Abstract
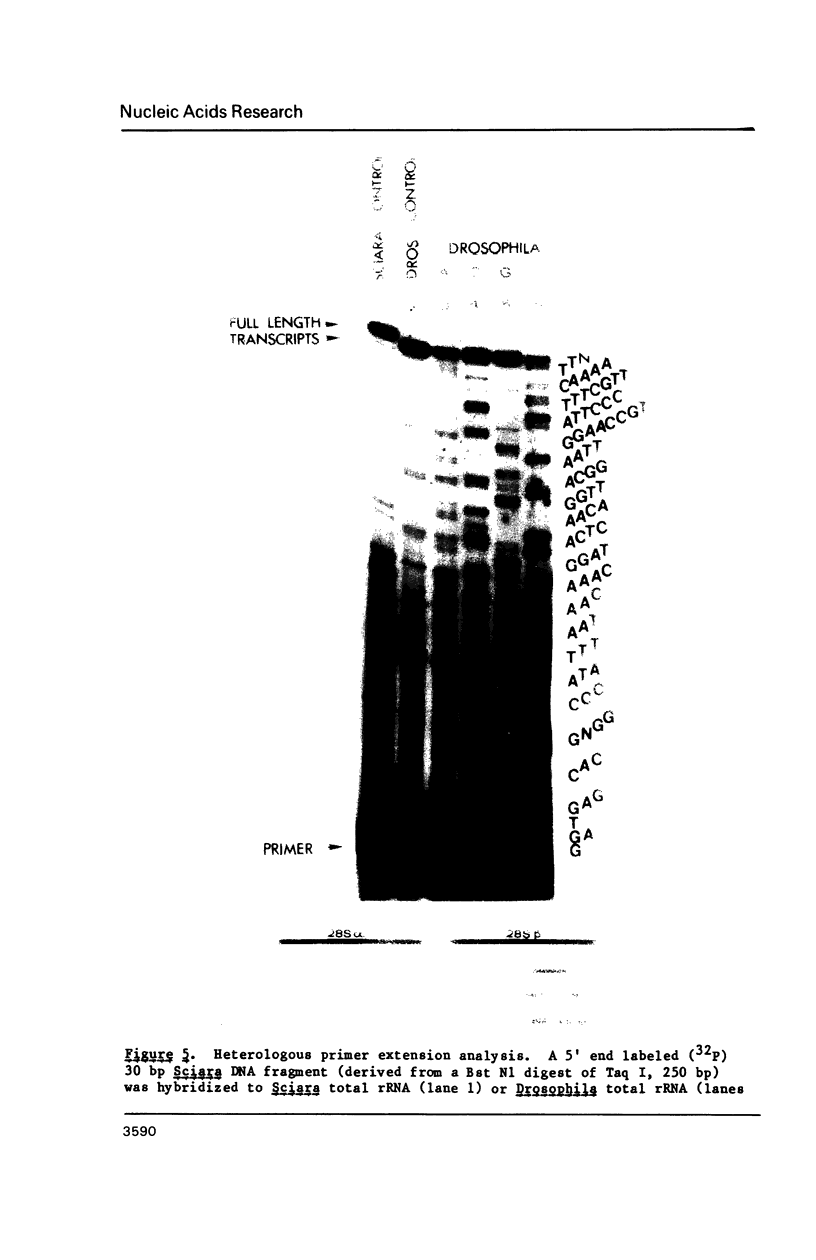
We have determined the sequence of the rDNA region between the 28S alpha and 28S beta rRNA coding segments (termed a "gap") in the insect Sciara coprophila, and have used S1 nuclease mapping and cDNA primer extension to define the 5' and 3' boundaries of the gap. Only 19 bases found in rDNA at the gap region are absent from mature 28S rRNA. Eukaryotic rRNAs contain stretches of nucleotides ("expansion segments") which are absent in E. coli rRNA. The gap region in Sciara is located within expansion segment V. Therefore, the excision of 19 bases in the Sciara gap suggests that a large portion of expansion segment V plays no function in mature ribosomes. Specific sequences conserved in Sciara and Drosophila are considered as candidates for recognition signals for the excision of the gap transcript.
Full text
PDF





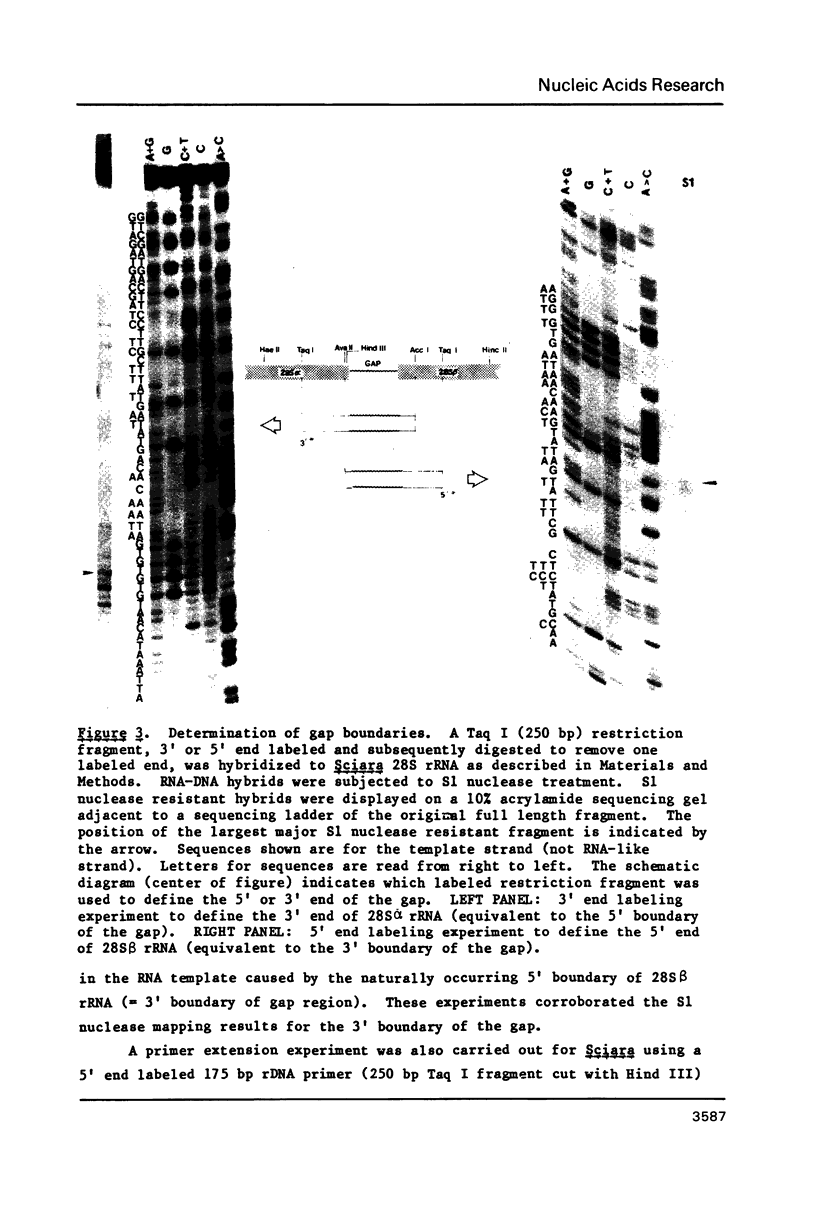
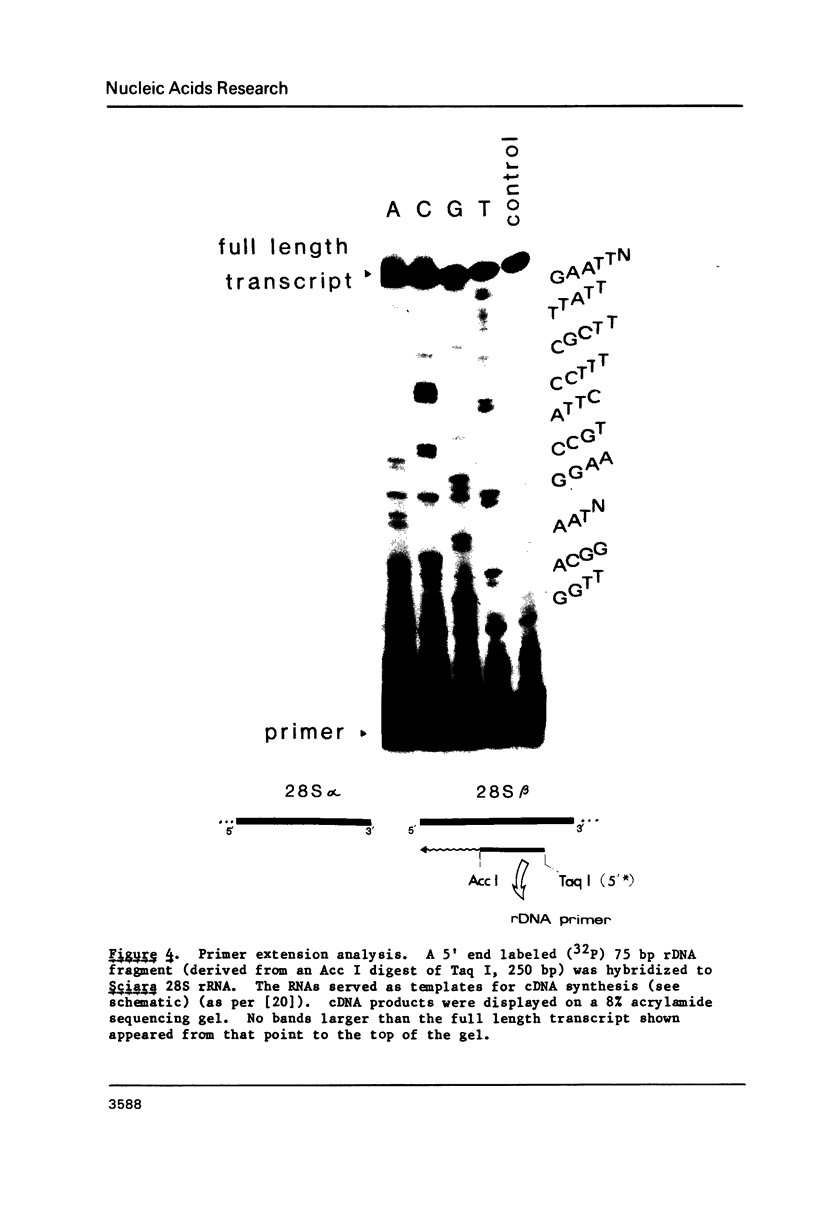








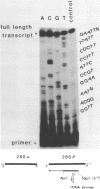
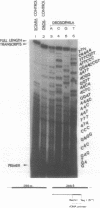
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Applebaum S. W., Ebstein R. P., Wyatt G. R. Dissociation of ribonucleic acid from silkmoth pupae by heat and dimethylsulfoxide: evidence for specific cleavage points. J Mol Biol. 1966 Oct 28;21(1):29–41. doi: 10.1016/0022-2836(66)90077-5. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Gerbi S. A. Fine structure of ribosomal RNA. II. Distribution of methylated sequences within Xenopus laevis rRNA. Nucleic Acids Res. 1979 Nov 24;7(6):1497–1511. doi: 10.1093/nar/7.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Rio D. C. Localization of transcribed regions on extrachromosomal ribosomal RNA genes of Tetrahymena thermophila by R-loop mapping. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5051–5055. doi: 10.1073/pnas.76.10.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C. G., Tague B. W., Ware V. C., Gerbi S. A. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984 Aug 10;12(15):6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K., Long E. O. Ribosomal DNA in Drosophila melanogaster. I. Isolation and characterization of cloned fragments. J Mol Biol. 1978 Dec 25;126(4):749–768. doi: 10.1016/0022-2836(78)90018-9. [DOI] [PubMed] [Google Scholar]
- Delanversin G., Jacq B. Séquence de la région de la coupure centrale du précurseur de l'ARN ribosomique 26S de Drosophile. C R Seances Acad Sci III. 1983;296(22):1041–1044. [PubMed] [Google Scholar]
- Din N., Engberg J., Kaffenberger W., Eckert W. A. The intervening sequence in the 26S rRNA coding region of T. thermophila is transcribed within the largest stable precursor for rRNA. Cell. 1979 Oct;18(2):525–532. doi: 10.1016/0092-8674(79)90069-2. [DOI] [PubMed] [Google Scholar]
- Eckert W. A., Kaffenberger W., Krohne G., Franke W. W. Introduction of hidden breaks during rRNA maturation and ageing in Tetrahymena pyriformis. Eur J Biochem. 1978 Jul 3;87(3):607–616. doi: 10.1111/j.1432-1033.1978.tb12413.x. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981 Jun 25;9(12):2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest M. J., Feigelson P. Increase in hepatic tyrosine aminotransferase mRNA during enzyme induction by N6,O2'-dibutyryl cyclic AMP. J Biol Chem. 1978 Jan 25;253(2):319–322. [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Georgiev O. I., Nikolaev N., Hadjiolov A. A., Skryabin K. G., Zakharyev V. M., Bayev A. A. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981 Dec 21;9(24):6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H. The fragments from the 28 S ribosomal RNA of Galleria mellonella with unesterified uridine at the 3'-termini. Biochim Biophys Acta. 1976 Mar 4;425(2):185–195. doi: 10.1016/0005-2787(76)90024-1. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Jourdan R., Jacq B. Late steps in the maturation of Drosophila 26 S ribosomal RNA: generation of 5-8 S and 2 S RNAs by cleavages occurring in the cytoplasm. J Mol Biol. 1976 Feb 15;101(1):85–105. doi: 10.1016/0022-2836(76)90067-x. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Coding and spacer sequences in the 5.8S-2S region of Sciara coprophila ribosomal DNA. Nucleic Acids Res. 1980 Aug 25;8(16):3565–3573. doi: 10.1093/nar/8.16.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lava-Sanchez P. A., Puppo S. Occurrence in vivo of "hidden breaks" at specific sites of 26 S ribosomal RNA of Musca carnaria. J Mol Biol. 1975 Jun 15;95(1):9–20. doi: 10.1016/0022-2836(75)90331-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Pene J. J., Knight E., Jr, Darnell J. E., Jr Characterization of a new low molecular weight RNA in HeLa cell ribosomes. J Mol Biol. 1968 May 14;33(3):609–623. doi: 10.1016/0022-2836(68)90309-4. [DOI] [PubMed] [Google Scholar]
- Qu H. L., Michot B., Bachellerie J. P. Improved methods for structure probing in large RNAs: a rapid 'heterologous' sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5' terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz-Pohl R., Matsumoto L., Gerbi S. A. Two distinct intervening sequences in different ribosomal DNA repeat units of Sciara coprophila. Nucleic Acids Res. 1981 Aug 11;9(15):3747–3764. doi: 10.1093/nar/9.15.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R., Gerbi S. A., Glätzer K. H. Ribosomal DNA of fly Sciara coprophila has a very small and homogeneous repeat unit. Mol Gen Genet. 1979 May 23;173(1):1–13. doi: 10.1007/BF00267685. [DOI] [PubMed] [Google Scholar]
- Shine J., Hunt J. A., Dalgarno L. Studies on the 3'-terminal sequences of the large ribosomal ribonucleic acid of different eukaryotes and those associated with "hidden" breaks in heart-dissociable insects 26S ribonucleic acid. Biochem J. 1974 Sep;141(3):617–625. doi: 10.1042/bj1410617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Wollenzien P. L., Youvan D. C., Hearst J. E. Structure of psoralen-crosslinked ribosomal RNA from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1642–1646. doi: 10.1073/pnas.75.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]