Abstract
The Ras association domain family (RASSF) comprises a group of tumor suppressors that are frequently epigenetically inactivated in various tumor entities and linked to apoptosis, cell cycle control and microtubule stability. In this work, we concentrated on the newly identified putative tumor suppressor RASSF10. Methylation analysis reveals RASSF10 promoter hypermethylation in lung cancer, head and neck (HN) cancer, sarcoma and pancreatic cancer. An increase in RASSF10 methylation from normal tissues, primary tumors to cancer cell lines was observed. Methylation was reversed by 5-aza-2'-deoxycytidine treatment leading to reexpression of RASSF10. We further show that overexpression of RASSF10 suppresses colony formation in cancer cell lines. In addition, RASSF10 is upregulated by cell–cell contact and regulated on promoter level as well as endogenously by forskolin, protein kinase A (PKA) and activator Protein 1 (AP-1), linking RASSF10 to the cAMP signaling pathway. Knockdown of the AP-1 member JunD interfered with contact inhibition induced RASSF10 expression. In summary, we found RASSF10 to be epigenetically inactivated by hypermethylation of its CpG island promoter in lung, HN, sarcoma and pancreatic cancer. Furthermore, our novel findings suggest that tumor suppressor RASSF10 is upregulated by PKA and JunD signaling upon contact inhibition and that RASSF10 suppresses growth of cancer cells.
Keywords: epigenetics, DNA methylation, RASSF10, tumor suppressor, gene regulation
Introduction
Ras association domain family 1A (RASSF1A) was identified as an epigenetically silenced tumor suppressor gene.1 The Ras association domain (RA) of RASSF1A displayed a high homology to the Ras effector Rassf5.2, 3 Since these initial studies the family has grown and to date comprises 10 members from RASSF1 to RASSF10.4 One characteristic feature of this family is the RA domain, which can be found in all members either C-terminally (RASSF1–RASSF6) or N-terminally (RASSF7–RASSF10).5, 6 The other characteristic feature is the SARAH domain, encoding a protein–protein interaction domain, which, however, is only found in RASSF1–6.7, 8 Whereas RASSF7–RASSF10 joined the family only recently and therefore little data exist.6, 9
Another common feature of the RASSFs is the epigenetic inactivation due to CpG island promoter hypermethylation in different cancer entities.4 The cellular functions of the RASSFs range from, for example apoptosis, cell cycle control to microtubule stabilization.4 Though little is known about the underlying mechanisms, tumor suppressing functions were reported for several members.4
RASSF10 is located at genomic region 11p15.2 and contains a CpG island promoter that covers >2 kb.10 We and others have shown that, similar to other RASSF family members, RASSF10 is frequently silenced by promoter hypermethylation in different tumor entities such as thyroid tumors,10 melanoma,11 leukemia12 and glioma.13 In secondary glioblastomas, RASSF10 methylation was said to be an independent prognostic factor associated with worst progression-free survival and overall survival and occurred at an early stage in their development.13 Regarding the regulation of RASSF10 almost nothing was known so far.
In our study, we demonstrate that the RASSF10 promoter is frequently methylated in lung and pancreatic cancer as well as in head and neck (HN) cancer and sarcoma. Demethylation of the RASSF10 promoter is accompanied by reexpression of RASSF10 in cancer cell lines. RASSF10 expression was further found upregulated upon cell–cell contact and we show that forskolin, protein kinase A (PKA) and the JunD pathway regulate RASSF10. Furthermore, ectopic expression of RASSF10 suppresses colony formation.
Results
Hypermethylation of RASSF10 in sarcoma and pancreas, lung and HN cancer
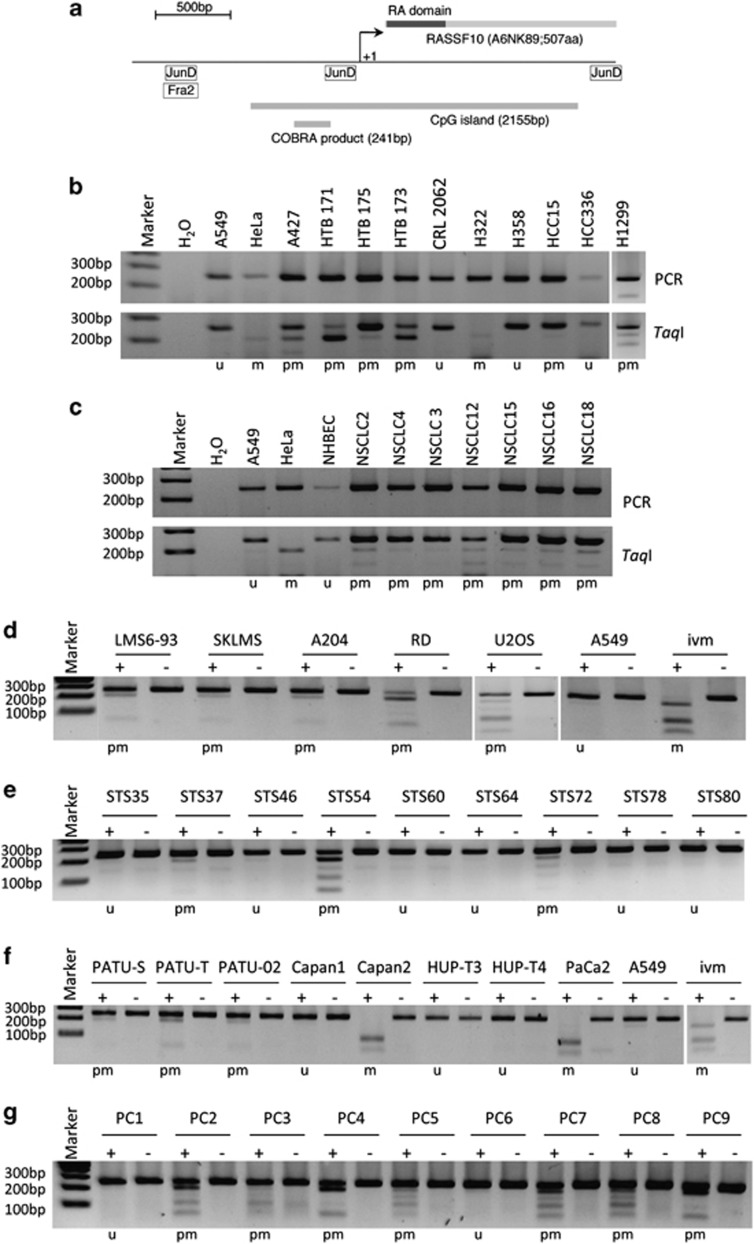
The RASSF10 locus and respective open reading frame is shown in Figure 1. The promoter lies within the CpG island (Figure 1a). ChIP-seq data from UCSC genome bioinformatics14 showed the binding of Activator Protein 1 (AP-1) members JunD and Fra2 in the RASSF10 locus (Figure 1a).
Figure 1.
Methylation status of the RASSF10 promoter in cancer cell lines and primary tumors. (a) Structure of RASSF10. Schematic representation of RASSF10 is shown with N-terminal RA domain (dark box). Arrows mark transcriptional (+1) start site for RASSF10. Relative localizations of CpG island (2155 bp; http://www.ebi.ac.uk) and COBRA PCR product (241 bp) are shown. JunD and Fra2 binding sites (http://genome.ucsc.edu) are indicated (JunD: −1279 to −1288; −186 to −235; +1537 to +1586 and Fra2: −1279 to −1328). COBRA is shown of (b) lung cancer cell lines, (c) primary non-small cell lung cancer (NSCLC) and normal human bronchial epithelial cells (NHBEC), (d) sarcoma cell lines, (e) primary soft-tissue sarcoma (STS), (f) pancreatic cancer cell lines and (g) primary pancreatic carcinoma (PC). In (b, c) upper panel shows PCR product of RASSF10 and lower panel shows TaqI digest. In (d–g) mock digest (−) and TaqI digest (+) are shown. In vitro methylated (ivm) HeLa DNA was used as control. PCR and digest were resolved on 2% gel with 100 bp marker. The classification of the methylation status (u, unmethylated, pm, partially methylated and m, methylated) is shown below each gel.
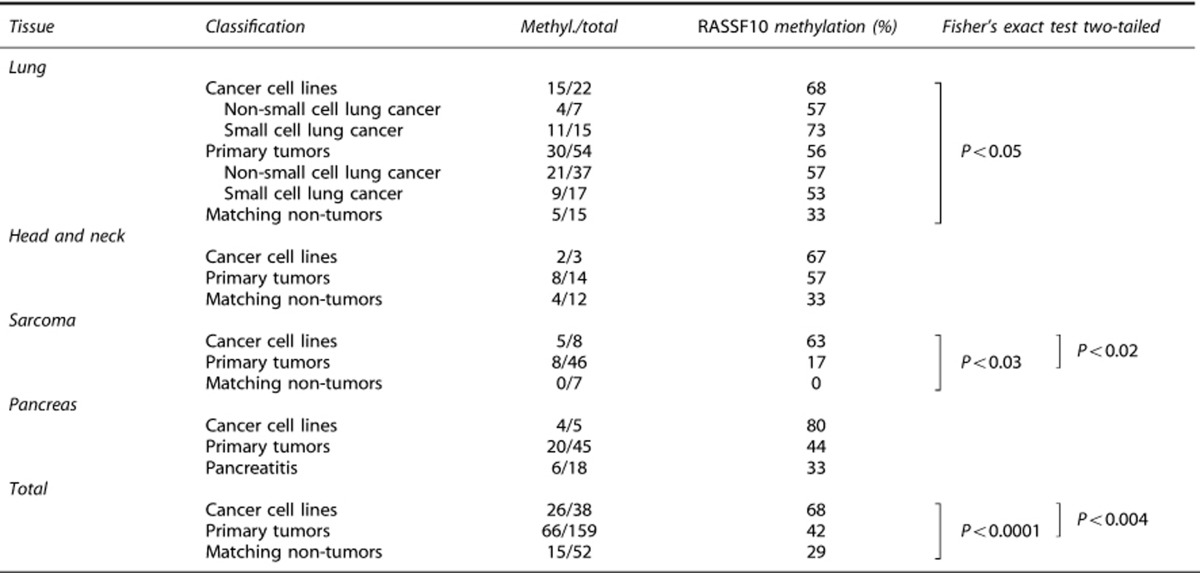
Methylation of the RASSF10 promoter in lung-, sarcoma-, pancreatic carcinoma cell lines, respective primary tumors and matching normal tissue was analyzed by combined bisulfite restriction analysis (COBRA) in Figure 1. In lung cancer cell lines the majority is methylated for RASSF10, especially A427, HTB171, HTB173 and H322 (Figure 1b), whereas primary tumors are rather slightly methylated (Figure 1c). Similar results are shown for sarcoma cell lines, primary sarcoma, pancreatic cancer cell lines and primary tumors (Figure 1). For lung cancer a total 22 cell lines were analyzed, of which 15 were methylated for RASSF10 (68%), consisting of 57% non-small cell lung cancer cell lines and 73% small cell lung cancer cell lines. For HN cancer cell lines methylation was 67%, sarcoma cell lines 63% and pancreatic cancer cell lines 80% (Table 1 and Supplementary Table S1).
Table 1. Summary of RASSF10 methylation in cancer.
Furthermore, 54 primary lung tumors were analyzed, of which 30 are methylated for RASSF10 (56%), comprising 57% non-small cell lung cancer and 53% small cell lung cancer. Overall, 57% primary HN tumors, 17% primary sarcomas and 44% primary pancreatic cancers are methylated for RASSF10 (Table 1). Matching lung control tissue was only methylated to 33% for RASSF10 and methylation of normal HN control tissue was also 33%. Matching non-tumor tissue for sarcoma is unmethylated and methylated at 33% in pancreatitis. Mean of RASSF10 promoter methylation significantly increases from 29% matching non-tumors, 42% primary tumors to 68% cancer cell lines (P<0.05; Table 1). In detail, lung cancer cell lines showed a significant increase in methylation of RASSF10 compared with non-tumors (P<0.05) (Table 1). Regarding sarcoma RASSF10 methylation increased significantly from normal tissue to cancer cell lines (P<0.03) and from primary tumors to cell lines (P<0.02) (Table 1). RASSF10 was methylated in 8 of 46 primary sarcomas (17% Supplementary Table S3). Methylation of RASSF10 occurred predominantly in myogenic sarcomas (rhabdomyosarcomas and leiomyosarcomas) whereas in other entities it occurred rather rarely. Interestingly, none of the eight stage I tumors showed a RASSF10 methylation, this number increased to 2 of 17 stage II and to 5 of 15 stage III tumors. However, only 1 of 6 stage IV tumors possessed a RASSF10 methylation, but process of metastasis might be related to other molecular factors.
Epigenetic silencing of RASSF10 occurs in cancer
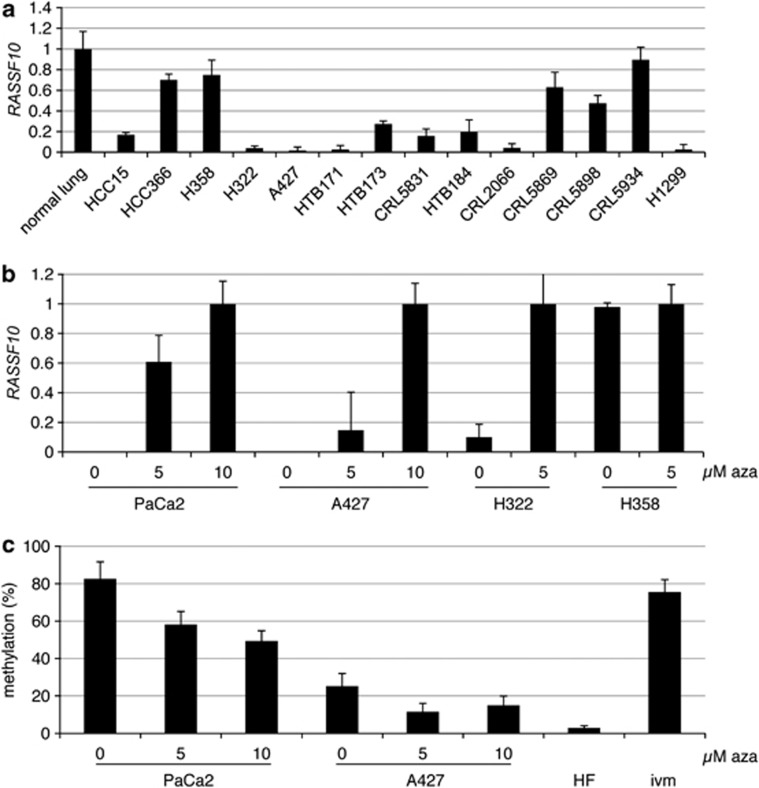
Using qRT–PCR we analyzed the altered expression of RASSF10 in lung cancer (Figure 2a). We earlier showed that RASSF10 is widely expressed normal tissues.10 Highest expression of RASSF10 was seen in normal lung, HCC366, H358 and CRL5934 (Figure 2a). Accordingly, COBRA revealed that RASSF10 was unmethylated for HCC366 and H358 (Figure 1b and Supplementary Table S1). Lower expression of RASSF10 was observed for CRL5869 and CRL5898 and both were partially methylated for RASSF10 (Supplementary Table S1). HCC15 and HTB173 showed about 20% expression of RASSF10 compared with normal lung and were partially methylated for RASSF10 (Figures 1 and 2a). Lowest expression of RASSF10 was seen in A427, HTB171, CRL2066, H1299 and H322 (Figure 2a) and all were partially methylated and fully methylated for RASSF10, respectively (Figure 1b and Supplementary Table S1).
Figure 2.
RASSF10 expression in lung cancer, reexpression and demethylation under aza treatment. (a) Expression analysis of RASSF10 is shown in lung cancer cell lines vs normal lung (=1) and was normalized to ACTB expression using qRT–PCR. (b) Aza treatment (0, 5 and 10 μℳ) was performed of the pancreatic cancer cell line PaCa2 and lung cancer cell lines A427, H322 and H358. RASSF10 was analyzed by RT–PCR after 4 days and normalized to ACTB. 10 μℳ PaCa2, 10 μℳ A427, 5 μℳ H322 and 5 μℳ H358 was set to 1. (c) RASSF10 promoter methylation analysis was performed in PaCa2 and A427 and quantified by pyrosequencing. Seven CpGs are included in analyzed region and respective mean and SD are shown. In vitro methylated (ivm) genomic DNA and human fibroblasts (HF) are used as control.
5-aza-2'-deoxycytidine (Aza) inhibits de novo methylation and is known to reverse epigenetic silencing of tumor suppressor genes.1 Therefore, cancer cell lines (PaCa2, pancreatic carcinoma; A427 and H322, both lung cancers) with methylated RASSF10 promoter were treated with aza and RASSF10 expression and methylation were analyzed by qRT–PCR and pyrosequencing, respectively (Figures 2b and c). Aza treatment of these cells induced RASSF10 expression (Figure 2b). RASSF10 reexpression was accompanied by its promoter demethylation (Figure 2c). PaCa2 were methylated by 83% and under aza treatment methylation decreased to 58 (5 μℳ) and 49% (10 μℳ) (Figure 2c). A427 showed methylation of RASSF10 by 25%, which was decreased to 12% under 5 μℳ aza (Figure 2c). As controls the lung cancer cell line H358 and human fibroblasts were utilized (Figure 2).
Regulation of RASSF10 promoter by forskolin, PKA and AP-1
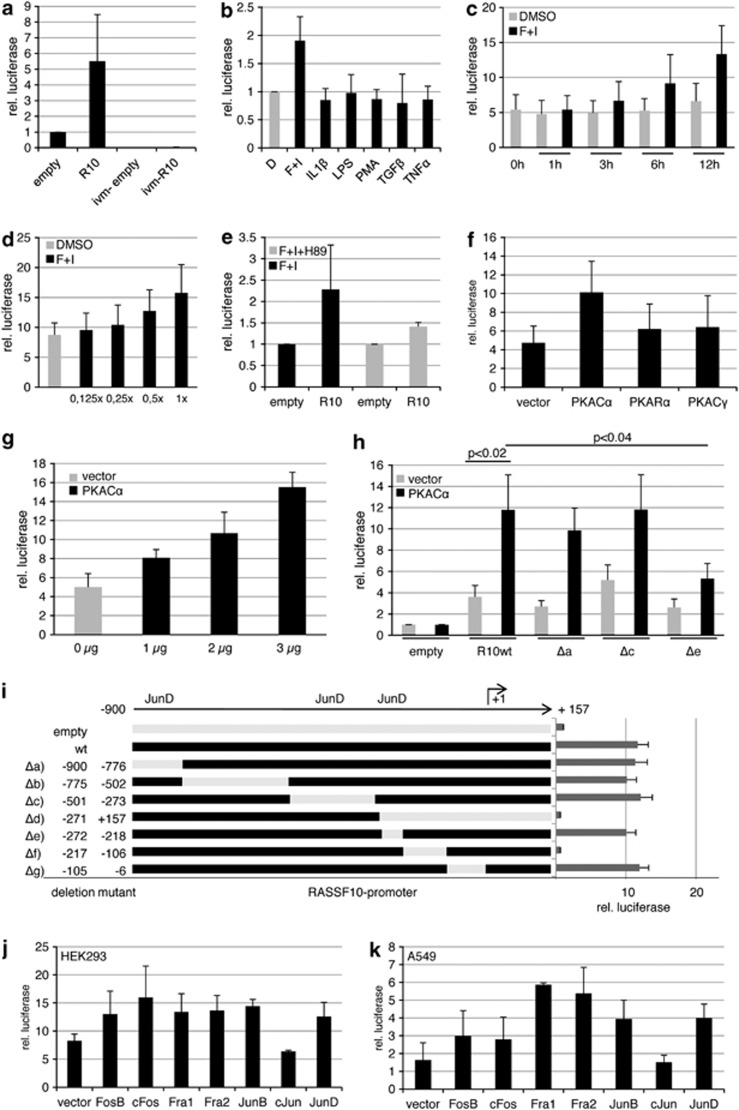
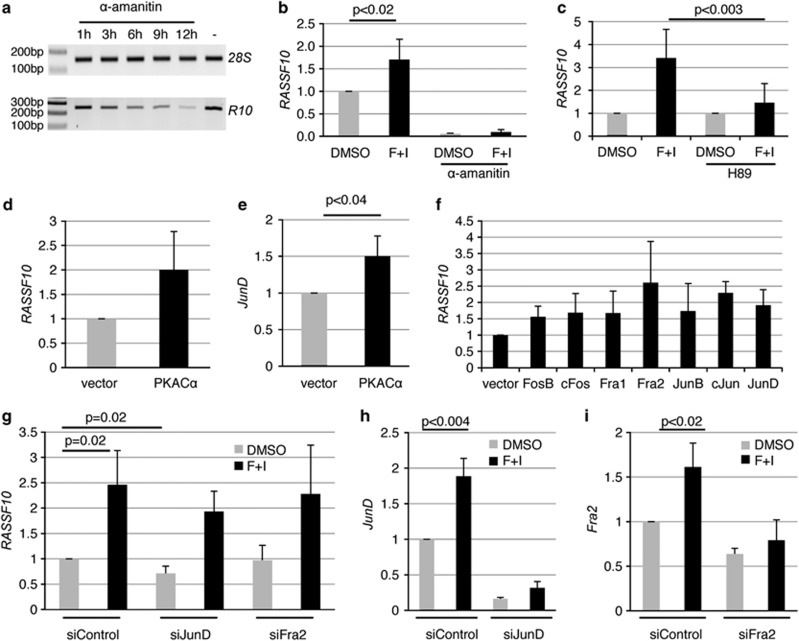
To clarify what kind of stimuli could induce RASSF10 expression, we used a luciferase assay and cloned the RASSF10 promoter (−900 to +157 bp) into the pRLnull vector (Figure 3). Compared with the empty vector we observed a five-fold induction by the RASSF10 promoter (Figure 3a). Upon in vitro methylation of the RASSF10 plasmid that signal is lost (Figure 3a). Various cytokines (forskolin/IBMX (3-isobutyl-1-methylxanthine), IL1β, LPS, PMA, TGF1β and TNFα) were used to narrow down possible upstream signals that could induce RASSF10 signaling (Figure 3b). Forskolin/IBMX treatment further induced the RASSF10 promoter by almost two-fold and we observed a time (Figure 3c) and dose (Figure 3d)-dependent activation. Next, we tested a set of inhibitors (LY294002, SB203580, SP600125, PD98059, Staurosporine and H89) to block the signaling of forskolin/IBMX on RASSF10, and found that the PKA inhibitor H89 reduces RASSF10 promoter induction (Figure 3e and data not shown). Similarly, we found that PKA overexpression induced RASSF10 promoter activation (Figure 3f). This was especially observed for the catalytic subunit alpha (PKACα) whereas for other subunits (PKARα and PKACγ) reporter activation was reduced (Figure 3f). PKACα shows a dose-dependent activation of the RASSF10 promoter (Figure 3g).
Figure 3.
Forskolin, PKA and AP-1 family members induce the RASSF10 promoter. Luciferase assay was performed in HEK293 cells transfected with pRLnull (empty), RASSF10 promoter construct and pGL3 for transfection control. (a) The relative luciferase activity is shown 12 h after transfection in empty or RASSF10-pRLnull (R10) and in vitro methylated (ivm) plasmids. (b) The RASSF10 promoter response to cytokines (20 μℳ forskolin, 500 μℳ IBMX, 10 ng/ml IL1β, 1 μg/ml LPS, 50 nℳ PMA, 2 ng/ml TGFβ, 20 ng/ml TNFα) in comparison with dimethyl sulfoxide (DMSO) (D; set 1) is shown. R10 was normalized to similarly treated empty vector for each condition. (c) Time-dependent induction of R10 under mock (DMSO) or forskolin/IBMX (F+I) treatment from 1 to 12 h was analyzed. The empty vector for each time point and stimulation was set 1. (d) Dose-dependent induction of R10 under mock (DMSO) or increasing amounts of F+I is shown (1x: 20 μℳ forskolin and 500 μℳ IBMX). Empty vector for each dose was set 1. (e) Forskolin induction of the RASSF10 promoter was inhibited by 10 μℳ H89. Empty was set 1. (f) The PKA effect on RASSF10 promoter induction was analyzed by cotransfecting empty vector or PKA isoforms (Cα, Rα and Cγ). Empty pRLnull was transfected accordingly and set 1. (g) Dose-dependent induction under increasing amounts of PKACα in comparison with vector control is shown. Empty pRLnull was transfected accordingly and set 1. (h) The effect of PKACα on RASSF10 deletion constructs (Δa: -900/-776; Δc: -501/-273; Δe: -272/-218) and wt promoter is depicted. Empty pRLnull was set 1 under vector or PKACα cotransfection. (i) RASSF10 wt promoter activity is shown in comparison with deletion mutants from –900 to +157 bp relative to transcription start site (+1). JunD binding sites are indicated. (j, k) Cotransfected AP-1 members affect the RASSF10 promoter in HEK293 or A549 cells. Empty pRLnull was transfected with AP-1 members and was set 1. Results represent mean of three different experiments (SD is indicated). In (i) a representative experiment is shown (SD is shown). P values were calculated using two-tailed t-test.
We next were interested in what factors downstream of PKA activated the RASSF10 promoter. UCSC browser data indicated that the AP-1 members Fra2 and JunD could bind to the RASSF10 promoter (Figure 1a). We generated deletion mutants of the RASSF10 promoter construct (Figure 3i). For validation we found that PKACα significantly (P<0.02) activates the wild-type RASSF10 promoter in comparison with control transfection (Figure 3h). However, we observed that a deletion from −272 to −218 (Δe), covering a JunD binding site, lead to a significant reduction in RASSF10 promoter activation by PKACα compared with deletions −900 to −776 (Δa) and −501 to −273 (Δc), both containing potential JunD binding sites predicted by Matinspector Genomatix software (http://www.genomatix.de) (Figure 3h). Next, we cotransfected the AP-1 members FosB, cFos, Fra1, Fra2, JunB, cJun, JunD and empty vector in HEK293 or A549 (Figures 3j and k). Though there are slight differences in the activation of the RASSF10 promoter, which are likely due to cell line-specific characteristics, we found that, except for cJun, all members activate the promoter (Figures 3j and k). Serial deletion constructs of the RASSF10 promoter allowed us to determine the core promoter region, where luciferase activity is down to empty vector control level at first between −271 to +157 (Δd) and later narrowed down to −217 up to −106 (Δf) (Figure 3i). These results indicate that RASSF10 is activated by forskolin and blocked by the PKA inhibitor H89. Its promoter is induced by PKACα and different AP-1 members.
Regulation of endogenous RASSF10 expression by forskolin, PKA and AP-1
We next sought to verify these results analyzing the endogenous expression of RASSF10 in A549 cancer cell line. At first inhibition of RNA polymerase II by α-amanitin was used to determine the half life of RASSF10 mRNA (Figure 4a). The RASSF10 transcript is relatively unstable and after 12 h strongly reduced. We next questioned whether forskolin treatment induces RASSF10 expression as concluded from the promoter studies (Figure 3). After 12 h of forskolin RASSF10 is significantly upregulated by 1.7-fold (Figure 4b). To test whether this upregulation is due to increased transcription or prolonged mRNA stability we used α-amanitin. We found RASSF10 expression by α-amanitin down by more than 90% when mock- or forskolin-treated (Figure 4b). These results indicate that RASSF10 is transcriptionally upregulated. We further show that the PKA inhibitor H89 endogenously blocks upregulation of RASSF10 by forskolin (Figure 4c). Similar to the promoter studies, we overexpressed PKACα and RASSF10 expression was upregulated by 2-fold (Figure 4d). Interestingly, JunD is also significantly upregulated by PKA overexpression (Figure 4e). The overexpression of the AP-1 members FosB, cFos, Fra1, Fra2, JunB, cJun, JunD showed that all induced RASSF10 expression (Figure 4f). UCSC browser data indicated that JunD and Fra2 might be most important to RASSF10 and we therefore concentrated on these and aimed to block forskolin-induced RASSF10 expression by downregulation of JunD and Fra2 (Figures 4g). Under siControl knockdown forskolin induction significantly upregulated RASSF10 almost 2.5-fold (Figure 4g). When JunD is knocked down there was a reduced activation of RASSF10 (Figure 4g). RASSF10 activation, however, is not fully inhibited, which can be explained by the fact that endogenous JunD is elevated by forskolin (Figure 4h). The knockdown of Fra2 did not downregulate endogenous RASSF10 expression (Figure 4g).
Figure 4.
Forskolin and downstream effectors PKA and JunD affect RASSF10 expression. (a) Stability of RASSF10 mRNA was determined after 10 μℳ α-amanitin treatment from 1 to 12 h. 28S rRNA was used as control. (b) Cells were treated with 20 μℳ forskolin and 500 μℳ IBMX (F+I) or DMSO for 12 h with or without 10 μℳ α-amanitin. RASSF10 expression was analyzed by qRT–PCR and normalized to 28S rRNA. DMSO alone was set 1. (c) H89 (10 μℳ) inhibition of RASSF10 induction by F+I is shown. F+I is depicted relative to DMSO and F+I+H89 is shown relative DMSO+H89. Cells were transfected with PKACα or empty vector and (d) RASSF10 and (e) JunD expression were analyzed after 24 h. Expression was normalized to ACTB. (f) Cells were transfected with AP-1 members or empty vector. RASSF10 expression was analyzed after 24 h by qRT–PCR (normalized to ACTB). (g) Cells were transfected with siControl, siJunD and siFra2. After 72 h cells were trypsinized, seeded at 10% density and upon attachment serum-starved overnight. Then cells were stimulated with F+I or DMSO for 12 h. RASSF10 expression was normalized to ACTB. (h) JunD and (i) Fra2 knockdown were verified for assay described in (g). All experiments were performed in A549 cells at least in triplicates and mean as well as respective SD are shown. P values were calculated using two tailed t-test. DMSO treatment, vector alone or siControl are set 1.
Cell–cell contact induces RASSF10 expression
We tested RASSF10 on the basis of the known tumor suppressive functions that were known for other RASSF members.4 The family members are linked to cell cycle regulation: especially RASSF1A blocking the cell cycle at different stages.15, 16, 17 To test a possible cell cycle-dependent expression of RASSF10 we synchronized A549 cells. However, we found no alteration in the RASSF10 expression level during cell cycle (data not shown). Next, we aimed to study the possible contribution of RASSF10 to cell cycle progression by fluorescence-activated cell sorting analysis (Supplementary Figure S1). Therefore, we transfected A549 cells with siRASSF10 or siControl. Then cells were split at equal cell numbers (siRASSF10 and siControl) to achieve about 80% density at day of isolation. After 4 days, RASSF10 was downregulated by about 70% (Supplementary Figure S1A). We observed a slight and reproducible increase of cells entering the cell cycle in S/G2/M phase by a mean of 2.7% after RASSF10 knockdown (Supplementary Figure S1B). Number of cells in G1 was decreased by the same amount (Supplementary Figure S1B).
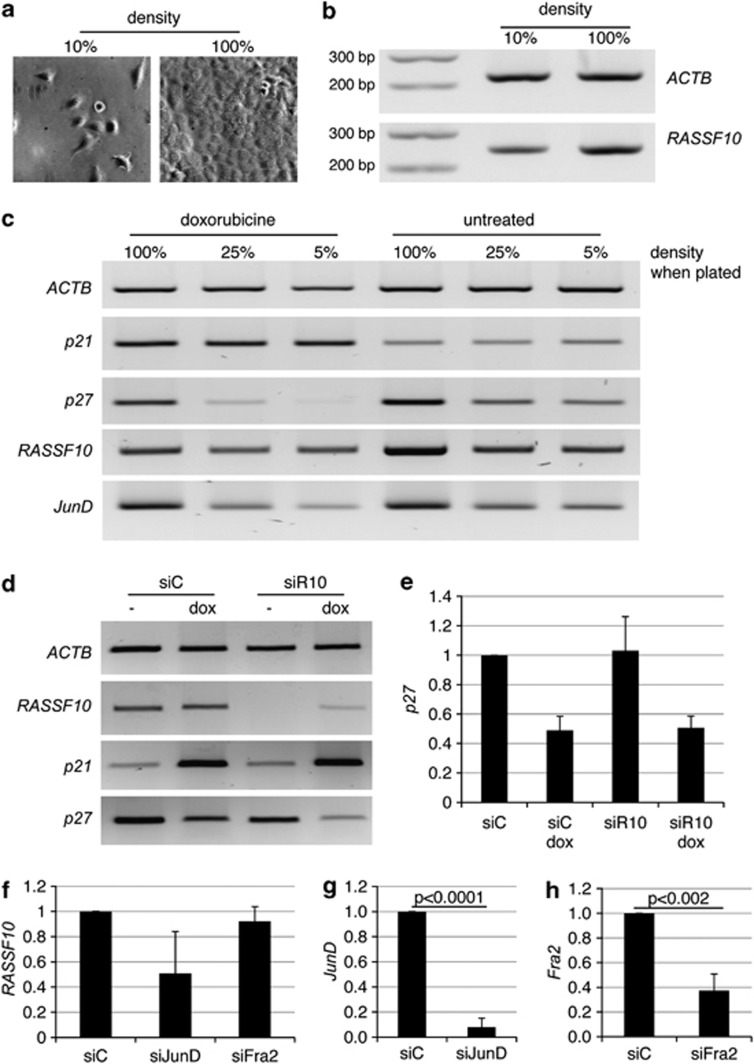
Interestingly, we observed that confluent cells showed increased expression of RASSF10 (Figures 5a and b). We found that expression depends on density of the cells, with increasing RASSF10 levels from lowest to highest density (Figure 5). To test, whether RASSF10 was linked to senescence, we treated the cells with doxorubicin (Figure 5c and Supplementary Figure S2). Doxorubicin was shown to promote senescence in A549 cells.18 We used the senescence-associated β-galactosidase staining19, 20 and measured p21 expression as a control.21 Doxorubicin treatment delays cell growth (Supplementary Figure S2A), whereas untreated A549 proliferate during the treatment and end up with higher cell numbers and density (Supplementary Figure S2A). Therefore, untreated cells show higher RASSF10 expression than doxorubicin-treated cells (Figure 5c). Senescence staining was positive when cells grew dense or at even 5% density after doxorubicin treatment (Supplementary Figure S2B). Effectiveness of doxorubicin treatment was verified by an increase in p21 expression (Figure 5c). However, doxorubicin does not induce RASSF10 expression (Figure 5c). We presumed that RASSF10 upregulation is not directly linked to senescence, but could rather be due to contact inhibition of densely growing cells. It is known that p27 (CDKN1B) is upregulated upon contact inhibition22 and as expected its expression is unaffected by doxorubicin, but increased when cells grew dense (Figure 5c). Interestingly, that expression pattern rather resembles that of RASSF10 (Figure 5c). As reviewed by Hernandez et al.23 JunD acts as a positive regulator of cellular maturation and JunD accumulates when fibroblasts become quiescent.24 We therefore analyzed JunD expression and we found its expression was similar to RASSF10, with increasing levels upon increasing density of cells (Figure 5c).
Figure 5.
RASSF10 levels are increased upon contact inhibition and JunD knockdown interferes with this upregulation. Cells were plated at (a) 10% or 100% density and (b) expression of RASSF10 and ACTB was determined. Marker of 100 bp is shown. (c) Cells were plated at indicated densities, treated with 200 nM doxorubicin for 72 h and RT–PCR analysis of ACTB, p21, p27, RASSF10 and JunD is shown. (d) Cells were transfected with siRASSF10 (siR10) or siControl (siC). After 24 h cells were treated with doxorubicin (dox) for another 72 h. Expression of ACTB, RASSF10, p21 and p27 is shown. (e) Accordingly qRT–PCR of p27 expression after normalization to ACTB is depicted and mean of three independent experiments is shown with SD. (f) Cells were transfected with siC, siJunD and siFra2. RASSF10 expression was analyzed after 96 h when cells were 100% dense. Expression was normalized to ACTB. Knockdown was confirmed for (g) JunD and (h) Fra2. Experiments were performed in triplicate in A549.
Furthermore, knockdown of RASSF10 was performed to test its endogenous influence on senescence or contact inhibition (Figure 5d). A549 cells were doxorubicin- or mock-treated and RASSF10, p21 and p27 expression was analyzed (Figure 5d). Cells were about 100% dense when cells were stained for senescence-associated β-galactosidase (Supplementary Figure S2C). Doxorubicin treatment induces p21 (Figure 5d). However, downregulation of RASSF10 under doxorubicin neither affects p21 induction (Figure 5d) nor the staining of β-galactosidase (Supplementary Figure S2D). Contact inhibition-associated upregulation of p27 is also unaltered upon knockdown of RASSF10 (Figure 5e).
In addition, we were interested in the contribution of JunD or Fra2 in upregulation of RASSF10 in contact-inhibited cells (Figure 5f). We therefore transfected A549 cells with siRNA against JunD or Fra2 when cells were almost 100% dense (Figures 5g and h). RASSF10 expression was reduced by 50% in contact-inhibited cells upon knockdown of JunD, but not after downregulation of Fra2 (Figure 5f). We further observed a correlation between the degree of JunD knockdown and decrease in RASSF10 expression (Supplementary Figure S3).
RASSF10 induces tumor suppressive growth
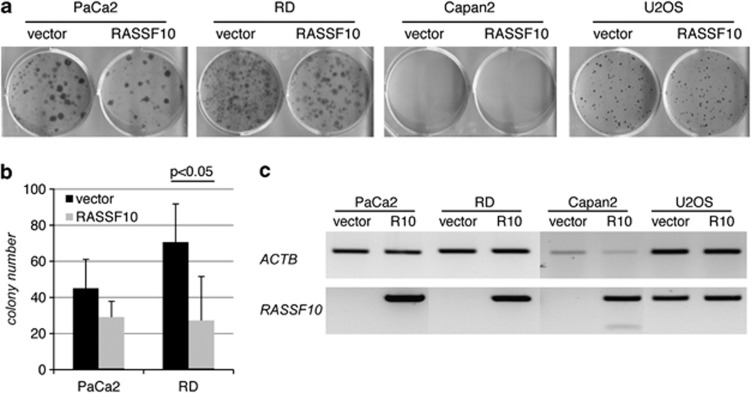
To functionally test RASSF10 and its ability to suppress tumor growth, we performed colony formation assays (Figure 6). Therefore, we transfected sarcoma (RD and U2OS) and pancreatic carcinoma cell lines (PaCa2 and Capan2) with a RASSF10 expression- or control construct and selected for 3 weeks (Figure 6a). Capan2 cells did not form colonies that were large enough and U2OS showed no differences between RASSF10 and vector overexpression. This is consistent with the weak promoter methylation for U2OS (Figure 1d) and its detectable endogenous RASSF10 mRNA level (Figure 6c). In PaCa2, Capan2 and RD cell lines the RASSF10 promoter is methylated (Figures 1f and d) and RASSF10 mRNA level is undetectable (Figure 6c). RASSF10 overexpression in PaCa2 and RD reduced the number of colonies (Figures 6a and b). RASSF10 overexpression in the colony formation assay was verified (Figure 6c).
Figure 6.
RASSF10 overexpression reduces colony formation in PaCa2 and RD cell lines. (a) PaCa2 (pancreas), RD (sarcoma), Capan2 (pancreas) and U2OS (sarcoma) cancer cell lines were transfected in triplicates with RASSF10 or empty vector, selected with G418 for 18 days and Giemsa stained. (b) Experiment was repeated three times and mean colony numbers with respective SD are shown for PaCa2 and RD (P<0.05; two-tailed t-test). (c) Expression of RASSF10 and ACTB is shown by RT–PCR.
Discussion
Epigenetic inactivation of the RASSF members has been shown in a variety of tumor entities.8, 25, 26, 27 Regarding RASSF10, which has been only recently discovered, research has just begun. Its hypermethylation was shown in thyroid tumors,10 melanoma,11 childhood leukemia12 and glioma.13 In the present study, we further add lung cancer, sarcoma, pancreatic carcinoma and HN cancer. We observed that RASSF10 methylation increases from normal tissue to primary tumors to cancer cell lines (Table 1), that matches the idea of progressive hypermethylation of tumor suppressors genes during tumorigenesis.28, 29 Using aza treatment we were able to demethylate the RASSF10 promoter and reexpress RASSF10, to prove that silencing of RASSF10 in cancer is due to its promoter hypermethylation (Figure 2).
An additional aim of this study was, to elucidate the regulation of this tumor suppressor in the cellular context. Therefore, we were interested in what kind of upstream signals would influence RASSF10 expression. First we found the RASSF10 transcript to be relatively unstable, and therefore assumed that regulation could occur relatively quickly. We observed that activation of PKA by forskolin/IBMX treatment induces RASSF10 after about 12 h (Figures 3 and 4). By directly blocking the PKA with H89 we observed an inhibition of RASSF10 activation (Figures 3 and 4). Overexpression of the catalytic subunit alpha of PKA activates the RASSF10 promoter and increases RASSF10 expression. In other studies, forskolin/IBMX signaling via cAMP and PKA was shown to inhibit proliferation, induce differentiation and apoptosis in glioma.30 Usually forskolin signaling is fast31, 32, 33 and cellular cAMP levels then decrease by the activity of adenosine 3',5'-cyclic monophosphate phosphodiesterase.34, 35 However, RASSF10 activation response is delayed and we concluded that intermediate steps must exist.
We observed that PKA also upregulates JunD expression in accordance with literature.36, 37 It is further known that AP-1 is activated by PKA, but not by phosphorylation and therefore an indirect activation was suggested.38 The delay of activation of RASSF10 suggests that RASSF10 is not directly activated by the PKA and its common substrates, but PKA rather activates JunD in between. Furthermore promoter studies showed that PKA fails to activate the RASSF10 promoter when a certain JunD binding site is missing (Figure 3). Additionally, JunD overexpression induces RASSF10. Concentrating on JunD and Fra2, of which we knew were chromatin immunoprecipitated in the RASSF10 locus,14 we performed knockdown studies and we detected a decrease in RASSF10 expression when JunD expression was downregulated. Moreover, RASSF10 activation upon forskolin/IBMX stimulation is impaired when JunD is missing (Figure 4). However, forskolin signaling sufficiently increases expression of JunD and Fra2 within 12 h of stimulation to counteract knockdown by siRNA (Figure 4). Hence, JunD knockdown is not able to fully block RASSF10 activation.
Furthermore, we observed that cells growing dense showed elevated RASSF10 levels (Figure 5) leading us to the assumption that senescence or cell–cell contact could have a role in the activation of this tumor suppressor. We therefore induced senescence by doxorubicin treatment and found p21 upregulated. RASSF10 levels, however, remained unaffected (Figure 5c) and led to the conclusion that its expression is rather an effect of increasing cell–cell contact than senescence. We found that p27 induction, marking contact inhibition of cells,39 and RASSF10 upregulation resembled each other (Figure 5c). This observation proved to be essential for experiments in this study, to assay the effect of stimuli on the RASSF10 expression in not yet contact-inhibited cells. Otherwise, cell–cell contact would increase RASSF10 expression and therefore mask the induction of RASSF10. The link between upregulation of a tumor suppressor such as RASSF10 and contact inhibition seems plausible and will need further investigation. It might be that RASSF10 blocks further cell division by induction of quiescence or differentiation, when cells come in contact. In accordance with our observation, knockdown of RASSF10 increases mitosis but decreases G1 phase (undistinguishable from G0 using propidium iodide staining in fluorescence-activated cell sorting; Supplementary Figure S1). When RASSF10, however, is inactivated in cancer cells by promoter hypermethylation this inhibition is missing and cells exhibit the tumor phenotype, thereby growing beyond contact inhibition (Figure 7).
Figure 7.
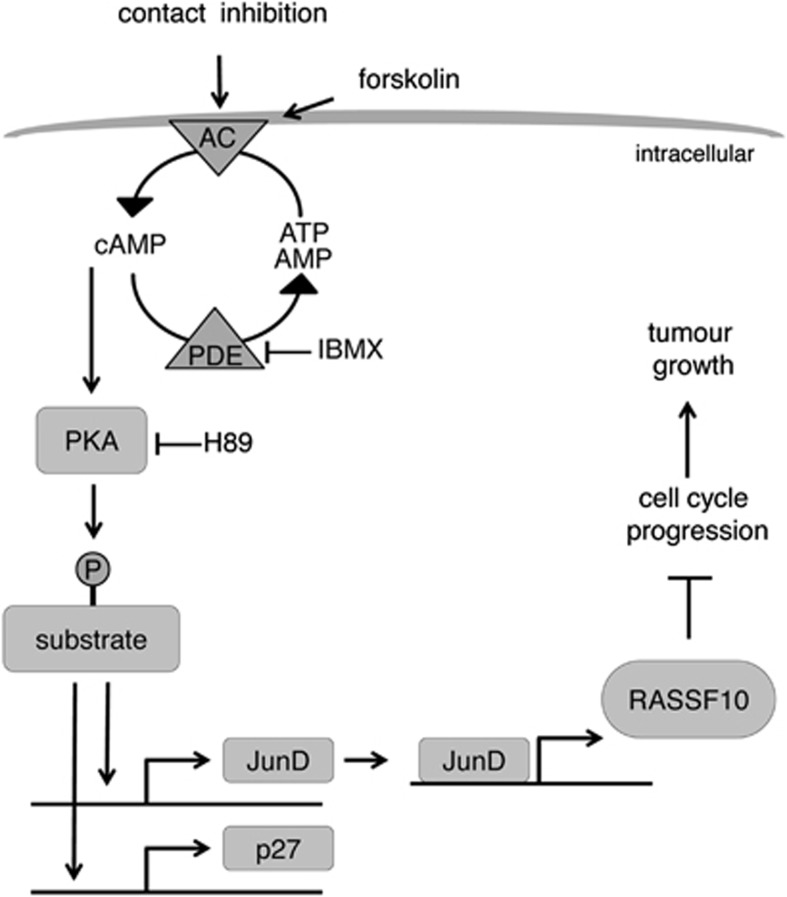
Model of RASSF10 regulation by contact inhibition via PKA. Contact inhibition (mimicked by forskolin) activates the adenylate cyclase (AC) and increases cAMP level. Cyclic AMP binds and activates PKA. PKA substrates become phosphorylated and activate JunD or p27 transcription. JunD (AP-1 member) binds and activates the RASSF10 promoter and increases its expression. RASSF10 acts as a tumor suppressor and inhibits tumor growth possibly by inhibiting cell cycle progression. IBMX inhibits adenosine 3',5'-cyclic monophosphate phosphodiesterase (PDE), leading to elevated cAMP levels. H89 blocks PKA signaling.
Most interestingly, JunD expression, which was known to be associated with quiescence,24 behaves similar to RASSF10 levels (Figure 5c). JunD is a member of AP-1, a family of factors that regulates early response genes and functions in tumor angiogenesis, cellular differentiation, proliferation and apoptosis.23 JunD was shown to be involved in osteoblast differentiation40, 41, 42 and suppressed the maturation of chondrocytes.43 It might be that in the A549 cancer cell line quiescence is induced upon contact inhibition whereby JunD and then the tumor suppressor RASSF10 are induced. When cells grew dense and we downregulated JunD using siRNA, we observed a decreased expression of RASSF10, which was in proportion to the degree of JunD downregulation (Figure 5 and Supplementary Figure S3). That further emphasizes a connection of the AP-1 member JunD and RASSF10. Dimerization of AP-1 members is required for specific and high-affinity binding to the consensus sequence.44 AP-1 is composed of a dimer formed between Jun and Fos proteins.44 Therefore, further experiments will reveal the contribution of other AP-1 members and their post-transcriptional modifications at the RASSF10 promoter.
Several studies support the link of contact inhibition and PKA signaling. Indolfi et al.45 showed that membrane-bound PKA inhibits smooth muscle cell proliferation by amplifying cAMP–PKA signals. The inverse relationship of cAMP levels and cell growth is known.46 It was stated that basal adenylate cyclase activity is low in rapidly growing cells and is enhanced as the population density increases.47 Levels of p27 increase in cAMP-treated cells48 and furthermore contact inhibition was said to act through p27.49 Forskolin or contact inhibition increase p27 levels in our study together with JunD and RASSF10. In addition, JunD knockdown interferes with RASSF10 upregulation. On the basis of the data presented here we conclude that the upregulation of RASSF10 by contact inhibition and forskolin are not separate events, but act through the same signaling pathway (Figure 7). Forskolin simply mimics contact inhibition in not yet dense and truly contact-inhibited cells and therefore we were able to modulate and inhibit single factors of this pathway under controlled conditions.
To verify the ability of RASSF10 to suppress tumor growth like other RASSFs, we performed colony formation assays in cancer cell lines. Our results show that RASSF10 suppresses colony growth in sarcoma and pancreatic cancer cells (Figure 6). In agreement with the tumor suppressive function of RASSF10 we observed that knockdown of RASSF10 increases mitosis in A549 lung cancer cells and decreases G1 phase (Supplementary Figure S1). Other studies demonstrated that RASSF10-depleted cells showed enhanced proliferation and viability in glioma cells.13
In summary, our model of RASSF10 regulation is depicted in Figure 7. Contact inhibition activates the adenylate cyclase and subsequent cAMP and PKA signaling is activated that lead to transcriptional upregulation of p27, a cell cycle inhibitor. In parallel JunD expression is increased and then binds and activates the RASSF10 promoter. RASSF10 negatively affects cell cycle progression and ultimately RASSF10 acts as a tumor suppressor and inhibits tumor growth. It will be fascinating to analyze the functional relationship of RASSF10 inactivation in cancer cell lines and their inability to respond appropriately to cell–cell contact.
Materials and methods
Methylation analysis
DNA was isolated by phenol–chloroform extraction. RASSF10 promoter methylation was analyzed by COBRA and bisulfite pyrosequencing as described previously.11, 50 Briefly, 200 ng of bisulfite-treated DNA were amplified in a PCR containing 0.2 mℳ dNTP mix, 1.5 mℳ MgCl2, 10 pmol of each primer (Supplementary Table S2) and 1.5 U Taq polymerase for 45 cycles. 20–50 ng of PCR products were digested with 10 U of TaqI (Fermentas, St Leon-Rot, Germany) and analyzed on a 2% Tris-borate EDTA agarose gel. Methylation status was quantified by using PyroMark Q24 (Qiagen, Hilden, Germany). Seven CpGs are included in analyzed region and mean methylation was calculated. For in vitro methylation SssI (NEB, Frankfurt, Germany) was used.
Tissue and cell lines
Primary tissues and cancer cell lines were previously published: sarcoma,51, 52 HN,53, 54 pancreas55 and lung1, 56 (Supplementary Table S1). All patients signed informed consent at initial clinical investigation. The study was approved by local ethic committees. Normal lung from cDNA panel (Clontech, Mountain View, CA, USA) was used.
Cell culture and stimulation/inhibition
Cell lines were cultured at 37 °C with 5% CO2 in respective medium. Cells were transfected with 4 or 10 μg of constructs for 3.5- or 10-cm plates using Polyethylenimine or Turbofect (Fermentas). SiRNA was purchased from Dharmacon (Lafayette, CO, USA) and was transfected using Lipofectamine RNAiMAX (Invitrogen, Karlsruhe, Germany). Aza was used at indicated concentrations for 4 days. TGF-β, LPS, PMA, TNF-α and IL-1β were a kind gift from Rajkumar Savai (MPI, Bad Nauheim, Germany). Treatment with 20 μℳ forskolin and 500 μℳ IBMX was performed on overnight serum-starved A549 for 12 h. To inhibit PKA 10 μℳ H89 was added 1 h before forskolin treatment. For senescence induction cells were treated with 200 nℳ doxorubicin. For β-galactosidase staining cells were grown on cover slips. After 72 h cells were fixed with 3.7% formaldehyde and stained. Cells were embedded in Mowiol 4-88-DABCO mix (Calbiochem, La Jolla, CA, USA) and analyzed. For RNA Polymerase II inhibition we used 10 μℳ of α-amanitin (Applichem, Darmstadt, Germany).
Constructs
Constructs were generated from cDNA vectors (RZPD, Berlin, Germany) and cloned into: PKACα-pCMVTag1 (IRAKp961P0684Q), PKARα-pCMVTag1 (IRAKp961P0312Q) and PKACγ-pCMVTag1 (IRAKp961C0782Q).17 RASSF10 was amplified from genomic DNA, cloned into pCMVTag1 and deletion mutations were generated with QuikChange Mutagenesis Kit (Promega, Heidelberg, Germany). The RASSF10 promoter was amplified from genomic DNA (Supplementary Table S2) cloned into pRLnull. Deletion mutants of RASSF10-pRLnull promoter were generated with mutagenesis primers listed in Supplementary Table S2. Mouse AP-1 family members FosB, cFos, Fra1, Fra2, JunB, cJun and JunD were a generous gift from Rolf Müller (IMT, Marburg, Germany).
Promoter studies
HEK293 or A549 cell lines were transfected with 1 μg of pRLnull and 0.35 μg of pGL3. Cells were isolated 24 h after transfection and studied using Dual-Luciferase Reporter Assay (Promega). Dose-dependent induction of RASSF10 by PKA was achieved with increasing amounts of PKACα-pCMVTag1. In each case total amount of cotransfected plasmid was 3 μg and if necessary filled up with empty pCMVTag1. For dose-dependent promoter induction we used increasing amount of forskolin/IBMX (1 × 20 μℳ forskolin/500 μℳ IBMX).
Expression analysis
RNA was isolated using TRIzol (Invitrogen). RNA was DNase (Fermentas) digested and then reversely transcribed.17 RT–PCR was performed with primers listed in Supplementary Table S2. qRT–PCR was performed in triplicate with SYBR Green (Quanta BioSciences, Gaithersburg, MD, USA) using Rotor-Gene 3000 (Qiagen).
Acknowledgments
This work was supported by grants from the DFG, LOEWE and Deutsche Krebshilfe. We would like to thank Rolf Müller, Kerstin Kaddatz and Rajkumar Savai for generous gift of materials and Nelli Baal for support with FACS.
Glossary
- RASSF10
Ras association domain family 10
- AP-1
activator Protein 1
- PKA
protein kinase A
- COBRA
combined bisulfite restriction analysis
- HN
head and neck
- aza
5-aza-2'-deoxycytidine
- IBMX
3-isobutyl-1-methylxanthine
- F
forskolin
- I
IBMX.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Benjamin DR. A novel family of Ras-binding domains. Trends Biochem Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J Biol Chem. 1998;273:5439–5442. doi: 10.1074/jbc.273.10.5439. [DOI] [PubMed] [Google Scholar]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem J. 2010;425:303–311. doi: 10.1042/BJ20091318. [DOI] [PubMed] [Google Scholar]
- Underhill-Day N, Hill V, Latif F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics. 2011;6:284–292. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagdarsurengin U, Richter AM, Hornung J, Lange C, Steinmann K, Dammann RH. Frequent epigenetic inactivation of RASSF2 in thyroid cancer and functional consequences. Mol Cancer. 2010;9:264. doi: 10.1186/1476-4598-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood V, Manbodh R, Sheppard C, Chalmers AD. RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol Biol Cell. 2008;19:1772–1782. doi: 10.1091/mbc.E07-07-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagdarsurengin U, Richter AM, Wohler C, Dammann RH. Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics. 2009;4:571–576. doi: 10.4161/epi.4.8.10056. [DOI] [PubMed] [Google Scholar]
- Helmbold P, Richter AM, Walesch S, Skorokhod A, Marsch W, Enk A, et al. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J Invest Dermatol. 2012;132:687–694. doi: 10.1038/jid.2011.380. [DOI] [PubMed] [Google Scholar]
- Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill VK, Underhill-Day N, Krex D, Robel K, Sangan CB, Summersgill HR, et al. Epigenetic inactivation of the RASSF10 candidate tumor suppressor gene is a frequent and an early event in gliomagenesis. Oncogene. 2010;30:978–989. doi: 10.1038/onc.2010.471. [DOI] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C, Wong N, Pagano M, Lun SW, Nakayama KI, Nakayama K, et al. Regulation of APC/C(Cdc20) activity by RASSF1A-APC/C(Cdc20) circuitry. Oncogene. 2012;31:1975–1987. doi: 10.1038/onc.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Song SJ, Kim SJ, Nakayama K, Nakayama KI, Lim DS. Skp2 regulates the antiproliferative function of the tumor suppressor RASSF1A via ubiquitin-mediated degradation at the G1-S transition. Oncogene. 2008;27:3176–3185. doi: 10.1038/sj.onc.1210971. [DOI] [PubMed] [Google Scholar]
- Richter AM, Schagdarsurengin U, Rastetter M, Steinmann K, Dammann RH. Protein kinase A-mediated phosphorylation of the RASSF1A tumour suppressor at Serine 203 and regulation of RASSF1A function. Eur J Cancer. 2010;46:2986–2995. doi: 10.1016/j.ejca.2010.06.128. [DOI] [PubMed] [Google Scholar]
- Litwiniec A, Grzanka A, Helmin-Basa A, Gackowska L, Grzanka D. Features of senescence and cell death induced by doxorubicin in A549 cells: organization and level of selected cytoskeletal proteins. J Cancer Res Clin Oncol. 2009;136:717–736. doi: 10.1007/s00432-009-0711-4. [DOI] [PubMed] [Google Scholar]
- Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi E, Palumbo G, Brady HJ. Roscovitine modulates DNA repair and senescence: implications for combination chemotherapy. Clin Cancer Res. 2005;11:8158–8171. doi: 10.1158/1078-0432.CCR-05-1042. [DOI] [PubMed] [Google Scholar]
- Dietrich C, Wallenfang K, Oesch F, Wieser R. Differences in the mechanisms of growth control in contact-inhibited and serum-deprived human fibroblasts. Oncogene. 1997;15:2743–2747. doi: 10.1038/sj.onc.1201439. [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Floyd DH, Weilbaecher KN, Green PL, Boris-Lawrie K. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene. 2008;27:4757–4767. doi: 10.1038/onc.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr CM, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, et al. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645–663. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26:7283–7301. doi: 10.1038/sj.onc.1210547. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TC, Hinton DR, Zidovetzki R, Hofman FM. Up-regulation of the cAMP/PKA pathway inhibits proliferation, induces differentiation, and leads to apoptosis in malignant gliomas. Lab Invest. 1998;78:165–174. [PubMed] [Google Scholar]
- Seamon KB, Padgett W, Daly JW. Forskolin: unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc Natl Acad Sci USA. 1981;78:3363–3367. doi: 10.1073/pnas.78.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson TH, Fain JN. Forskolin-activated adenylate cyclase. Inhibition by guanyl-5'-yl imidodiphosphate. J Biol Chem. 1983;258:9755–9761. [PubMed] [Google Scholar]
- Totsuka Y, Ferdows MS, Nielsen TB, Field JB. Effects of forskolin on adenylate cyclase, cyclic AMP, protein kinase and intermediary metabolism of the thyroid gland. Biochim Biophys Acta. 1983;756:319–327. doi: 10.1016/0304-4165(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Madelian V, La Vigne E. Rapid regulation of a cyclic AMP-specific phosphodiesterase (PDE IV) by forskolin and isoproterenol in LRM55 astroglial cells. Biochem Pharmacol. 1996;51:1739–1747. doi: 10.1016/0006-2952(96)00167-0. [DOI] [PubMed] [Google Scholar]
- Erdogan S, Houslay MD. Challenge of human Jurkat T-cells with the adenylate cyclase activator forskolin elicits major changes in cAMP phosphodiesterase (PDE) expression by up-regulating PDE3 and inducing PDE4D1 and PDE4D2 splice variants as well as down-regulating a novel PDE4A splice variant. Biochem J. 1997;321 (Pt 1:165–175. doi: 10.1042/bj3210165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobierski LA, Chu HM, Tan Y, Comb MJ. cAMP-dependent regulation of proenkephalin by JunD and JunB: positive and negative effects of AP-1 proteins. Proc Natl Acad Sci USA. 1991;88:10222–10226. doi: 10.1073/pnas.88.22.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson DJ, Zellmer E, Lewis EJ. AP1 proteins mediate the cAMP response of the dopamine beta-hydroxylase gene. J Biol Chem. 1998;273:24065–24074. doi: 10.1074/jbc.273.37.24065. [DOI] [PubMed] [Google Scholar]
- de Groot RP, Sassone-Corsi P. Activation of Jun/AP-1 by protein kinase A. Oncogene. 1992;7:2281–2286. [PubMed] [Google Scholar]
- Levenberg S, Yarden A, Kam Z, Geiger B. p27 is involved in N-cadherin-mediated contact inhibition of cell growth and S-phase entry. Oncogene. 1999;18:869–876. doi: 10.1038/sj.onc.1202396. [DOI] [PubMed] [Google Scholar]
- McCabe LR, Banerjee C, Kundu R, Harrison RJ, Dobner PR, Stein JL, et al. Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of Fra-2 and Jun D during differentiation. Endocrinology. 1996;137:4398–4408. doi: 10.1210/endo.137.10.8828501. [DOI] [PubMed] [Google Scholar]
- Naito J, Kaji H, Sowa H, Hendy GN, Sugimoto T, Chihara K. Menin suppresses osteoblast differentiation by antagonizing the AP-1 factor, JunD. J Biol Chem. 2005;280:4785–4791. doi: 10.1074/jbc.M408143200. [DOI] [PubMed] [Google Scholar]
- Akhouayri O, St-Arnaud R. Differential mechanisms of transcriptional regulation of the mouse osteocalcin gene by Jun family members. Calcif Tissue Int. 2007;80:123–131. doi: 10.1007/s00223-006-0102-7. [DOI] [PubMed] [Google Scholar]
- Kameda T, Watanabe H, Iba H. C-Jun and Jun Dsuppress maturation of chondrocytes. Cell Growth Differ. 1997;8:495–503. [PubMed] [Google Scholar]
- Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- Indolfi C, Stabile E, Coppola C, Gallo A, Perrino C, Allevato G, et al. Membrane-bound protein kinase A inhibits smooth muscle cell proliferation in vitro and in vivo by amplifying cAMP-protein kinase A signals. Circ Res. 2001;88:319–324. doi: 10.1161/01.res.88.3.319. [DOI] [PubMed] [Google Scholar]
- Abell CW, Monahan TM. The role of adenosine 3',5'-cyclic monophosphate in the regulation of mammalian cell division. J Cell Biol. 1973;59:549–558. doi: 10.1083/jcb.59.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WB, Russell TR, Carchman RA, Pastan I. Interrelationship between adenylate cyclase activity, adenosine 3':5' cyclic monophosphate phosphodiesterase activity, adenosine 3':5' cyclic monophosphate levels, and growth of cells in culture. Proc Natl Acad Sci USA. 1973;70:3802–3805. doi: 10.1073/pnas.70.12.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato JY, Matsuoka M, Polyak K, Massague J, Sherr CJ. Cyclic AMP-induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell. 1994;79:487–496. doi: 10.1016/0092-8674(94)90257-7. [DOI] [PubMed] [Google Scholar]
- Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, et al. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel C, Bartel F, Rastetter M, Bluemke K, Wurl P, Taubert H, et al. Alterations of cancer-related genes in soft tissue sarcomas: hypermethylation of RASSF1A is frequently detected in leiomyosarcoma and associated with poor prognosis in sarcoma. Int J Cancer. 2005;114:442–447. doi: 10.1002/ijc.20707. [DOI] [PubMed] [Google Scholar]
- Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S, et al. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22:1519–1526. doi: 10.3892/or_00000596. [DOI] [PubMed] [Google Scholar]
- Steinmann K, Richter AM, Dammann RH. Epigenetic silencing of erythropoietin in human cancers. Genes Cancer. 2011;2:65–73. doi: 10.1177/1947601911405043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, et al. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806–3812. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–1236. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.