore, Watford, UK) in 100% acetonitrile. After 2 h, the sample was viewed under a UV light and the spot corresponding to the labelled sugars was cut from the paper and the sugars were eluted using four sequential aliquots of water (0.5 mL) into a nitric acid-washed glass tube. The eluted N-glycans were vacuum dried and then redissolved in water (200 μL).
Glycan sequencing using exoglycosidase digestion
One to two picomoles of 2AB-labelled N-glycans were vacuum dried and incubated at 37 °C for 10 h in 50 mM sodium citrate buffer (pH 5.2) with exoglycosidases (Glyko) as follows: Arthrobacter ureafaciens sialidase (ABS, EC 3.2.1.18), a mixture of ABS and bovine testis β-galactosidase (BTG, EC 3.2.1.23), and a mixture of ABS, BTG and Streptococcus pneumoniae N-acetyl-hexosaminidase (GUH, EC 3.2.1.30) at the manufacturer's recommended concentrations. In a second experiment, glycans were incubated with Jack bean mannosidase (JBM, EC 3.2.1.24) 10 μL, 50 U/mL and Aspergillus saitoi α1-2-mannosidase (ASM), 2 μL, 1 mU/mL. The enzymes were removed after exoglycosidase digestions using washed (200 μL water, centrifugation at 7000 rpm for 10 min) nitrocellulose 0.45 μm microspin filters (Radleys, Saffron Walden, UK). The sample (10 μL) was added to the filter and centrifuged at 13,000 rpm for 3 min. The digestion tube was washed with water (20 μL) which was transferred to the filter and centrifuged at 13,000 rpm for 3 min. The filter was rinsed with water (100 μL) and centrifuged at 13,000 rpm for a further 3 min. The collected solution was vacuum dried and then redissolved in water (20 μL) for analysis by HPLC.
Normal-phase HPLC analysis
Acetonitrile (80 μL) was added to 10% of the sample (20 μL) and 95 μL was examined on a normal-phase (NP)-HPLC system (Waters Alliance 2690, Waters Corporation, Milford, MA, USA) fitted with a GlycoSep N column (Oxford GlycoSciences, Abingdon, UK) with a run time of 180 min (Guile et al., 1996). The N-linked sugars eluted from the column with a retention time of between 40 and 120 min. Comparison with a dextran ladder allowed assignment of glucose unit (GU) values to each peak, which facilitated identification of the glycans present in the released pool.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)
Positive ion MALDI-TOF mass spectra were obtained with a Waters-MS Technologies TofSpec 2E reflectron TOF mass spectrometer (Waters Ltd, Manchester). The pulse and acceleration voltages were 3 and 20 kV, respectively. Data acquisition and processing were performed with MassLynx software version 3.3. Samples were prepared by mixing an aqueous solution of the sample (0.5 μL) with a saturated solution of DHB in acetonitrile (0.3 μL) on the stainless steel MALDI target and allowing the mixture to dry under ambient conditions. The dried sample spot was then re-dissolved in ethanol (0.2 μL) and again allowed to dry.
Electrospray mass spectrometry
Electrospray mass spectrometry was performed with a Waters quadrupole-time-of-flight (Q-TOF) Ultima Global instrument in negative ion mode. Samples in 1:1 (v:v) methanol:water were infused through Proxeon nanospray capillaries (Proxeon Biosystems, Odense, Denmark). The ion source conditions were as follows: temperature, 120 °C; nitrogen flow, 50 L/h; infusion needle potential, 1.2 kV; cone voltage, 100 V; RF-1 voltage, 150 V. Spectra (2 s scans) were acquired with a digitization rate of 4 GHz and accumulated until a satisfactory signal:noise ratio had been obtained. The major ions were [M+H2PO4]−, the phosphate apparently arising from residual phosphate in the original sample, as observed previously from all glycan samples released as above. For MS/MS data acquisition, the parent ion was selected at low resolution (about 5 m/z mass window) to allow transmission of isotope peaks and fragmented with argon at a pressure (recorded on the instrument's pressure gauge) of 0.5 mBar. The voltage on the collision cell was adjusted with mass and charge to give an even distribution of fragment ions across the mass scale. Typical values were 80–120 V. Other voltages were as recommended by the manufacturer. Instrument control, data acquisition and processing were performed with MassLynx software version 4.0. Displayed spectra have been smoother using the Savitzky Golay algorithm (5 × 2) and abundant background ions at m/z 195, 291 and 344 have been removed for clarity.
Acknowledgments
We thank the Wellcome Trust and the Biotechnology and Biological Sciences Research Council for equipment grants to purchase the Q-TOF and TofSpec mass spectrometers, respectively. We thank the Public Health Agency of Canada (PHAC) for financial support.
References
- Bergeron E., Vincent M.J., Wickham L., Hamelin J., Basak A., Nichol S.T., Chrétien M., Seidah N.G. Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun. 2005;326:554–563. doi: 10.1016/j.bbrc.2004.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigge J.C., Patel T.P., Bruce J.A., Goulding P.N., Charles S.M., Parekh R.B. Nonselective and efficient fluorescent labeling of glycans using 2-aminobenzamide and anthranilic acid. Anal. Biochem. 1995;230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- Börnsen K.O., Mohr M.D., Widmer H.M. Ion exchange and purification of carbohydrates on a Nafion(R) membrane as a new sample pretreatment for matrix-assisted laser desorption-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1995;9:1031–1034. [Google Scholar]
- Chen J., Subbarao K. The immunobiology of SARS. Annu. Rev. Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- Domon B., Costello C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988;5:397–409. [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Eng. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H., Song H.C., Abrignani S., Covacci A., Rappuoli R. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Nichol S.T., Klenk H.D., Peters C.J., Sanchez A. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology. 1994;199:469–473. doi: 10.1006/viro.1994.1147. [DOI] [PubMed] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell–cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guile G.R., Rudd P.M., Wing D.R., Prime S.B., Dwek R.A. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal. Biochem. 1996;240:210–226. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey D.J. Fragmentation of negative ions from carbohydrates. Part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J. Am. Soc. Mass Spectrom. 2005;16:622–630. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Harvey D.J. Fragmentation of negative ions from carbohydrates. Part 2. Fragmentation of high-mannose N-linked glycans. J. Am. Soc. Mass Spectrom. 2005;16:631–646. doi: 10.1016/j.jasms.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Harvey D.J. Fragmentation of negative ions from carbohydrates. Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 2005;16:647–659. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Harvey D.J., Martin R.L., Jackson K.A., Sutton C.W. Fragmentation of N-linked glycans with a MALDI-ion trap time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2004;18:2997–3007. doi: 10.1002/rcm.1709. [DOI] [PubMed] [Google Scholar]
- Harvey D.J., Crispin M., Scanlan C., Singer B.B., Lucka L., Chang V.T., Radcliffe C.M., Thobhani S., Yuen C.-T., Rudd P.M. Differentiation between isomeric triantennary N-linked glycans by negative ion tandem mass spectrometry and confirmation of glycans containing galactose attached to the bisecting (β1-4-GlcNAc) residue in N-glycans from IgG. Rapid Commun. Mass Spectrom. 2008;22:1047–1052. doi: 10.1002/rcm.3470. [DOI] [PubMed] [Google Scholar]
- Harvey D.J., Royle L., Radcliffe C.M., Rudd P.M., Dwek R.A. Structural and quantitative analysis of N-linked glycans by MALDI and negative ion nanospray mass spectrometry. Anal. Biochem. 2008;376:44–60. doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Harvey D.J., Merry A.H., Royle L., Campbell M.P., Dwek R.A., Rudd P.M. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D.J., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., DeMartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci., USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V.N., Cheng K., Simonsen J.N., Perreault H., Wilkins J., Ens W., Plummer F., Standing K.G. Mass spectrometric characterization of proteins from the SARS virus. A preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Cannon A.D.L., Curtis M., Farrar B., Morgan L., Pezzanite L., Sanchez A.J., Slaughter K.A., Stevens T.L., Stockton P.C., Wagoner K.D., Sanchez A., Nichol S., Vincent M., Osborne J., Honig J., Erickson B.R., Holloway B., McCaustland K., Lingappa J., Lowe L., Scott S., Lu X., Villamarzo Y., Cook B., Chen Q., Birge C., Shu B., Pallansch M., Tatti K.M., Morken T., Smith C., Greer P., White E., McGlothen T., Bhatnagar J., Patel M., Bartlett J., Montague J., Lee W., Packard M., Thompson H.A., Moen A., Fukuda K., Uyeki T., Harper S., Klimov A., Lindstrom S., Benson R., Carlone G., Facklam R., Fields P., Levett P., Mayer L., Talkington D., Thacker W.L.T., Tondella M.L.C., Whitney C., Robertson B., Warnock D., Brooks J.T., Schrag S., Rosenstein N., Ganem D., Poutanen S.M., Chen T.-J., Hsiao C.-H., Wai-Fu N.G., Ho M., Keung T.-K., Nghiem K.H., Nguyen H.K.L., Le M.Q., Nguyen H.H.T., Hoang L.T., Vu T.H., Vu H.Q., Chunsuttiwat S. A novel coronavirus associated with severe acute respiratory syndrome. New Eng. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Küster B., Wheeler S.F., Hunter A.P., Dwek R.A., Harvey D.J. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high performance liquid chromatography. Anal. Biochem. 1997;250:82–101. doi: 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus p450. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M.F., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Lubas W.A., Spiro R.G. Golgi endo-alpha-d-mannosidase from rat liver, a novel N-linked carbohydrate unit processing enzyme. J. Biol. Chem. 1987;262:3775–3781. [PubMed] [Google Scholar]
- Lubas W.A., Spiro R.G. Evaluation of the role of rat liver Golgi endo-alpha-d-mannosidase in processing N-linked oligosaccharides. J. Biol. Chem. 1988;263:3990–3998. [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Marzi A., Gramberg T., Simmons G., Möller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pöhlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.E., Spiro R.G. Demonstration that Golgi endo-alpha-d-mannosidase provides a glucosidase-independent pathway for the formation of complex N-linked oligosaccharides of glycoproteins. J. Biol. Chem. 1990;265:13104–13112. [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powlesland A.S., Fisch T., Taylor M.E., Smith D.F., Tissot B., Dell A., Pohlmann S., Drickamer K. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J. Biol. Chem. 2008;283:593–602. doi: 10.1074/jbc.M706292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-h., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Shi Z., Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.C., Seo M.Y., Stadler K., Yoo B.J., Choo Q.L., Coates S.R., Uematsu Y., Harada T., Greer C.E., Polo J.M., Pileri P., Eickmann M., Rappuoli R., Abrignani S., Houghton M., Han J.H. Synthesis and characterization of a native oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2004;78:10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Lim S.G., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust D.J., Shepherd V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Eaton B.T. Bats, civets and the emergence of SARS. Curr. Top. Microbiol. Immunol. 2007;315:325–344. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W.-P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W., Dai S., Li X., Zhu Y., Li J., Wu S., Guo L., Dai J., Wang J., Wan P., Chen T., Du C., Li D., Wan J., Kuai X., Li W., Shi R., Wei H., Cao C., Yu M., Liu H., Dong F., Wang D., Zhang X., Qian X., Zhu Q., He F. Proteomic analysis on structural proteins of severe acute respiratory syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]