Abstract
Nucleophosmin (NPM) is a nucleolar phosphoprotein that is involved in many cellular processes and has both oncogenic and growth suppressing activities. NPM is localized primarily in nucleoli but shuttles between the nucleus and the cytoplasm, and sustained cytoplasmic distribution contributes to its tumor promoting activities. Plakoglobin (PG, γ-catenin) is a homolog of β-catenin with dual adhesive and signaling functions. These proteins interact with cadherins and mediate adhesion, while their signaling activities are regulated by association with various intracellular partners. Despite these similarities, β-catenin has a well-defined oncogenic activity, whereas PG acts as a tumor/metastasis suppressor through unknown mechanisms. Comparison of the proteomic profiles of carcinoma cell lines with low- or no PG expression with their PG-expressing transfectants has identified NPM as being upregulated upon PG expression. Here, we examined NPM subcellular distribution and in vitro tumorigenesis/metastasis in the highly invasive and very low PG expressing MDA-MB-231 (MDA-231) breast cancer cells and their transfectants expressing increased PG (MDA-231-PG) or NPM shRNA (MDA-231-NPM-KD) or both (MDA-231-NPM-KD+PG). Increased PG expression increased the levels of nucleolar NPM and coimmunoprecipitation studies showed that NPM interacts with PG. PG expression or NPM knockdown decreased the growth rate of MDA-231 cells substantially and this reduction was decreased further in MDA-231-NPM-KD+PG cells. In in vitro tumorigenesis/metastasis assays, MDA-231-PG cells showed substantially lower and MDA-231-NPM-KD cells substantially higher invasiveness relative to the MDA-231 parental cells, and the co-expression of PG and NPM shRNA led to even further reduction of the invasiveness of MDA-231-PG cells. Furthermore, examination of the levels and localization of PG and NPM in primary biopsies of metastatic infiltrating ductal carcinomas revealed coordinated expression of PG and NPM. Together, the data suggest that PG may regulate NPM subcellular distribution, which may potentially change the function of the NPM protein from oncogenic to tumor suppression.
Keywords: NPM, plakoglobin, tumor suppressor, metastasis suppressor
Introduction
Nucleophosmin (NPM; also known as B23, NO38 or Numatrin) is a 38-kDa estrogen-regulated nucleolar phosphoprotein that shuttles between the nucleus and cytoplasm. NPM has been implicated in many cellular processes including the shuttling of ribosomal precursor proteins between the cytoplasm and nucleus, nuclear protein chaperone activity, the maintenance of genomic stability and the indirect regulation of growth and proliferation.1, 2, 3 NPM interacts with a number of growth regulating proteins including the p53 and ARF tumor suppressors and the p53 inhibitor HDM2. The overexpression of NPM has been strongly correlated with tumor progression and may serve as a marker for certain types of carcinomas, such as gastric, ovarian and prostate,1 and Ewing's sarcoma.4 Similarly, chromosomal translocations involving the NPM gene give rise to acute myeloid leukemia (AML).5 Because of these observations, it was proposed that NPM acts as an oncogene. Recent studies with NPM knockout mice, however, have shown that heterozygous animals exhibit signs of myelodysplastic syndrome and exhibit genome instability,3, 6 implying that NPM can also act as a tumor suppressor. Whether NPM is oncogenic or acts as a tumor suppressor appears to depend on its subcellular localization. In the nucleolus, NPM serves as a histone chaperone during nucleosome disassembly and regulates the stabilization and localization of p14ARF.3, 7 DNA damage and stress induce the translocation of the NPM-p14ARF complex from the nucleolus to the nucleoplasm where p14ARF can bind to and dissociate the p53 inhibitor HDM2 from p53, thereby activating the tumor suppressor pathway.8 In the nucleolus, NPM also binds to and stabilizes the ubiquitin ligase FBW7γ, which has been implicated in the ubiquitination and degradation of the oncogenic transcription factor Myc.9, 10 In the absence of functional NPM, FBW7γ is mislocalized to the cytoplasm and degraded, leading to increased levels of Myc and cell proliferation. Similarly, in AML, the majority of NPM mutations introduce a de novo nuclear export signal, leading to the cytoplasmic localization of the protein.11
Plakoglobin (PG) is a structural component of the adherens junction and desmosomes.12 It is a member of the Armadillo family of proteins and a homolog of β-catenin. PG interacts with the same cellular partners as β-catenin and likewise has dual adhesive and signaling functions.13, 14 Despite their similarity, β-catenin has known oncogenic potential via its role in the Wnt signaling pathway,15, 16 whereas PG generally functions as a tumor/metastasis suppressor.17, 18, 19, 20, 21, 22 How PG functions as a tumor suppressor has yet to be determined. To investigate the underlying mechanism(s) of the tumor/metastasis suppressing activities of PG, we have developed an experimental model system using human squamous and mammary carcinoma cell lines that have very little or no PG expression, and exhibit transformed morphologies and different degrees of invasiveness. These cells were used to develop transfectants with different levels of expression and subcellular localization of PG to specifically assess its role at the membrane, in the cytoplasm and in the nucleus. These transfectants were then characterized by a combination of morphological, molecular and biochemical approaches, including proteomics and transcriptome analyses.18, 21, 22, 23, 24, 25, 26 These studies have provided strong evidence that PG can function as a growth regulator and tumor/metastasis suppressor in both the cytoplasm and nucleus by modifying the expression/function of genes involved in cell proliferation, differentiation, migration and apoptosis. NPM was one such molecule that was identified among the differentially expressed proteins/genes in both squamous cell and mammary carcinoma cell lines and was further characterized. In this study, we describe our observations in mammary carcinoma cells and show that PG interacts with NPM in normal mammary epithelial cells in various subcellular compartments. Furthermore, we show that increased expression of PG in the highly invasive and very low PG expressing mammary carcinoma cell line MDA-MB-231 (hereafter called MDA-231) changes the morphology of these cells from fibroblastoid to epithelioid, concurrent with increased nucleolar and decreased cytoplasmic pools of NPM, as well as a decrease in cell proliferation, migration and invasion in vitro.
Results
Plakoglobin expression in MDA-231 cells increases NPM protein levels, induces an epithelioid phenotype and decreases growth
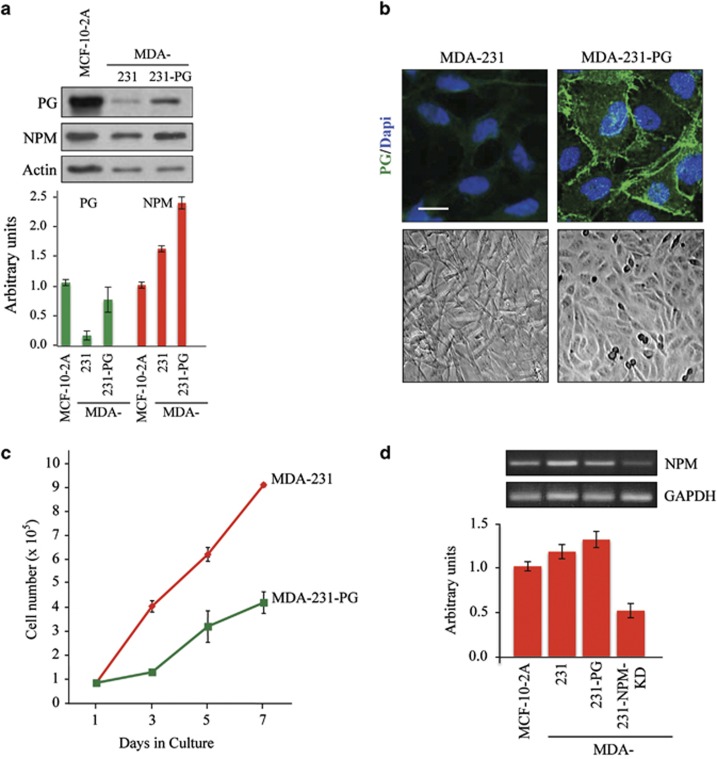
PG expression and subcellular localization in MDA-231-PG transfectants were verified by western blot and immunofluorescence. In Supplementary Figure 1, equivalent amounts of total proteins from 10 single-cell-isolated clones from three independent transfections were processed for western blot using PG and NPM antibodies. Several clones were analyzed, producing similar, if not identical results in various assays, and data from a representative clone are presented throughout this study. In PG transfectants, NPM levels increased in all MDA-231-PG clones relative to the parental MDA-231 cells (Figure 1a, Supplementary Figure 1). In all PG expressing clones, PG was primarily localized to the areas of cell–cell contact (Figure 1b, top). Furthermore, PG expression induced a morphological change from fibroblastoid to epithelioid (Figure 1b, bottom). Concurrent with this change in morphology, the growth rate of the PG expressing cells was substantially reduced (Figure 1c). RT–PCR was used to assess the effect of PG on NPM expression and NPM transcript levels were not noticeably changed relative to the parental MDA-231 and the normal MCF-10-2A cell lines (Figure 1d). Therefore, PG expression in MDA-231 cells induced an epithelioid morphology and decreased growth rate concurrent with increased levels of NPM protein.
Figure 1.
Characterization of plakoglobin expressing MDA-231 cells (MDA-231–PG). (a) Total cell lysates from MDA-231, MDA-231-PG and MCF-10-2A (control) were processed for western blot using PG, NPM and actin antibodies. Histograms were generated by normalizing the amount of each protein in each cell line to the amount of actin in the same line relative to MCF-10-2A cells. Quantitation for the PG transfectants represents the average of multiple independent clones (Supplementary Figure 1). Each experiment was repeated three to five times and a typical western blot is shown. (b) MDA-231 and MDA-231-PG cells were grown to confluence, viewed under a phase contrast microscope and photographed using the × 20 objective. To visualize the subcellular distribution of PG, confluent cultures grown on glass coverslips were extracted with CSK buffer, fixed with formaldehyde and stained with PG antibodies. Nuclei were counterstained using DAPI. Images were captured using a × 63 objective of a laser scanning confocal microscope, enlarged × 2 and photographed. Bar, 10 μm. (c) Replicate cultures of MDA-231 and MDA-231-PG were established at single cell density and cells were counted at 1, 3, 5 and 7 days. Each time point represents the average of three independent experiments. For MDA-231-PG transfectants the values are the average of multiple independent clones. The absence of error bars at some time points is because of the small differences among the experiments. (d) Total cellular RNA was isolated from MCF-10-2A, MDA-231, -231-PG and -231-NPM-KD (MDA-231 cells expressing NPM shRNA) cells, reverse transcribed and processed for PCR using primers specific to NPM and GADPH. The histogram represents the NPM/GAPDH ratio for each cell line normalized to MCF-10-2A cells. Quantitations for theMDA-231-PG and MDA-231-NPM-KD transfectants represent the average of multiple independent clones (Supplementary Figure 5). Each experiment was repeated at least three times and a typical blot is presented.
Subcellular distribution of NPM and PG in MDA-231 parental and MDA-231-PG transfectants
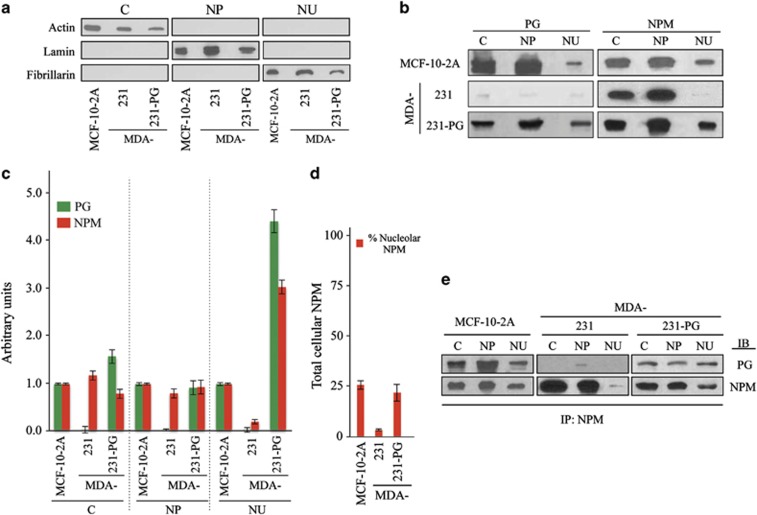
NPM shuttles between the cytoplasm and nucleus and in non-transformed cells is primarily detected in nucleoli.3 As PG expression increased NPM protein levels only, we determined if PG influences the levels of NPM in various subcellular compartments. To this end, cultures of MCF-10-2A (control), MDA-231 and MDA-231-PG cells were processed for subcellular fractionation and nucleoli isolation according to the Lamond's protocol followed by western blot (Figure 2a). The purity of the cytoplasmic, nucleoplasmic and nucleolar fractions were verified by western blot using actin, lamin B and fibrillarin antibodies, respectively, (Figure 2a). The three fractions were then processed for western blots using NPM and PG antibodies (Figure 2b). For quantitation, the amount of each protein detected in the cytoplasmic, nucleoplasmic and nucleolar fractions of each cell line was first normalized to the amount of actin, lamin B and fibrillarin in each cell line, respectively, and then to the amount of each protein in MCF-10-2A cells as depicted in the histogram (Figure 2c). PG and NPM were detected in all three fractions in MCF-10-2A and MDA-231-PG cells. In MDA-231 cells, however, PG was barely detectable, and although NPM was present in all three fractions of these cells with similar amounts in the cytoplasmic and nucleoplasmic fractions, the amount in the nucleolar fraction was negligible (Figures 2b and c). In MDA-231-PG cells, we observed a small but consistent decrease in the amount of cytoplasmic NPM relative to the MDA-231 cells, whereas in these cells, the amount of NPM was highly increased in the nucleolar fraction (>10 × , Figures 2c and d). These observations indicate that PG expression increases the amount of the nucleolar NPM.
Figure 2.
Subcellular distribution of PG and NPM and their association in MDA-231 and MDA-231-PG cells. (a, b) MCF-10-2A (control), MDA-231 and -231-PG cells were grown in 150 mm culture dishes until confluent. Cell pellets (total of 108 cells) were processed for subcellular fractionation according to the Lamond's protocol to isolate the cytoplasmic (C), nucleoplasmic (NP) and nucleolar (NU) fractions. The purity of C, NP and NU fractions were verified by immunoblotting with actin, lamin and fibrillarin antibodies, respectively (a). Fractions were processed for immunoblotting with PG and NPM antibodies (b). The experiment was repeated five times with similar results and the data is representative of one such experiment. (c) Quantitation of the data presented in b. The amount of each protein in the cytoplasmic (C), nucleoplasmic (NP) and nucleolar (NU) fraction was normalized to the amounts of actin, lamin and fibrillarin, respectively, in the same sample extract relative to MCF-10-2A cells. PG and NPM values for MDA-231-PG transfectants are the average of multiple independent clones. (d) The ratio of the nucleolar/total cellular NPM detected in various cell lines. The value for MDA-231-PG transfectants is the average of multiple independent clones. (e) Cytoplasmic (C), nucleoplasmic (NP) and nucleolar (NU) fractions from MCF-10-2A (control), MDA-231 and -231-PG cells were prepared as described in Figure 2a and processed for immunoprecipitation using NPM antibodies. Immune complexes were resolved by SDS–PAGE and immunoblotted with antibodies to PG and NPM. Each experiment was repeated at least five times with similar results and the data is presented for one typical experiment.
PG associates with NPM in the cytoplasmic, nucleoplasmic and nucleolar fractions
The change in the levels of NPM in various subcellular fractions in MDA-231-PG led us to examine the possible association of these two proteins. In Figure 2e, cytoplasmic, nucleoplasmic and nucleolar fractions of MCF-10-2A, MDA-231 and -231-PG were processed for immunoprecipitation with NPM antibodies followed by immunoblotting with PG and NPM antibodies. PG was coimmunoprecipitated with NPM from all the subcellular fractions of MCF-10-2A and MDA-231-PG cells. In MDA-231 cells, a very small amount of PG was coprecipitated with NPM from the nucleoplasmic fraction (Figure 2e). In this assay, lower amounts of PG were coprecipitated with NPM from the nucleoplasmic fraction of MDA-231-PG relative to MCF-10-2A cells. In contrast, higher amounts of PG were coprecipitated with NPM from the nucleolar fraction of MDA-231-PG cells relative to that of MCF-10-2A (Figure 2e, Supplementary Figure 2). The relative amount of the nucleolar to the total cellular NPM was similar in MCF-10-2A and MDA-231-PG as was demonstrated earlier by western blot of various fractions with NPM antibodies (Figure 2d). Reciprocal immunoprecipitation and immunoblotting of the cytoplasmic, nucleoplasmic and nucleolar fractions of MDA-231-PG with PG and NPM antibodies further confirmed the association of these proteins (Supplementary Figure 3). Together, these results suggest that PG expression increases the nucleolar pool of NPM and that, at least, some of this pool is present in association with PG (Supplementary Figure 4).
NPM knockdown induces epithelioid morphology and decreases growth rate in MDA-231 cells
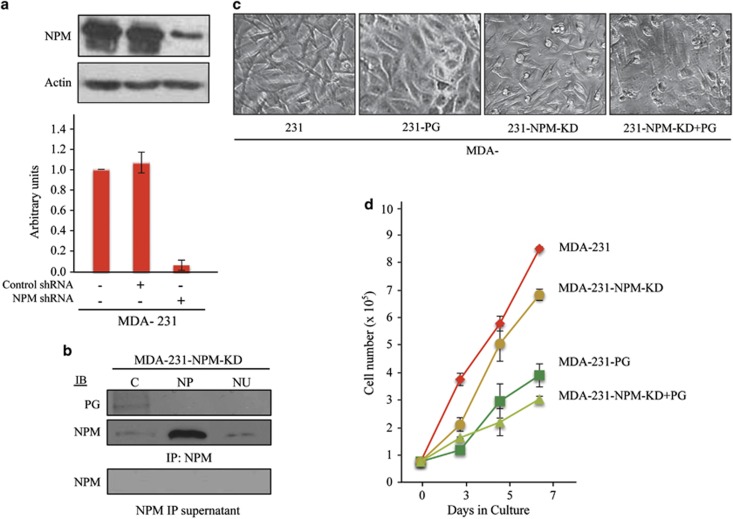
The results of the coimmunoprecipitation experiments suggest that PG interactions with NPM in the cytoplasm and nucleoplasm may sequester the excess NPM pools in these compartments, which are associated with its tumorigenic activities.3, 5, 27, 28 To assess whether NPM knockdown leads to changes in its subcellular distribution and the morphology and growth properties of MDA-231 cells, we knocked down NPM using NPM shRNA and generated stable transfectants (MDA-231-NPM-KD). In Figure 3a, MDA-231, MDA-231 control shRNA transfected and MDA-231-NPM-KD cells were processed for immunoblotting with NPM antibodies, which showed the reduction of NPM protein by 90% in MDA-231-NPM-KD cells relative to MDA-231 and -231-shRNA control cells. Furthermore, there was >60% reduction in NPM transcripts in MDA-231-NPM-KD cells relative to MDA-231cells (Figure 1d). Subcellular fractionation of the MDA-231-NPM-KD cells showed reductions in the cytoplasmic and nucleoplasmic pools of NPM relative to MDA-231 cells (compare MDA-231-NPM-KD in Figure 3b with MDA-231 in Figure 2e). This reduction was particularly drastic in the cytoplasmic fraction, which also coprecipitated the small amount of PG expressed in these cells. Additionally, the morphology of MDA-231-NPM-KD cells was somewhat different from MDA-231 cells. Unlike MDA-231 cells, which have long cellular processes, consistent with their migratory and invasive behavior, MDA-231-NPM-KD cells had shorter processes (Figure 3c; compare MDA-231 with 231-NPM-KD). However, MDA-231-NPM-KD cells did not exhibit the extensive cell-to-cell contact that was present in MDA-231-PG cells (Figure 3c; MDA-231-PG). Finally MDA-231-NPM-KD cells had a significantly lower growth rate relative to MDA-231 cells although the growth rate of these cells was still significantly higher than that of MDA-231-PG cells (Figure 3d and Supplementary Table 1).
Figure 3.
Characterization of the NPM knockdown MDA-231 cells. (a) MDA-231 cells were transfected with NPM shRNA or control shRNA to generate stable NPM knockdown cells (MDA-231-NPM-KD) as described in materials and methods. Total cell lysates from MDA-231, -231-control shRNA and -231-NPM-KD cells were processed for western blot using NPM and actin antibodies. NPM levels were quantitated by normalizing the amount NPM in each cell line to the amount of actin in the same line relative to MDA-231 cells. The values for MDA-231-control shRNA and -231-NPM-KD transfectants are the average of multiple independent clones. The experiment was repeated at least three times and the displayed blot is from one typical experiment. (b) MDA-231-NPM-KD cells were processed for subcellular fractionation according to the Lamond's protocol and the cytoplasmic (C), nucleoplasmic (NP) and nucleolar (NU) fractions processed for immunoprecipitation with NPM antibodies followed by western blot with NPM and PG antibodies. Three independent clones were tested and the experiment was repeated at least three times. The data presented is for one typical experiment. (c) MDA-231, -231-NPM-KD and –231-NPM-KD+PG cells were grown to confluence, viewed under a phase contrast microscope and photographed using the × 20 objective. (d) Replicate cultures of MDA-231, -231-PG, -231-NPM-KD and -231-NPM-KD+PG cells were established at single cell density and cells were counted at 3, 5 and 7 days. Each time point represents the average of three independent experiments. The values for MDA-231-PG, -231-NPM-KD and -231-NPM-KD+PG transfectants are the average of at least three independent clones. The absence of error bars at some time points is because of the small differences among the experiments.
Effects of PG expression and NPM knockdown on the migratory and invasive properties of MDA-231 cells in vitro
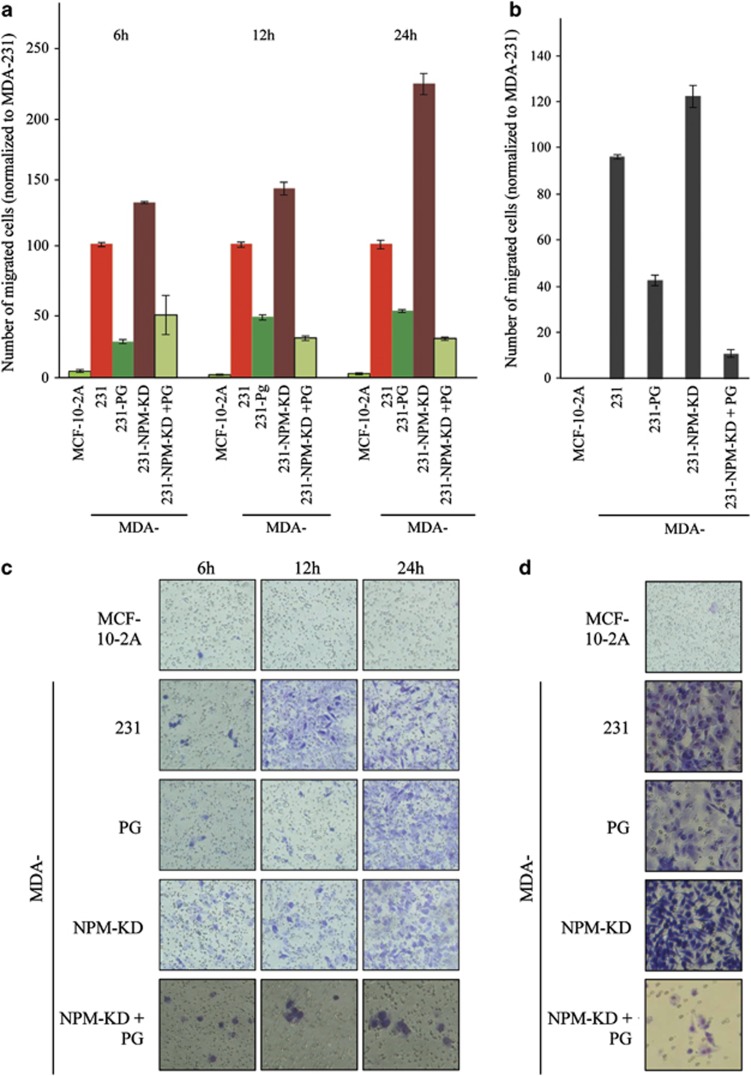
MDA-231 cells are highly invasive. To examine the effect of PG expression and NPM knockdown on the in vitro metastasis properties of these cells we performed migration and invasion assays (Figure 4). Cell migration was assessed using a transwell assay as described in materials and methods. MCF-10-2A (control), MDA-231, -231-PG and -231-NPM-KD cells were allowed to migrate through transwell filters for 6–24 h, the number of migrated cells were counted using ImageJ Cell Counter software (National Institutes of Health, Bethesda, MD, USA, http://imagej.nih.gov/ijNIH) and normalized to MDA-231 cells (Figures 4a and c). As expected, very few, if any MCF-10-2A cells had migrated through the membrane. The number of cells that had migrated was reduced by ∼75%, 55% and 60% for 6, 12 and 24 h, respectively, for MDA-231-PG cells relative to MDA-231 cells. In contrast, the numbers of migrated cells were increased by ∼30%, 40% and 120% for 6, 12 and 24 h, respectively, for MDA-231-NPM-KD cells relative to MDA-231 cells (Figure 4a).
Figure 4.
PG and NPM act synergistically to decrease motility and invasiveness of MDA-231 cells. (a, c) Transwell migration assays were performed in triplicates for MCF-10-2A (control), MDA-231, -231-PG, -231-NPM-KD and -231-NPM-KD+PG cell lines. At 6, 12 and 24 h time points the membranes were fixed, stained, cut, mounted on slides, viewed under an inverted microscope and six random fields counted for each membrane. Histograms represent the average±s.d. of the number of migrated cells for each cell line normalized to that of MDA-231 cells. (b, d) Twenty-four hour invasion assays were performed as described in A with the exception that membranes were coated with Matrigel.
Invasion through Matrigel was used to assess the invasive properties of various cell lines. Consistent with the results of the migration assays, there was a 50% reduction in the number of cells invaded through the Matrigel in MDA-231-PG cultures relative to MDA-231 cells, whereas the number of invaded cells had increased by ∼30% in MDA-231-NPM-KD cells (Figures 4b and d). Together, the results of these studies and the growth assays suggested that although both PG expression and NPM knockdown decreased the growth rate of MDA-231 cells, only PG expression decreased the in vitro migration and invasiveness of these cells. To further confirm this observation, we transfected MDA-231-NPM-KD cells with PG cDNAs and assessed cell migration and invasion in the MDA-231-NPM-KD+PG transfectants.
Supplementary Figure 5 shows the western blot of total cell lysates from MDA-231-NPM-KD cells transfected with empty vector (lane 2) or PG cDNAs (lanes 3–8; MDA-231-NPM-KD+PG). The six MDA-231-NPM-KD+PG clones originated from single cell isolates from three independent transfections. Different clones were used to assess the growth, migratory and invasive properties of MDA-231-NPM-KD+PG cells relative to MDA-231 and –231-NPM-KD cells and data in Figures 3d and 4 are representative of these clones. MDA-231-NPM-KD+PG cells showed a significant decrease in growth rate relative to both MDA-231-NPM-KD and MDA-231-PG cells (Figure 3d; Supplementary Table 1). Similarly, the results of the transwell migration assays showed >60 and ∼30% reduction in the number of migrated cells for the 12 and 24 h time points in MDA-231-NPM-KD+PG cells relative to those of MDA-231 and MDA-231-PG cells, respectively (Figures 4a and c). The differences among these cell lines were greater in Matrigel invasion assays. Here, the average number of invaded cells in MDA-231-NPM-KD+PG cultures was decreased by ∼90%, 75% and 115% relative to those detected in MDA-231, -231-PG and -231-NPM-KD cells, respectively (Figures 4b and d). Collectively, these results suggest that PG expression alone decreases the growth rate as well as the migration and invasiveness of MDA-231 cells, but when cellular/cytoplasmic levels of NPM are reduced in MDA-231-PG cells (NPM-KD+PG), these effects are more pronounced.
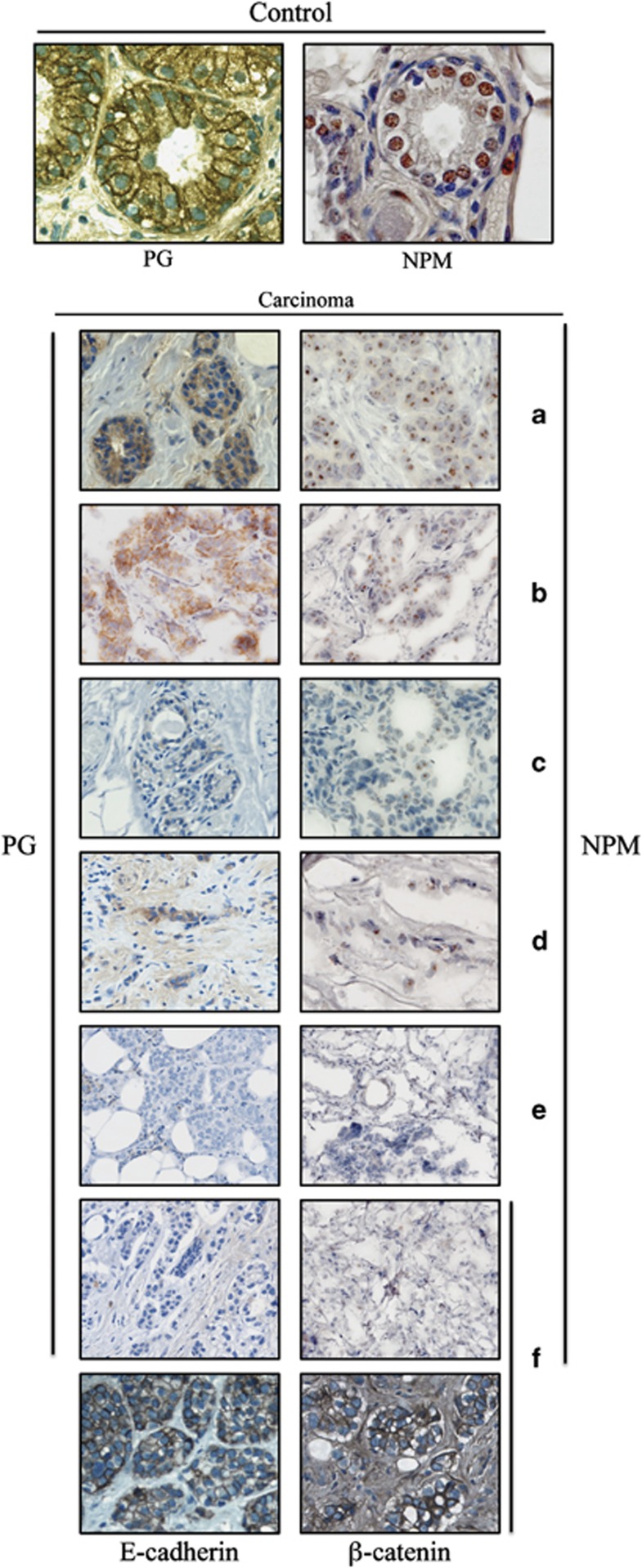
Coordinated expression of NPM and PG in normal breast and breast carcinomas
To further confirm the results of the in vitro metastasis studies, we examined the levels and localization of NPM and PG in normal breast tissue and a number of primary biopsies from metastatic infiltrating ductal carcinomas (Figure 5). In normal breast, PG distribution was primarily membrane-associated while small amounts of this protein were also detected in the cytoplasm and nuclei. NPM distribution in the normal breast tissue was primarily nucleolar with some staining in the nucleoplasm (Figure 5, top). In the tumor tissues, NPM and PG were expressed coordinately as depicted in the representative images in Figure 5a–f. In the biopsies in which PG levels were reduced and/or it's localization altered, NPM levels were also decreased and it's distribution modified (Figures 5a–f). NPM was not detected in the tumor tissues in which PG was not present (Figure 5f). Furthermore, the expression of other adherens junction markers such as E-cadherin and β-catenin in the tumors lacking PG and NPM supported the specificity of coordinated expression of PG and NPM (Figure 5f). Together, these results suggest a concordant expression of NPM and PG in vivo and support the results of in vitro metastasis studies.
Figure 5.
Concordant expression of PG and NPM in normal breast tissue and breast carcinomas. Representative IHC images ( × 63) of normal breast tissue (top) and primary biopsies of metastatic infiltrating ductal carcinomas (a–f) stained with PG and NPM antibodies as described in materials and methods.
Discussion
NPM is a multifunctional protein that is involved in many key cellular processes3, 29 and possesses both growth promoter and suppressor activities. By proteomic analysis, we identified NPM as a protein whose levels were increased upon exogenous PG expression in squamous and breast carcinoma cells that expressed little or no PG, concurrent with a fibroblastoid to epithelioid morphological change. To investigate the underlying mechanism(s) of increased NPM levels, here, we characterized the subcellular distribution of NPM in and its contribution to the migration and invasiveness of the highly invasive and very low PG-expressing MDA-231 mammary carcinoma cell line and their transfectants expressing PG (MDA-231-PG). In MDA-231-PG cells, NPM protein, but not transcript, levels were increased and increased protein levels were particularly prominent in its nucleolar pool. NPM was detected in association with PG and NPM and PG were colocalized in the nucleoli. Furthermore, PG expression decreased the growth rate as well as the in vitro migration and invasiveness of MDA-231 cells, whereas down regulation of NPM in MDA-231 cells decreased growth rate but increased their in vitro migration and invasiveness. However, the simultaneous expression of PG and downregulation of NPM not only decreased the growth rate of transfectants, but also decreased their in vitro migration and invasiveness relative to MDA-231, -231-PG and -231-NPM-KD cells. The results of these in vitro studies were in concert with the concordant expression (or lack thereof) of PG and NPM in normal and invasive breast carcinoma tissues.
NPM was initially identified as a tumor promoter based on its increased levels in highly proliferating and malignant cells.3, 30, 31 NPM is the most frequently mutated gene in acute myeloid leukemia and its overexpression has been reported in various carcinomas.3, 31 However, studies with NPM+/− mice and NPM null cultured cells have demonstrated that NPM may act as a tumor suppressor by its roles in limiting centrosome duplication to once per cell cycle and maintenance of genomic stability, as well as the regulation of p53 and p14ARF tumor suppressors.3, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 Although considered a nucleolar protein, NPM shuttles between the nucleus and cytoplasm and its overexpression and/or excess cytoplasmic and nucleoplasmic pools have been implicated in its oncogenic function.28, 42, 43 The increased NPM levels in MDA-231-PG cells was unexpected, as although PG increased NPM protein levels, PG expression in these cells also induced an epithelioid morphology concurrent with decreased growth rate. This phenotypic change by PG is consistent with its tumor suppressor potential as we and others had previously reported.17, 18, 21, 22 The increased NPM protein levels in MDA-231-PG cells were not associated with increased NPM transcript levels, which suggested that PG may regulate NPM levels post-translationally. PG may potentially increase NPM stability by associating with NPM and/or modifying its subcellular distribution. The increased amount of nucleolar NPM from ∼2% in MDA-231 to ∼20% MDA-231-PG cells and the nucleolar colocalization of NPM and PG in these cells further supported this possibility. Coimmunoprecipitation studies verified the association of NPM with PG in all subcellular compartments. Studies have suggested that high/excess cytoplasmic and nucleoplasmic pools of NPM contribute to its oncogenic function.27, 28, 31 Therefore, we developed stable MDA-231-NPM-KD cells in which NPM protein levels were decreased by >90%, with almost a complete disappearance of the cytoplasmic pool and a considerable reduction in the nucleoplasmic pool.
NPM knockdown was associated with decreased growth rate, in keeping with previous studies.7, 28, 43, 44, 45, 46 Notably, although the growth rate of MDA-231-NPM-KD cells was clearly lower than MDA-231 cells, it was still higher than that of MDA-231-PG cells. Furthermore, the assessment of the in vitro metastasis properties of the NPM-KD cells showed higher rates of migration and invasion relative to MDA-231 cells. In contrast to NPM-KD cells, MDA-231-PG cells had considerably lower migration and invasiveness relative to MDA-231 cells. These observations are consistent with the role of PG in inhibiting cellular motility and its potential tumor/metastasis suppressor function.19, 22, 47, 48, 49, 50 These results suggest that whereas the high levels of extra nucleolar NPM contribute to its growth promoting activities, its nucleolar pool may have metastasis suppressor function.
In leukemia, the oncogenic mutations in NPM are generally in exon 12 and generate a novel nuclear export signal, which leads to the cytoplasmic accumulation of NPM.5, 21, 51, 52, 53 In contrast, no mutations in NPM have been found in solid tumors such as lung, thyroid, hepatocellular, colorectal, gastric and breast carcinomas.54 However, Shao et al.55 have shown that the migration and invasiveness of NIH3T3 cells are significantly increased upon expression of a mutant NPM, which localized to the cytoplasm. Studies that have reported the oncogenic function of NPM in solid tumors generally described its overexpression and have not determined the subcellular distribution of the protein.4, 56, 57, 58, 59 Recently, a few studies have examined the subcellular localization of NPM in solid cancers. Pianta et al.60 showed decreased nucleolar NPM in follicular adenomas and papillary carcinomas of the thyroid as well as tumorigenic thyroid cancer cell lines. Coutinho-Camillo et al.61 reported that although overexpression of NPM was associated with local recurrence in oral squamous carcinomas, <10% of the protein was detected in the nuclei. In solid tumors, the cellular distribution and function of NPM may be regulated by its interactions with other intracellular partners. NPM has been shown to interact with a number of proteins that contribute to its oncogenic versus tumor suppressor activities, including 14-3-3, AKT, Bax, p120-catenin, HLJ1, Plk2 and so on.45, 62, 63, 64, 65, 66 Our data suggest that PG may be one such protein. In addition to the mammary carcinoma cell line, we have observed NPM-PG interactions in other cell lines including the PG-deficient squamous carcinoma cell line SCC9 after the exogenous expression of PG (unpublished). The higher levels of NPM in MDA-231 cells upon PG expression concurrent with decreased in vitro tumorigenic and metastatic properties of these cells are consistent with a recent in vivo study by Karhemo et al.67 in which an analysis of 1160 breast carcinomas showed a substantial decrease in the NPM levels of the tumors relative to normal breast tissue. These results are also consistent with the decreased levels of NPM in a number of metastatic breast carcinoma tissues examined in this study.
In summary, the data presented here demonstrates that PG associates either directly or indirectly with NPM and increases its nucleolar pool, concurrent with a decrease in cell growth rate and in vitro invasiveness. PG interactions with NPM may alter NPM interactions with its tumor promoting partners and its subcellular distribution, thus modifying its oncogenic versus tumor suppressor activities. Analysis of the NPM intracellular interactions warrants further investigation and is currently in progress in our laboratory.
Materials and methods
Cell lines and culture conditions
All tissue culture reagents were purchased from Invitrogen (Burlington, ON, Canada) unless stated otherwise. The human mammary carcinoma cell line MCF-10-2A has been described.68 The highly invasive human mammary carcinoma cell line MDA-MB-231 was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), and maintained in RPMI media supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) L-glutamine, non-essential amino acids, sodium pyruvate and penicillin–streptomycin–kanamycin (PSK) antibiotics. MDA-231-PG transfectants were maintained in RPMI media containing geneticin (G418; 1 mg/ml). The MDA-231-NPM-KD and -231-NPM-KD+PG transfectants were maintained in RPMI containing (4 μg/ml) puromycin and both puromycin and geneticin, respectively.
Plasmid construction and transfection
The construct containing the full-length PG cDNA (pBK-CMV-PG) has been described.18 MDA-231 cells were transfected with 4 μg of either pBK-CMV or pBK-CMV-PG and LipofectAMINE reagent (Life Technologies, Inc., Grand Island, NY, USA) and G418-resistant colonies were selected and screened for PG expression using immunofluorescence and immunoblot assays. Positive colonies expressing PG were subcloned by limiting dilution to obtain single-cell isolated clones.
NPM shRNA lentiviral particle transduction
Control, scrambled shRNA (sc-108080) and human NPM shRNA (sc-29771-SH) lentiviral particles were obtained from Santa Cruz Biotechnology (Santa Cruz Biotechnology, CA, USA) and used to transduce MDA-231 cells according to the manufacturer's protocol. Puromycin-resistant stable cell lines expressing the scrambled or NPM shRNAs (NPM-KD) were isolated and the decreased expression of NPM was verified by RT–PCR and western blot.
MDA-231-NPM-KD cells were further transfected with 4 μg of either pBK-CMV or pBK-CMV-PG as described earlier to generate the MDA-231-NPM-KD+PG cell line expressing PG.
Antibodies
A list of antibodies and their respective dilutions in specific assays is presented in Table 1.
Table 1. Antibodies, fluorescent probes and their respective dilutions in specific assays.
| Antibodies | Assay |
Source/reference | |||
|---|---|---|---|---|---|
| IP | WB | IF | IHC | ||
| Primaries | |||||
| Nucleophosmin (m)a | 1:100 | 1:1000 | Abcam (AB10530) | ||
| Nucleophosmin (r)a | 1:50 | Santa Cruz (SC-5564) | |||
| Plakoglobin (m) | 1:100 | 1:1000 | 1:200 | 1:400 | Transduction Laboratories (610254) |
| Plakoglobin (r) | 1:100 | 1:1000 | Reference18 | ||
| E-cadherin (m) | 1:800 | Transduction Laboratories (610182) | |||
| β-catenin (r) | 1:1200 | Sigma (C-2206) | |||
| Actin (m) | 1:1000 | Sigma (A-4700) | |||
| Lamin B1 (r) | 1:1000 | Santa Cruz (SC-20682) | |||
| Fibrillarin (m) | 1:1000 | Abcam (AB18380) | |||
| Secondaries | |||||
| Alexa Fluor 488 | 1:750 | Molecular Probes (A11035) | |||
| Anti-mouse HRP | 1:5000 | Sigma (054H-8914) | |||
| Anti-rabbit HRP | 1:5000 | Sigma (054H-8918) | |||
m, mouse; r, rabbit.
RNA isolation and RT–PCR
All chemicals and reagents were purchased from Sigma-Aldrich (Oakville, Ontario, Canada) unless stated otherwise RNA was isolated using the RNeasy Plus Mini Kit (QIAGEN, Valencia, CA, USA) and reverse transcribed as described.22 PCR was performed using the following primers: NPM, forward: 5′-GCACTTAGTAGCTGTGGAGGA-3′ reverse: 5′-TTCTTCACTGGCGCTTTTTCT-3′ and GAPDH, forward: 5′-GAAGGTGAAGGTCGGAGTC-3′ reverse: 5′-GAAGATGGTGATGGGATTTC-3′. RT–PCR products were resolved on 2% agarose gels and visualized by ethidium bromide staining.
Preparation of total cell extracts, western blotting and immunoprecipitation
Confluent 150 mm culture dishes were extracted with RIPA buffer and processed for western blot and immunoprecipitation as described in Supplementary Materials and Methods.
Immunohistochemistry and indirect immunofluorescence
Immunohistochemistry was performed as previously described69 and is detailed in Supplementary Materials and Methods. For indirect immunofluorescence, confluent cultures of cells grown on glass coverslips and were extracted either with cytoskeleton extraction buffer (CSK;70) or with modified RIPA buffer (Lamond's protocol) and processed for staining with PG and NPM antibodies. Nuclei were counterstained with DAPI for 5 min and coverslips were viewed using a × 63 objective of an LSM510 META (Zeiss, Thornwood, NY, USA) laser scanning confocal microscope.
Nucleolar isolation
This procedure was performed according to the Lamond's protocol (http://www.lamondlab.com/f7nucleolarprotocol.htm) as described in the Supplementary Materials and Methods.
Transwell cell migration and invasion assays
Each assay was performed in duplicate and repeated three independent times. The numbers of migrated/invaded cells were calculated using the ImageJ Cell Counter program and averaged. Histograms were constructed after normalizing the average numbers of migrated/invaded cells in each cell line to those of MDA-231 cells.
For cell migration assays, 2 × 105 cells were resuspended in 100 μl serum-free media and plated in the upper chamber of transwells (3 μm pore, 6.5 mm diameter; BD Biosciences, Sparks, MD, USA). Normal media containing 10% FBS (0.75 ml) was added to the lower chamber. Cultures were incubated at 37 °C for 6–24 h to allow cell migration. At the end of each time point, the inserts were removed from the chambers, gently submerged in PBS to remove the unattached cells and then fixed and stained using Diff Quick (IHC World, Woodstock, MD, USA). Following staining, membranes were cut, mounted on slides using permount (Fisher, Ottawa, ON, Canada), viewed under an inverted microscope using a × 20 objective and photographed. The migrated cells on the underside of the membrane were counted in six random fields for each transwell filter from the photographs.
Matrigel invasion assays were performed according to the manufacturer's protocol (BD Bioscience). For each cell line, 5 × 105 cells in 0.5 ml serum-free media were plated in the top compartment of Matrigel-coated invasion chambers (8 μm pore membrane). Fibroblast conditioned media (0.75 ml) was added to the bottom chambers and plates were incubated overnight at 37 °C in 5% CO2. After 24 h, the membranes were recovered, fixed, stained with DiffQuick, viewed under an inverted microscope using a × 20 objective and photographed. The invaded cells were counted in six random fields for each membrane.
Acknowledgments
We are grateful to Dr E Shibuya for the critical review of the manuscript. This work is supported by the Canadian Breast Cancer Foundation- Prairies/NWT Chapter (MP) and by the Killam Trusts Izaak Walton Killam Memorial Graduate Scholarship (ZA). JM is supported by the Alberta Cancer Research Institute and Alberta Cancer Foundation. ZA currently holds a University of Alberta Dissertation Fellowship.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Yung BY. Oncogenic role of nucleophosmin/B23. Chang Gung Med J. 2007;30:285–293. [PubMed] [Google Scholar]
- Skaar TC, Prasad SC, Sharareh S, Lippman ME, Brünner N, Clarke R. Two-dimensional gel electrophoresis analyses identify nucleophosmin as an estrogen regulated protein associated with acquired estrogen-independence in human breast cancer cells. J Steroid Biochem Mol Biol. 1998;67:391–402. doi: 10.1016/s0960-0760(98)00142-3. [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Kikuta K, Tochigi N, Shimoda T, Yabe H, Morioka H, Toyama Y, et al. Nucleophosmin as a candidate prognostic biomarker of Ewing's sarcoma revealed by proteomics. Clin Cancer Res. 2009;15:2885–2894. doi: 10.1158/1078-0432.CCR-08-1913. [DOI] [PubMed] [Google Scholar]
- Falini B, Gionfriddo I, Cecchetti F, Ballanti S, Pettirossi V, Martelli MP. Acute myeloid leukemia with mutated nucleophosmin (NPM1): any hope for a targeted therapy. Blood Rev. 2011;25:247–254. doi: 10.1016/j.blre.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, et al. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hann SR. The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle. 2009;8:2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerset RA. DNA damage, p14ARF, nucleophosmin (NPM/B23), and cancer. J Mol Histol. 2006;37:239–251. doi: 10.1007/s10735-006-9040-y. [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J Cell Biol. 2008;182:19–26. doi: 10.1083/jcb.200711040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano AR, Colombo E, Luzi L, Martinelli P, Volorio S, Bernard L, et al. Cytoplasmic localization of NPM in myeloid leukemias is dictated by gain-of-function mutations that create a functional nuclear export signal. Oncogene. 2006;5:4376–4380. doi: 10.1038/sj.onc.1209453. [DOI] [PubMed] [Google Scholar]
- Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harb Perspect Biol. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol. 2000;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey M, Dawson C. Desmosomes: a role in cancer. Br J Cancer. 2007;96:1783–1787. doi: 10.1038/sj.bjc.6603808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1:a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roarty K, Rosen JM. Wnt and mammary stem cells: hormones cannot fly wingless. Curr Opin Pharmacol. 2010;10:643–649. doi: 10.1016/j.coph.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcha I, Geiger B, Yehuda-Levenberg S, Salomon D, Ben-Ze'ev A. Suppression of tumorigenicity by plakoglobin: an augmenting effect of N-cadherin. J Cell Biol. 1996;133:199–209. doi: 10.1083/jcb.133.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HR, Li Z, Sheinin H, Lauzon G, Pasdar M. Plakoglobin induces desmosome formation and epidermoid phenotype in N-cadherin-expressing squamous carcinoma cells deficient in plakoglobin and E-cadherin. Cell Motil Cytoskeleton. 1998;40:87–100. doi: 10.1002/(SICI)1097-0169(1998)40:1<87::AID-CM8>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Rieger-Christ KM, Ng L, Hanley RS, Durrani O, Ma H, Yee AS, et al. Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Br J Cancer. 2005;92:2153–2159. doi: 10.1038/sj.bjc.6602651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldi T, Bonvini P, Sartori F, Marrone A, Iolascon A, Rosolen A. Plakoglobin is differentially expressed in alveolar and embryonal rhabdomyosarcoma and is regulated by DNA methylation and histone acetylation. Carcinogenesis. 2006;27:1758–1767. doi: 10.1093/carcin/bgl008. [DOI] [PubMed] [Google Scholar]
- Li L, Chapman K, Hu X, Wong A, Pasdar M. Modulation of the oncogenic potential of beta-catenin by the subcellular distribution of plakoglobin. Mol Carcinog. 2007;46:824–838. doi: 10.1002/mc.20310. [DOI] [PubMed] [Google Scholar]
- Aktary Z, Chapman K, Lam L, Lo A, Ji C, Graham K, et al. Plakoglobin interacts with and increases the protein levels of metastasis suppressor Nm23-H2 and regulates the expression of Nm23-H1. Oncogene. 2010;29:2118–2129. doi: 10.1038/onc.2009.495. [DOI] [PubMed] [Google Scholar]
- Li Z, Gallin WJ, Lauzon G, Pasdar M. L-CAM expression induces fibroblast-epidermoid transition in squamous carcinoma cells and down-regulates the endogenous N-cadherin. J Cell Sci. 1998;111:1005–1019. doi: 10.1242/jcs.111.7.1005. [DOI] [PubMed] [Google Scholar]
- Hakimelahi S, Parker HR, Gilchrist AJ, Barry M, Li Z, Bleackley RC, et al. Plakoglobin regulates the expression of the anti-apoptotic protein BCL-2. J Biol Chem. 2000;275:10905–10911. doi: 10.1074/jbc.275.15.10905. [DOI] [PubMed] [Google Scholar]
- Ji C, Li L, Gebre M, Pasdar M, Li L. Identification and quantification of differentially expressed proteins in E-cadherin deficient SCC9 cells and SCC9 transfectants expressing E-cadherin by dimethyl isotope labeling, LC-MALDI MS and MS/MS. J Proteome Res. 2005;4:1419–1426. doi: 10.1021/pr050094h. [DOI] [PubMed] [Google Scholar]
- Aktary Z, Pasdar M.Plakoglobin: role in tumorigenesis and metastasis Int J Cell Bio(in press). [DOI] [PMC free article] [PubMed]
- Oelschlaegel U, Koch S, Mohr B, Schaich M, Falini B, Ehninger G, et al. flow cytometric detection of aberrant cytoplasmic localization of nucleophosmin (NPMc) indicating mutant NPM1 gene in acute myeloid leukemia. Leukemia. 2010;24:1813–1816. doi: 10.1038/leu.2010.178. [DOI] [PubMed] [Google Scholar]
- Brady SN, Maggi LB, Jr, Winkeler CL, Toso EA, Gwinn AS, Pelletier CL. Nucleophosmin protein expression level, but not threonine 198 phosphorylation, is essential in growth and proliferation. Oncogene. 2009;28:3209–3220. doi: 10.1038/onc.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore PP. Playing both sides: nucleophosmin between tumor suppression and oncogenesis. J Cell Biol. 2008;182:7–9. doi: 10.1083/jcb.200806069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WY, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, et al. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989;28:1033–1039. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- Lim MJ, Wang XW. Nucleophosmin and human cancer. Cancer Detect Prev. 2006;30:481–490. doi: 10.1016/j.cdp.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein N, Chan PK, Mond JJ. Identification of numatrin, the nuclear matrix protein associated with induction of mitogenesis, as the nucleolar protein B23. Implication for the role of the nucleolus in early transduction of mitogenic signals. J Biol Chem. 1988;263:10608–10612. [PubMed] [Google Scholar]
- Subong EN, Shue MJ, Epstein JI, Briggman JV, Chan PK, Partin AW. Monoclonal antibody to prostate cancer nuclear matrix protein (PRO: 4–216) recognizes nucleophosmin/B23. Prostate. 1999;39:298–304. doi: 10.1002/(sici)1097-0045(19990601)39:4<298::aid-pros11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sportoletti P, Grisendi S, Majid SM, Cheng K, Clohessy JG, Viale A, et al. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111:3859–3862. doi: 10.1182/blood-2007-06-098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, et al. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- Maiguel DA, Jones L, Chakravarty D, Yang C, Carrier F. Nucleophosmin sets a threshold for p53 response to UV radiation. Mol Cell Biol. 2004;24:3703–3711. doi: 10.1128/MCB.24.9.3703-3711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Falini B, Bolli N, Liso A, Martelli MP, Manucci R, Pileri S, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23:1731–1743. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
- Shandilya J, Swaminathan V, Gadad SS, Choudhari R, Kodaganur GS, Kundu TK. Acetylated NPM1 localizes in the nucleoplasm and regulates transcriptional activation of genes implicated in oral cancer manifestation. Mol Cell Biol. 2009;29:5115–5127. doi: 10.1128/MCB.01969-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, St Clair DK. Nucleophosmin blocks mitochondrial localization of p53 and apoptosis. J Biol Chem. 2009;284:16409–16418. doi: 10.1074/jbc.M109.005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Nguyen TL, Lee SB, Park SB, Lee KH, Cho SW, et al. Akt2 and nucleophosmin/B23 function as an oncogenic unit in human lung cancer cells. Exp Cell Res. 2011;317:966–975. doi: 10.1016/j.yexcr.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Qin FX, Shao HY, Chen XC, Tan S, Zhang HJ, Miao ZY, et al. Knockdown of NPM1 by RNA interference inhibits cells proliferation and induces apoptosis in leukemic cell line. Int J Med Sci. 2011;8:287–294. doi: 10.7150/ijms.8.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Heasman J, Lang SA, Wylie CC. Plakoglobin is required for maintenance of the cortical actin skeleton in early Xenopus embryos and for cdc42-mediated wound healing. J Cell Biol. 2002;158:695–608. doi: 10.1083/jcb.200202123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T, Getsios S, Caldelari R, Kowalczyk AP, Müller EJ, Jones JC, et al. suppresses keratinocyte motility through both cell-cell adhesion-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2005;102:5420–5425. doi: 10.1073/pnas.0501676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiei F, Rahnama F, Pawella L, Mitchell MD, Gluckman PD, Lobie PE. DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells. Oncogene. 2008;27:2602–2612. doi: 10.1038/sj.onc.1210917. [DOI] [PubMed] [Google Scholar]
- Todoroviæ V, Desai BV, Patterson MJ, Amargo EV, Dubash AD, Yin T, et al. regulates cell motility through Rho- and fibronectin-dependent Src signaling. J Cell Sci. 2010;123:3576–3586. doi: 10.1242/jcs.070391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, et al. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67:6230–6237. doi: 10.1158/0008-5472.CAN-07-0273. [DOI] [PubMed] [Google Scholar]
- Falini B, Sportoletti P, Martelli MP. Acute myeloid leukemia with mutated NPM1: diagnosis, prognosis and therapeutic perspectives. Curr Opin Oncol. 2009;21:573–581. doi: 10.1097/CCO.0b013e3283313dfa. [DOI] [PubMed] [Google Scholar]
- Luo J, Qi C, Xu W, Kamel-Reid S, Brandwein J, Chang H. Cytoplasmic expression of nucleophosmin accuratety predicts mutation in the nucleophosmin gene in patients with acute myeloid leukemia and normal karyotype. Am J Clin Pathol. 2010;133:34–40. doi: 10.1309/AJCPCI1FFE2DRXIV. [DOI] [PubMed] [Google Scholar]
- Jeong EG, Lee SH, Yoo NJ, Lee SH. Absence of nucleophosmin 1 (NPM1) gene mutations in common solid cancers. APMIS. 2007;115:341–346. doi: 10.1111/j.1600-0463.2007.apm_592.x. [DOI] [PubMed] [Google Scholar]
- Shao HY, Miao ZY, Hui-Chen, Qin FX, Chen XC, Tan S, et al. Nucleophosmin gene mutations promote NIH3T3 cell migration and invasion through CXCR4 and MMPs. Exp Mol Pathol. 2011;90:38–44. doi: 10.1016/j.yexmp.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sasaki H, Kino I, Sugimura T, Terada M. Genes preferentially expressed in embryo stomach are predominantly expressed in gastric cancer. Cancer Res. 1992;52:3372–3377. [PubMed] [Google Scholar]
- Nozawa Y, Van Belzen N, Van der Made AC, Dinjens WN, Bosman FT. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J Pathol. 1996;178:48–52. doi: 10.1002/(SICI)1096-9896(199601)178:1<48::AID-PATH432>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Yun JP, Miao J, Chen GG, Tian QH, Zhang CQ, Xiang J, et al. expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br J Cancer. 2007;96:477–484. doi: 10.1038/sj.bjc.6603574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui KH, Juang HH, Lee TH, Chang PL, Chen CL, Yung BY. Association of nucleophosmin/B23 with bladder cancer recurrence based on immunohistochemical assessment in clinical samples. Acta Pharmacol Sin. 2008;29:364–370. doi: 10.1111/j.1745-7254.2008.00747.x. [DOI] [PubMed] [Google Scholar]
- Pianta A, Puppin C, Passon N, Franzoni A, Romanello M, Tell G, et al. Nucleophosmin delocalization in thyroid tumour cells. Endocr Pathol. 2011;22:18–23. doi: 10.1007/s12022-011-9147-x. [DOI] [PubMed] [Google Scholar]
- Coutinho-Camillo CM, Lourenço SV, Nishimoto IN, Kowalski LP, Soares FA. Nucleophosmin, p53, and Ki-67 expression patterns on an oral squamous cell carcinoma tissue microarray. Hum Pathol. 2010;41:1079–1086. doi: 10.1016/j.humpath.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Pozuelo-Rubio M. Proteomic and biochemical analysis of 14-3-3-binding proteins during C2-ceramide-induced apoptosis. FEBS J. 2010;277:3321–3342. doi: 10.1111/j.1742-4658.2010.07730.x. [DOI] [PubMed] [Google Scholar]
- Kerr LE, Birse-Archbold JL, Short DM, McGregor AL, Heron I, Macdonald DC, et al. Nucleophosmin is a novel Bax chaperone that regulates apoptotic cell death. Oncogene. 2007;26:2554–2562. doi: 10.1038/sj.onc.1210044. [DOI] [PubMed] [Google Scholar]
- Chartier NT, Oddou CI, Lainé MG, Ducarouge B, Marie CA, Block MR, et al. Cyclin-dependent kinase 2/cyclin E complex is involved in p120 catenin (p120ctn)-dependent cell growth control: a new role for p120ctn in cancer. Cancer Res. 2007;67:9781–9790. doi: 10.1158/0008-5472.CAN-07-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TP, Yu SL, Lin SY, Hsiao YJ, Chang GC, Yang PC, et al. Tumor suppressor HLJ1 binds and functionally alters nucleophosmin via activating enhancer binding protein 2alpha complex formation. Cancer Res. 2010;70:1656–1667. doi: 10.1158/0008-5472.CAN-09-2453. [DOI] [PubMed] [Google Scholar]
- Krause A, Hoffmann I. Polo-like kinase 2-dependent phosphorylation of NPM/B23 on serine 4 triggers centriole duplication. PLoS One. 2010;5:e9849. doi: 10.1371/journal.pone.0009849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhemo PR, Rivinoja A, Lundin J, Hyvönen M, Chernenko A, Lammi J, et al. An extensive tumor array analysis supports tumor suppressive role for nucleophosmin in breast cancer. Am J Pathol. 2011;179:1004–1014. doi: 10.1016/j.ajpath.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam L, Hu X, Aktary Z, Andrews DW, Pasdar M. Tamoxifen and ICI 182,780 increase Bcl-2 levels and inhibit growth of breast carcinoma cells by modulating PI3K/AKT, ERK and IGF-1R pathways independent of ERalpha. Breast Cancer Res Treat. 2009;118:605–621. doi: 10.1007/s10549-008-0231-y. [DOI] [PubMed] [Google Scholar]
- Asgarian N, Hu X, Aktary Z, Chapman KA, Lam L, Chibbar R, et al. Learning to predict relapse in invasive ductal carcinomas based on the subcellular localization of junctional proteins. Breast Cancer Res Treat. 2010;121:527–538. doi: 10.1007/s10549-009-0557-0. [DOI] [PubMed] [Google Scholar]
- Pasdar M, Nelson WJ. Kinetics of desmosome assembly in Madin-Darby canine kidney epithelial cells: temporal and spatial regulation of desmoplakin organization and stabilization upon cell-cell contact. II. Morphological analysis. J Cell Biol. 1988;106:687–695. doi: 10.1083/jcb.106.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.